Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites (original) (raw)
Abstract
Analysis of the human genome sequence has identified ≈25,000–30,000 protein-coding genes, but little is known about how most of these are regulated. Mapping DNase I hypersensitive (HS) sites has traditionally represented the gold-standard experimental method for identifying regulatory elements, but the labor-intensive nature of this technique has limited its application to only a small number of human genes. We have developed a protocol to generate a genome-wide library of gene regulatory sequences by cloning DNase HS sites. We generated a library of DNase HS sites from quiescent primary human CD4+ T cells and analyzed ≈5,600 of the resulting clones. Compared to sequences from randomly generated in silico libraries, sequences from these clones were found to map more frequently to regions of the genome known to contain regulatory elements, such as regions upstream of genes, within CpG islands, and in sequences that align between mouse and human. These cloned sites also tend to map near genes that have detectable transcripts in CD4+ T cells, demonstrating that transcriptionally active regions of the genome are being selected. Validation of putative regulatory elements was achieved by repeated recovery of the same sequence and real-time PCR. This cloning strategy, which can be scaled up and applied to any cell line or tissue, will be useful in identifying regulatory elements controlling global expression differences that delineate tissue types, stages of development, and disease susceptibility.
One major goal of current genome research is to identify the location of all cis-acting gene regulatory elements (1). This will be necessary if we are to understand global gene regulation in different tissues and identify regulatory variants that make individuals more susceptible to common diseases. Although computational and experimental methods for identifying exons are becoming quite powerful, systematic identification of non-coding functional elements remains a daunting task.
A number of approaches are available that attempt to identify gene regulatory elements on a genomewide scale. Comparative genomics is well suited for identifying evolutionarily conserved sequences (2, 3), but not all such regions are involved in gene regulation, and some regulatory elements will be missed because they are small or lineage/species-specific (4). Another method uses chromatin immunoprecipitated material, isolated with antibodies to specific transcription factors, to probe microarrays that contain DNA from intergenic regions (5, 6). However, this procedure does not yet have the resolution to discriminate precise cis-regulatory elements and cannot yet be readily applied to the entire human genome because of its large size (7). Computational methods, such as the TRANSFAC database (8), are designed to search for known cis-binding motifs, but most transcription factors only recognize short 6- to 10-bp motifs, making the false-positive rate extremely high (9). In addition, many DNA-binding proteins have not yet had their cognate sequences identified.
Mapping DNase hypersensitive (HS) sites has been used to identify the precise location of many different regulatory elements in specific, well studied genes. Promoters, enhancers, suppressors, insulators, and locus control regions all have been shown to be associated with DNase HS sites (10). This approach has the advantage of taking chromatin context into account by digesting nucleosome-free regions of the genome, allowing for identification of both ubiquitous and tissue-specific regulatory elements. Mapping DNase HS sites has also identified a number of inducible gene regulatory elements, such as those associated with genes that are regulated by steroid hormones or cellular differentiation (11, 12). However, this procedure is technically demanding and thus has not been previously considered applicable on a genomewide scale.
Here, we report a method for rapidly recovering all gene regulatory elements in a specific tissue or cell line by the cloning of DNase HS sites. Sequence analysis from a library of DNase HS sites generated from primary human CD4+ T cells has verified extensive enrichment for regions of the genome known to contain regulatory elements. Clones from this library also represent regions of the genome significantly more hypersensitive to DNase digestion than randomly chosen regions. We believe this technique will be a valuable addition to the growing collection of tools that can be used to globally identify gene regulatory elements.
Materials and Methods
Cloning Strategy. Primary CD4+ T cells were purified from human peripheral blood lymphocytes (National Institutes of Health Blood Bank, Institutional Review Board exemption issued by National Institutes of Health Office of Human Subjects) by using a magnetic cell sorting CD4+ T cell isolation kit (Miltenyi Biotec, Auburn, CA). This protocol uses a negative selection procedure that produces >95% pure CD4+ T cells, as tested by fluorescence-activated cell sorting analysis. Cells were washed with RSB buffer (10 mM Tris, pH 7.4/10 mM NaCl/3 mM MgCl2) and gently lysed with 0.1% Nonidet P-40 in RSB buffer. Intact nuclei were resuspended in RSB buffer and digested with increasing concentrations (0–40 units of DNase) of DNase I (Roche Molecular Biochemicals) for 5 min at 37°C. DNase digestion was stopped with 0.1 M EDTA. DNA was immediately embedded in 1% InCert (BioWhittaker) low-melt agarose to protect the DNA from shearing forces. DNase and proteins were removed by incubating the resulting plugs in LIDS buffer (1% lauryl sulfate/10 mM Tris·Cl/100 mM EDTA) overnight at 37°C. Plugs were washed three times in 0.2× NDS (0.1 M Na2 EDTA-2H2O/2 mM Tris base/0.2% _N_-lauroylsarcosine) and three times with 50 mM EDTA, and then stored in 50 mM EDTA at 4°C. Pulsed-field gel electrophoresis (20–60 switch time, 18 h, 6 V/cm; Bio-Rad) was used to assess the DNase digestion. DNase-digested ends were made blunt with T4 DNA polymerase, and the DNA were purified by heating the agarose, phenol extraction, and ethanol precipitation. The purified DNA was digested with _Bam_HI and _Bgl_II, phenol/chloroform-extracted, and ethanol-precipitated. Fragments with one blunt end (from the DNase digestion and T4 DNA polymerase treatment) and one sticky end (from the _Bam_HI or _Bgl_II digestion) were directionally cloned into pBluescript KS that had been digested with _Eco_RV (to generate a blunt end) and _Bam_HI (to generate a sticky end compatible with both _Bam_HI- and _Bgl_II-digested ends).
Sequencing, Eliminating Artifacts, and Aligning to the Human Genome. DNase HS clones (10,368 total) from the optimal DNase concentration (1.2 units) were sequenced by using a universal T7 primer adjacent to the blunt (DNase-treated) end of the insert. Low-quality and vector sequences were identified by phred and cross_match, respectively, and removed. Sequences down-stream of _Bam_HI and _Bgl_II sites were removed to eliminate concatamerized sequences. Approximately 40% of clones resulted from a _Bam_HI or _Bgl_II half-site ligating illegitimately to the blunt end of the cloning vector. These inserts invariably represented full _Bam_HI or _Bgl_II sites within the genome and were removed from the analysis because they do not represent genuine DNase HS sites. The remaining 5,635 sequences were aligned to the April 2003 (build 33) human genome assembly by using the ssaha sequence alignment program (13). We chose ssaha because it uses both sequence and phred quality scores to ensure high-quality alignments. Only those inserts with a single high-quality ssaha alignment were retained. To further reduce alignment artifacts, we required a high-stringency alignment between the genome and the 20 nucleotides immediately adjacent to the DNase HS site (>80% sequence identity, with exact matches at positions 4–6 within the insert). The genomic position of the blunt (DNase-digested) end of the remaining 4,864 inserts was recorded and used in further analysis.
Generation of a Control Set of 1,000 in Silico Libraries. As a control, we computationally generated 1,000 libraries, each composed of sequences directly matched in length and quality scores to sequences in the experimental library. To pick sequences at random in the human genome, we concatenated all human chromosome sequences into a single long sequence. We then used the random number generator function in perl to pick a number between 1 and the total number of nucleotides in the genome (3,110,847,713), identified this position in the concatenated sequence, and correlated this position back to its chromosomal origin. If this coordinate fell within a sequencing gap, a new number was picked. For each randomly chosen genomic coordinate, we extracted the correct amount of 3′ sequence to emulate one sequence in the experimental dataset. This sequence was also assigned the phred quality values from that experimental sequence. To ensure even better emulation with the experimental data, positions in the random sequences were mutated at a rate consistent with their phred scores. Each of the 1,000 in silico datasets contained 5,635 sequences, which were then realigned to the human genome by using the same protocol as that used for the 5,635 experimental sequences. This method of selecting in silico libraries ensured that the control coordinates were derived from regions of the genome that, like the experimental data, were computationally accessible. After performing ssaha alignments with the random sequences, the 5′ coordinates were determined and used for further analysis.
Genomic Context. We developed a bioinformatics pipeline to compare the positions of the DNase HS and random in silico library coordinates to annotated features of the human genome. Specifically, we were interested in three tracks from the University of California, Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu). (i) The refseq Gene track showing the intron and exon coordinates of 18,604 National Center for Biotechnology Information mRNA reference sequences (RefSeqs) mapped by blat (14). We determined the number and position (upstream, downstream, exon, and intron) of coordinates that mapped within 2,000 bp of an annotated refseq mRNA. Some refseq sequences align only partially to the genome; we disregarded cases where a coordinate mapped “upstream” of a RefSeq not completely aligned at its 5′ end or “downstream” of a RefSeq not completely aligned at its 3′ end. To simplify the analysis, we also disregarded any DNase HS sites that mapped within 2 kb of more than one RefSeq. (ii) The CpG Islands track depicts regions where a cytosine followed by a guanine is present more frequently than is typical for the genome as a whole. We identified all coordinates that mapped within a CpG Island. (iii) The Mouse Cons track displays sequence conservation between the human and mouse genomes for 50-bp windows in the human genome, of which at least 15 bp align to mouse (15). We determined the number of coordinates that mapped within such regions. We also obtained Affymetrix U133A gene expression data from human CD4+ T cells from the Children's National Medical Center Microarray Center web site (http://microarray.cnmcresearch.org). The results from random libraries were used to assign probability scores to the results from the DNase HS sites. We performed an ANOVA on all of the analyses from the in silico libraries to demonstrate that 1,000 random libraries were sufficient (data not shown).
Real-Time PCR. Real-time PCR was used to confirm coordinates that truly represent DNase HS sites (16). Nuclei from CD4+ T cell DNA were digested with 11 different concentrations of DNase I (0–40 units), phenol-extracted, and ethanol-precipitated. Three hundred base pairs of flanking, repeat-masked sequence were selected from each coordinate, and primer sets were designed to amplify 200- to 300-bp products by using the program primer3 (17). Real-time PCR detects the number of additional cycles for PCR primers flanking a DNase HS site to amplify a PCR product from a DNase-treated DNA template. Regions insensitive to DNase digestion, however, do not require as many additional cycles. PCR was performed by using SYBR green PCR kits (Qiagen, Valencia, CA), and real-time PCR was performed on a 7900 real-time PCR machine (Applied Biosystems). ΔCt values were determined by subtracting the Ct value from each DNase concentration from the no DNase control Ct value for each primer set. ΔCt values for no DNase were determined by performing the no-DNase experiments in duplicate. Genomic DNA was quantitated twice by using a fluorimeter (Molecular Devices) and a PicoGreen dsDNA quantification kit (Molecular Probes). Eight nanograms of genomic DNA digested with 11 different concentrations of DNase was stamped onto 384-well optical PCR plates by using a Quadra 384 machine (Tomtec, Orange, CT). Pilot PCRs performed in triplicate generated highly reproducible results (SD < 0.2). Only dissociation curves with single peaks, which indicate specific amplification, were used in the analysis.
Results
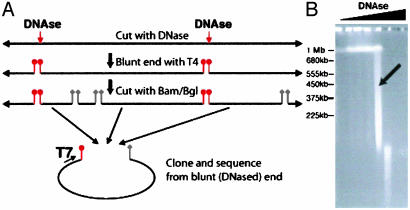
Cloning and Bioinformatics Strategy. A library of human DNase HS sites was generated from primary CD4+ T cell lymphocytes (Fig. 1_A_), because this cell type is well characterized, is easily purified from blood, is a relatively homogenous cell population, and has none of the major chromosomal abnormalities often present in immortalized cell lines. In addition, the CD4+ T cells are not undergoing cell division, a potentially important point because cells in S phase may expose DNA randomly to DNase. Genomic DNA purified from cells not treated with DNase was extremely high in molecular weight, indicating minimal amounts of shearing (Fig. 1_B_). To select for the most hypersensitive sites, we chose a smear of 100-kb to 1-Mb fragments.
Fig. 1.
Cloning of DNase HS sites. (A) Cloning strategy. Intact nuclei are digested with DNase, and the DNase-digested ends are made blunt with T4 DNA polymerase. After digestion with _Bam_HI and _Bgl_II, blunt/sticky fragments were ligated into pBluescript SK(+) and sequenced from the blunt (DNase-digested) end. (B) Pulsed-field electrophoresis of genomic CD4+ T cell DNA treated with increasing amounts of DNase. An arrow marks the concentration of DNase (1.2 units) from which the library was made.
After cloning, sequencing, and removal of known cloning artifacts, we had 5,635 high-quality sequences from DNase HS clones. Over 86% (4,864) of these sequences mapped to a single position in the human genome (a complete list of DNase HS site coordinates within the genome is available at http://research.nhgri.nih.gov/DNaseHS). We tested whether this library was enriched for potential regulatory regions by comparing it to 1,000 randomly generated in silico libraries, each containing 5,635 sequences. Using the same alignment strategy, almost 91% of the computationally selected sequences mapped to a unique genomic position. The positions of the unique coordinates from the DNase HS and random in silico libraries were analyzed relative to features displayed on the UCSC genome browser, including known genes, CpG islands, GC content, and mouse–human conservation. We also determined the percentage of sequences that map near genes expressed in CD4+ T cells.
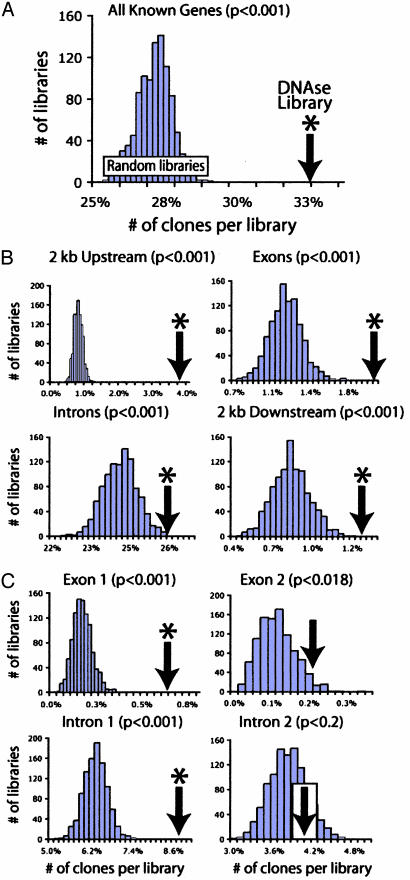
Enrichment of DNase HS Sites Nearby or Within Known Genes. We first assessed whether the isolated DNase HS sites were more likely to reside near genes than randomly generated sites. Here, a gene is defined as one of the 18,604 National Center for Biotechnology Information mRNA RefSeqs aligned to the human genome by the UCSC genome browser. For the DNase HS site library, 33% of the coordinates mapped within a window that spanned the transcriptional unit as well as 2 kb upstream and downstream (Fig. 2_A_). This is significantly higher than the random in silico libraries, which mapped within 2 kb of a gene ≈27% of the time (P < 0.001). We further subdivided each gene into four regions: 2 kb upstream, exons, introns, and 2 kb downstream (Fig. 2_B_). A 5-fold enrichment of DNase HS sites was detected within regions 2 kb upstream of genes (P < 0.001). A significant enrichment was also observed for DNase HS sites falling within exons, introns, and regions 2 kb downstream of genes (P < 0.001). A significant enrichment was detected within the first exon and first intron of known genes (Fig. 2_C_). At the P < 0.01 level, there was no significant enrichment in any other exons or introns (Fig. 2_C_ and data not shown).
Fig. 2.
Comparison of DNase HS site library (marked with arrows) relative to 1,000 random in silico libraries, which show a seminormal distribution. *, Significant differences (P < 0.01) between DNase HS site and random libraries. (A) Alignment to known genes. Thirty-three percent of DNase HS site coordinates map to a 2-kb window surrounding all known genes. (B) Each 2-kb gene window surrounding all known genes was divided into four segments: 2 kb upstream of genes, exons, and introns, and 2 kb downstream of genes. The DNase library is significantly enriched for all four regions (P < 0.001). (C) DNase HS site and random libraries were mapped to first and second exons and introns, and show that DNase HS site coordinates are significantly enriched for first exons and first introns but less so for second exons and not at all for second introns.
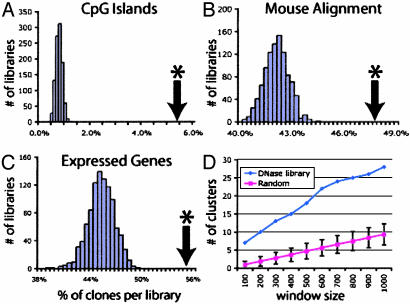
Enrichment of DNase HS Sites Within CpG Islands. Many CpG islands have been associated with the 5′ end of housekeeping genes (18), indicating that they may contain regulatory elements. Approximately 6% of the DNase HS site coordinates mapped within a CpG island (Fig. 3_A_). This was ≈6-fold higher than with the random in silico libraries (P < 0.001). We also detected an enrichment of DNase HS site coordinates in regions of the genome with high GC content. This was most likely due to an overlap of such regions with CpG islands (data not shown).
Fig. 3.
Comparison of DNase HS site and random libraries. (A) Alignment of libraries to CpG islands. A significant enrichment (*) of sequences was detected in the DNase HS library, compared to the random libraries. (B) Alignment of DNase HS and random in silico libraries to regions of the genome that align between mouse and human. Approximately 48% of the DNase HS library is within regions that align to mouse, a significantly different percentage than that of the random libraries. (C) Alignment of libraries to genes that are expressed in CD4+ T cells. The percentage of clones that land near or within a gene with detectable transcripts was determined for each library. (D) The number of clustered coordinates from DNase HS site and random libraries was determined within 100- to 1,000-bp windows.
Enrichment of DNase HS Sites Within Regions of Human–Mouse Conservation. Because many regulatory elements are expected to be conserved among mammals (3), we asked whether the DNase HS site coordinates are enriched within regions that can be aligned between the human and mouse genomes. Approximately 48% of the DNase HS site coordinates mapped to regions that can be aligned, whereas on average only 42% of the random in silico library coordinates mapped to those regions (Fig. 3_B_). This enrichment was significantly different from random (P < 0.001). Of all of the coordinates that map to regions that align between human and mouse, DNase HS site coordinates displayed significantly higher conservation scores than the random coordinates (data not shown).
It should be mentioned that the observations reported here about enrichment of DNase HS sites in 5′ flanking regions, CpG islands, and regions of human–mouse conservation are not entirely independent, because these features are correlated in the genome.
Enrichment of DNase HS Sites Within and Around Genes Expressed in CD4+ T Cells. We obtained data about the gene expression levels of primary resting CD4+ T cells, based on analysis performed with an Affymetrix U133A chip spotted for 14,049 mRNAs. Of all of the DNase HS site and random library coordinates that mapped within 2 kb of a gene that is spotted on the Affymetrix U133A chip, the percentage of those genes with detectable transcripts was determined. In the DNase HS site library, 56% of the arrayed mRNAs lying within 2 kb of a DNase HS site were expressed, whereas in the random libraries, this number was 46% (P < 0.001; Fig. 3_C_). The average expression level of genes that map within 2 kb of a DNase HS site coordinate displayed a significantly higher expression level, indicating enrichment for active regions of the genome (data not shown).
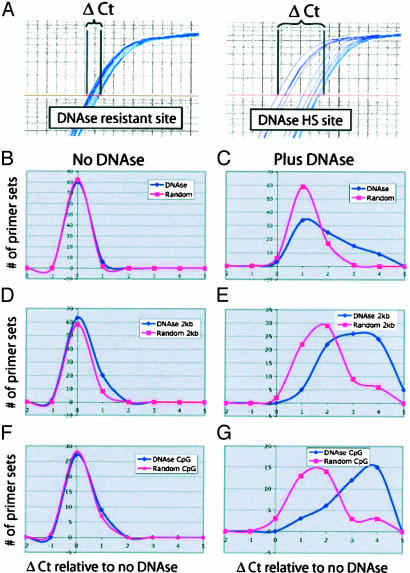
Confirmation of DNase HS Sites. Real-time PCR was performed to confirm that DNase HS site coordinates were more likely to be hypersensitive to DNase digestion than those from randomly generated libraries. Primer sets were designed to flank putative DNase HS sites or random coordinates. Genuine DNase HS sites should require additional PCR cycles (ΔCt) to reach the same quantity of PCR product as the controls (not digested with DNase), whereas regions of the genome resistant to DNase will not require as many additional cycles to reach the same threshold value (Fig. 4_A_). This protocol for confirming DNase HS sites is much more sensitive, quantitative, and rapid than are Southern blots (16).
Fig. 4.
Real-time PCR of putative DNase HS site and random coordinates. Intact CD4+ T cell nuclei were digested with increasing amounts of DNase and used for real-time PCR. (A) Amplification plot. Each curve represents amplification from lowest concentration of DNase to highest (left to right). The y axis represents the amount of PCR product, and the x axis is the number of cycles. The middle line is the threshold set during the linear phase of the PCR used to calculate the Ct value. Primer sets flanking DNase HS sites have larger ΔCt values when amplified from DNase-sensitive sites than from DNase-resistant sites. (B and C) Primer sets flanking nonbiased DNase HS site and random coordinates amplified from genomic DNA treated with no DNase (B) or plus DNase (C). The y axis represents the number of primer sets studied, and the x axis represents the additional number of cycles (ΔCt) required to amplify the same amount of template as with no DNase. (D and E) PCR primers flanking DNase HS site and random coordinates that are <2 kb upstream from genes amplified genomic DNA treated with no DNase (_D_) or plus DNase (_E_). (_F_ and _G_) PCR primers flanking DNase and random libraries that map to CpG islands >2 kb from a gene-amplified genomic DNA treated with no DNase (F) or plus DNase (G).
To determine the efficiency of the DNase HS site library as a whole, flanking PCR primers were first designed in a nonbiased manner from 200 putative DNase HS sites and 200 random coordinates. With no DNase, the ΔCt for both random and DNase coordinates were indistinguishable (Fig. 4_B_). However, at a higher concentration of DNase, primer sets flanking the DNase HS site coordinates required more cycles to amplify a product than the random controls, indicating that the DNase HS site clones contain regions digested by DNase (Fig. 4_C_). Setting a threshold at a ΔCt value of 2, ≈30% of DNase HS site coordinates were more sensitive to DNase digestion than random coordinates. Only a single primer set designed against a randomly generated coordinate produced a ΔCt value of greater than two, and this coordinate mapped within 2 kb upstream of a gene, possibly indicating that this may be a true DNase HS site.
Because a significant enrichment of DNase HS site coordinates was detected within a 2-kb window upstream of genes, we wanted to verify that most of these coordinates represented genuine DNase HS sites. Real-time PCR was performed with primer sets that flank 96 DNase HS site coordinates and 96 random coordinates located <2 kb upstream of genes. With no DNase (performed in duplicate), the results were identical for both sets (Fig. 4_D_). However, at the higher DNase concentration, the primer sets from the DNase HS site library required significantly more cycles to amplify than the random primer sets (Fig. 4_E_). With ΔCt set at a threshold of 2, ≈70% of DNase HS site primer sets were above the threshold, whereas only 20% of random primer sets were above the threshold.
Real-time PCR was also performed with 96 primer sets that flank DNase HS site and random coordinates that map to CpG islands. For this, we only analyzed coordinates within CpG islands located >2 kb from genes. Approximately 80% of DNase HS site coordinates that mapped to CpG islands were significantly more hypersensitive to DNase digestion than random coordinates, indicating there may be subsets or subregions of CpG islands that have functional significance (Figs. 4 F and G). We have also noted that randomly generated coordinates that were both within 2 kb upstream of a gene and within a CpG island tended to be more sensitive to DNase digestion than randomly generated coordinates that were chosen in a nonbiased manner. However, this was hardly an unexpected observation as CpG islands and regions near genes are likely sites for regulatory signals, and some of these random generated clones are probably true positives.
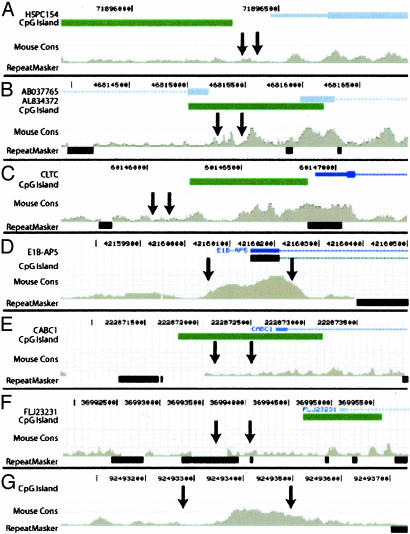
Clustering of DNase HS Sites. We expected genuine DNase HS sites to cluster within a deep library, whereas clones that result from background (e.g., shearing) would appear only once. Because DNase HS sites in the genome vary in size, we looked at the location of clones in window sizes ranging from 100 to 1,000 bp to find evidence of pairs of clones that might derive from the same DNase HS site. At all window sizes, significantly more paired coordinates within the DNase HS site library were identified than within the random libraries (Fig. 3_D_). Of 15 DNase HS coordinate pairs that mapped to within 400 bp, we were able to successfully assay nine by real-time PCR. Seven were confirmed to be genuine DNase HS sites (data not shown). We looked in more detail at the genomic context of these experimentally confirmed DNase HS sites by using the UCSC genome browser (Fig. 5). Many mapped near or within CpG islands, are within regions of human–mouse sequence conservation, or are upstream of genes expressed in CD4+ T cells (Fig. 5 A–F). Interestingly, one pair of DNase HS coordinates was located >250 kb away from any known gene but mapped to a region that was highly conserved between human and mouse (Fig. 5_G_). Note that not all validated DNase HS sites mapped precisely to the most highly conserved regions between mouse and human.
Fig. 5.
Seven pairs of DNase HS site clones that map <400 bp from each other. Positions of DNase HS sites (arrows) are indicated relative to known genes, CpG islands, regions of conservation between human and mouse, and repetitive elements using the UCSC genome browser. Six pairs map within 2 kb upstream of a gene (A–F) and nearby or in CpG islands. Note that not all of these six pairs map to the most highly conserved regions between human and mouse. One pair maps 250 kb from any known gene (G) but is in a region that is highly conserved with mouse.
Discussion
We have developed a method for identifying DNase-hypersensitive sites on a genome-wide scale. Unlike approaches based on comparative genomics, the strategy described here enriches for segments within the genome that correspond to open chromatin and should be able to define tissue specificity of functional elements. The isolated DNase HS sites are significantly enriched for sequences that map within regions 2 kb upstream and downstream of genes, CpG islands, first exons, first introns, and segments conserved between mouse and human. These regions correspond to where a large number of gene regulatory elements are generally thought to reside, but our approach is not limited to searching in regions of known genes. In fact, some of the most interesting DNase HS sites may be located long distances from any known gene, because they may represent locus control regions.
An experimental validation step was important to assess whether the sequences captured in this protocol were truly representative of regions of the genome that are hypersensitive to DNase. After all, random shearing during the nuclear preparation stage could generate clones that are indistinguishable from genuine DNase HS sites. Furthermore, DNase does not digest exclusively at hypersensitive sites (19). Less sensitive regions, such as genomic regions undergoing transcription or DNA replication, are likely to be digested also, albeit at a slower rate. Preliminary data suggest that DNA replication may introduce background, because we have sequenced a small number of clones from a DNase HS library generated from the K562 erythroleukemia cell line and do not detect as robust an enrichment of sequences that map to regions 2 kb upstream of genes.
Even with the use of a quiescent cell type, there was a modest level of background within the library. High-throughput real-time PCR confirmed that ≈30% of the CD4+ T cell DNase HS site coordinates were significantly more sensitive to DNase digestion than randomly chosen regions of the genome. However, >70% of DNase HS site coordinates that mapped within 2 kb upstream of genes and CpG islands were more sensitive to DNase digestion than randomly selected coordinates within the same regions. This emphasizes how validation of this library can be used to finely map and identify putative regulatory elements within these regions of the genome.
How many actual DNase HS sites are present in a given cell type? Our observation of clustered clone sets in CD4+ T cells allows an estimation, using the capture–recapture method for estimating population size (20). The number of DNase sites is given by N = [(Ym)2]/n, where Y is the estimated fraction of clones that are real versus background (0.3), m is the number of clones that were aligned to the genome (4,864), and n is the number of clones that clustered within 500 bp of each other (18). Thus, our data suggest there are ≈100,000 DNase HS sites within quiescent CD4+ T cells, which corresponds to roughly six DNase HS sites for every expressed gene.
Although presented on a pilot scale, the data presented here validate the general approach to identification of functional elements on a genomewide basis. But for full utility of the method, much larger data sets will need to be generated. In a fully optimized realization of this approach, multiple sampling of all genomic DNase HS sites in a particular tissue would provide a means of distinguishing true signals from noise and would even provide a means of quantitating the degree of hypersensitivity of a particular site, which might allow interesting comparisons between tissues. To enable a complete characterization of all of the DNase HS sites in a particular tissue or cell line for an affordable cost, other approaches to high-throughput sequence tagging of DNase HS sites will need to be pursued. One option would be a SAGE-like approach, where short 20-bp tags adjacent to DNase HS sites are isolated, concatemerized, and sequenced (21). An even higher throughput option would be to adapt the method of massively parallel signature sequencing, which utilizes a bead-based approach to produce up to a million short 20-base reads in a single sequencing run (22).
In conclusion, the method described here holds the promise of providing useful genome-wide information on regulatory sites in the genome of virtually any eukaryote. In the future, it would be interesting to apply this approach to multiple cell types, different species, various stages of development, and healthy and diseased tissue, to understand in greater molecular detail how changes in chromatin structure dictate cell function and fate.
Acknowledgments
We thank Stacie Anderson, Christiane Robbins, and Tracy Moses for excellent technical assistance, and Mike Erdos, Larry Brody, and Dave Bodine for helpful suggestions.
Abbreviations: HS, hypersensitive; RefSeq, reference sequence; UCSC, University of California, Santa Cruz.
References
- 1.Collins, F. S., Green, E. D., Guttmacher, A. E. & Guyer, M. S. (2003) Nature 422**,** 835–847. [DOI] [PubMed] [Google Scholar]
- 2.Ureta-Vidal, A., Ettwiller, L. & Birney, E. (2003) Nat. Rev. Genet. 4**,** 251–262. [DOI] [PubMed] [Google Scholar]
- 3.Thomas, J. W., Touchman, J. W., Blakesley, R. W., Bouffard, G. G., Beckstrom-Sternberg, S. M., Margulies, E. H., Blanchette, M., Siepel, A. C., Thomas, P. J., McDowell, J. C., et al. (2003) Nature 424**,** 788–793. [DOI] [PubMed] [Google Scholar]
- 4.Slightom, J. L., Bock, J. H., Tagle, D. A., Gumucio, D. L., Goodman, M., Stojanovic, N., Jackson, J., Miller, W. & Hardison, R. (1997) Genomics 39**,** 90–94. [DOI] [PubMed] [Google Scholar]
- 5.Lieb, J. D., Liu, X., Botstein, D. & Brown, P. O. (2001) Nat. Genet. 28**,** 327–334. [DOI] [PubMed] [Google Scholar]
- 6.Ren, B., Robert, F., Wyrick, J. J., Aparicio, O., Jennings, E. G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., et al. (2000) Science 290**,** 2306–2309. [DOI] [PubMed] [Google Scholar]
- 7.Weinmann, A. S., Yan, P. S., Oberley, M. J., Huang, T. H. & Farnham, P. J. (2002) Genes Dev. 16**,** 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wingender, E., Dietze, P., Karas, H. & Knuppel, R. (1996) Nucleic Acids Res. 24**,** 238–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pennacchio, L. A. & Rubin, E. M. (2001) Nat. Rev. Genet. 2**,** 100–109. [DOI] [PubMed] [Google Scholar]
- 10.Gross, D. S. & Garrard, W. T. (1988) Annu. Rev. Biochem. 57**,** 159–197. [DOI] [PubMed] [Google Scholar]
- 11.Zaret, K. S. & Yamamoto, K. R. (1984) Cell 38**,** 29–38. [DOI] [PubMed] [Google Scholar]
- 12.Szabo, G., Jr., Damjanovich, S., Sumegi, J. & Klein, G. (1987) Exp. Cell Res. 169**,** 158–168. [DOI] [PubMed] [Google Scholar]
- 13.Ning, Z., Cox, A. J. & Mullikin, J. C. (2001) Genome Res. 11**,** 1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kent, W. J. (2002) Genome Res. 12**,** 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. (2002) Nature 420**,** 520–562. [DOI] [PubMed] [Google Scholar]
- 16.McArthur, M., Gerum, S. & Stamatoyannopoulos, G. (2001) J. Mol. Biol. 313**,** 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozen, S. & Skaletsky, H. (2000) Methods Mol. Biol. 132**,** 365–386. [DOI] [PubMed] [Google Scholar]
- 18.Larsen, F., Gundersen, G., Lopez, R. & Prydz, H. (1992) Genomics 13**,** 1095–1107. [DOI] [PubMed] [Google Scholar]
- 19.Jantzen, K., Fritton, H. P. & Igo-Kemenes, T. (1986) Nucleic Acids Res. 14**,** 6085–6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seber, G. A. (1986) Biometrics 42**,** 267–292. [PubMed] [Google Scholar]
- 21.Velculescu, V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. (1995) Science 270**,** 484–487. [DOI] [PubMed] [Google Scholar]
- 22.Brenner, S., Johnson, M., Bridgham, J., Golda, G., Lloyd, D. H., Johnson, D., Luo, S., McCurdy, S., Foy, M., Ewan, M., et al. (2000) Nat. Biotechnol. 18**,** 630–634. [DOI] [PubMed] [Google Scholar]