Personalizing cancer treatment in the age of global genomic analyses: PALB2 gene mutations and the response to DNA damaging agents in pancreatic cancer (original) (raw)
. Author manuscript; available in PMC: 2012 Mar 19.
Abstract
Metastasis and drug resistance are the major causes of mortality in patients with pancreatic cancer. Once developed, the progression of pancreatic cancer metastasis is virtually unstoppable with current therapies. Here we report the remarkable clinical outcome of a patient with advanced, gemcitabine-resistant, pancreatic cancer who was later treated with DNA damaging agents, based on the observation of significant activity of this class of drugs against a personalized xenograft generated from the patient’s surgically resected tumor. Mitomycin C treatment, selected based on its robust preclinical activity in a personalized xenograft generated from the patient’s tumor, resulted in long lasting (36+ months) tumor response. Global geneomic sequencing revealed biallelic inactivation of the gene encoding PalB2 protein in this patient’s cancer, the mutation is predicted to disrupt BRCA1 and BRCA2 interactions critical to DNA double strand break repair. This work suggests that inactivation of the PALB2 gene is a determinant of response to DNA damage in pancreatic cancer and a new target for personalizing cancer treatment. Integrating personalized xenografts with unbiased exomic sequencing led to customize therapy, tailored to the genetic environment of patient’s tumor and identification of a new biomarker of drug response in a lethal cancer.
Keywords: PALB2, pancreatic cancer, personalized xenograft, mitomycin C
Introduction
Pancreatic cancer is an aggressive malignancy with one of the worst outcomes among all solid malignancies (1). At advanced, metastatic stages, pancreatic cancer can almost never be controlled by any of the available therapeutic options, mirrored by an extremely low estimated 5-year survival rate of <2% (2). Clinical benefit of gemcitabine as a systemic agent in the treatment of advanced pancreatic cancer results in a median survival of less than six months (3). Improvements in therapy have been modest with the addition of erlotinib to gemcitabine in combination, resulting in improved median survival on the order of weeks (4).
One strategy actively sought to improve outcome is to personalize cancer treatment. The development of molecular profiling technologies to assess DNA, RNA, protein, and metabolites has fueled efforts to tailor medical care, both at tumor and patient levels. Indeed, validated molecular tests assessing tumor tissue or patient germline DNA already drive therapeutic decision making. These approaches have the potential to fulfill the promise of delivering the right dose for the right indication to the right patient at the right time (5). With the ability to interrogate the entire human cancer genome, it is becoming apparent that some cancers can be effectively treated by targeting specific somatic alterations present in these cancers. This is perhaps best exemplified by the observation that patients with lung cancer harboring mutations in the epidermal growth factor (EGFR) gene respond rather dramatically to agents that target this receptor (6). This relationship was discovered only after thousands of patients had been treated with the agents in the clinic (7).
The pancreatic cancer genome project identified heterogeneity in the molecular alterations of pancreatic cancer indicating the need for personalized cancer therapy (8). The recent complete sequencing of the coding genomes of several cancer types, together with the dramatically reduced cost of whole genome sequencing, provides an unprecedented opportunity to discover novel targets for personalized gene-specific cancer therapy (8). Here we present a case of a patient with advanced pancreatic cancer who responded dramatically to mitomycin C (MMC). The molecular basis for this response - biallelic inactivation of the PALB2 gene - was discovered by sequencing of virtually all of the coding genes in this patient’s cancer (8).
Materials and Methods
Patient
The patient described in this report was enrolled in the J0507 Johns Hopkins Medical Institute clinical trial (NCT00276744). This was a pilot prospective clinical trial in which patients with resectable pancreatic cancer signed a written consent to have a portion of their resected tumor implanted and propagated in nude mice. These xenografted tumors are treated with a set of anticancer agents with the goal to identify the most effective agents that can be used to treat the patient’s cancer.
Xenograft establishment and in vivo tumor therapy studies
Female nu/nu athymic mice (Harlan) were used for the study. Animals were maintained under pathogen-free conditions and a 12-hour light/12-hour dark cycle. Animal experiments were conducted following approval and in accordance with the Animal Care and Use Committee guidelines of the Johns Hopkins University. Fresh pancreatic tumor specimens resected from patients at the time of surgery, with informed written patient consent, were implanted s.c. into the flanks of 6-week-old mice. Grafted tumors were subsequently transplanted from mouse to mouse and maintained as a live PancXenoBank according to an Institutional Review Board–approved protocol (9). JH033 xenograft tumor (originated from the patient described here) from the PancXenoBank collection at the exponential growth phase were resected aseptically and used as the source of tumor for subcutaneous implantation. Cohorts of mice with tumor size of ~200 mm3 were randomized to four treatment groups (6 mice; 10 tumors per group): (a) vehicle (control); (b) 5 mg/kg mitomycin C (MMC) i.p. single dose; (c) 5 mg/kg cisplatin i.p. once a week for 4 weeks; (d) 100 mg/kg gemcitabine i.p. twice a week for 4 weeks. As a negative control, Panc185 xenograft which has wild type PALB2 was treated with MMC and gemcitabine. Tumor size was evaluated twice weekly by caliper measurements and tumor volume was calculated using the following formula: tumor volume = [length X width2]/2.
Genomic analysis
The sequences of 23,219 transcripts representing 20,661 protein-coding genes in the patient’s cancer were determined as has been published in detail elsewhere. Whenever a variant was identified in the cancer, the patient’s germline DNA was also sequenced, revealing information about germline variations in this patient (10).
Co-immunoprecipitation
To investigate the BRCA1 and BRCA2 protein nuclear binding, a co-immunoprecipitation assay was performed using a commercially available kit (Thermo Scientific). Samples from the index patient’s tumor (JH033), which was sensitive to MMC, as well as samples of Panc185, a patient pancreatic tumor resistant to MMC, were used. The monoclonal antibody (mAB) OP107 against the BRCA1 protein, purchased from Calbiochem, was used to immunoprecipitate the BRCA1/2 protein complex. After the OP107 antibody was stably bound to the resin by a covalent union, lysates of JH033 or Panc185 were added and incubated for 24 h. Samples were eluted, electrophoresed and further immunoblotted with mAB against BRCA1 (OP107) and BRCA2 (OP95) purchased from Calbiochem.
Protein extraction and Western blot analysis
Protein extracts from tumors were prepared according to previously published methods (11). Briefly, tumors (75 mg) were minced on ice in prechilled lysis buffer. The minced tissue was homogenized and Protein lysates (30 µg) were fractionated by SDS-PAGE, electrotransferred onto nitrocellulose membranes, and blotted with primary antibodies against BRCA1 (OP107), BRCA2 (OP95), or PALB2 (2134.00.02) from (Strategic Scientific Inc.) and FANCD2 (4945) from (Cell Signaling Technology Inc.). The membranes were probed with horseradish peroxidase–conjugated secondary antibody (Santa Cruz Biotechnology Inc.) and the antibody binding was detected by enhanced chemiluminescence (GE Healthcare) as previously reported (12).
Results
Clinical case
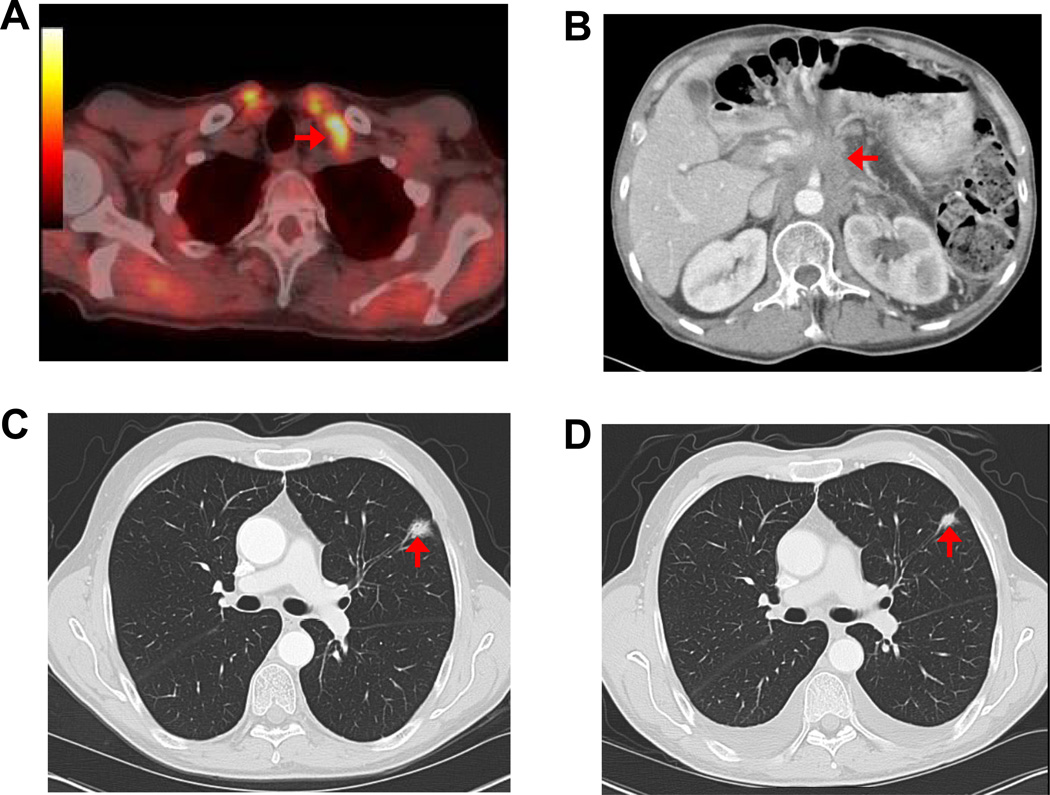
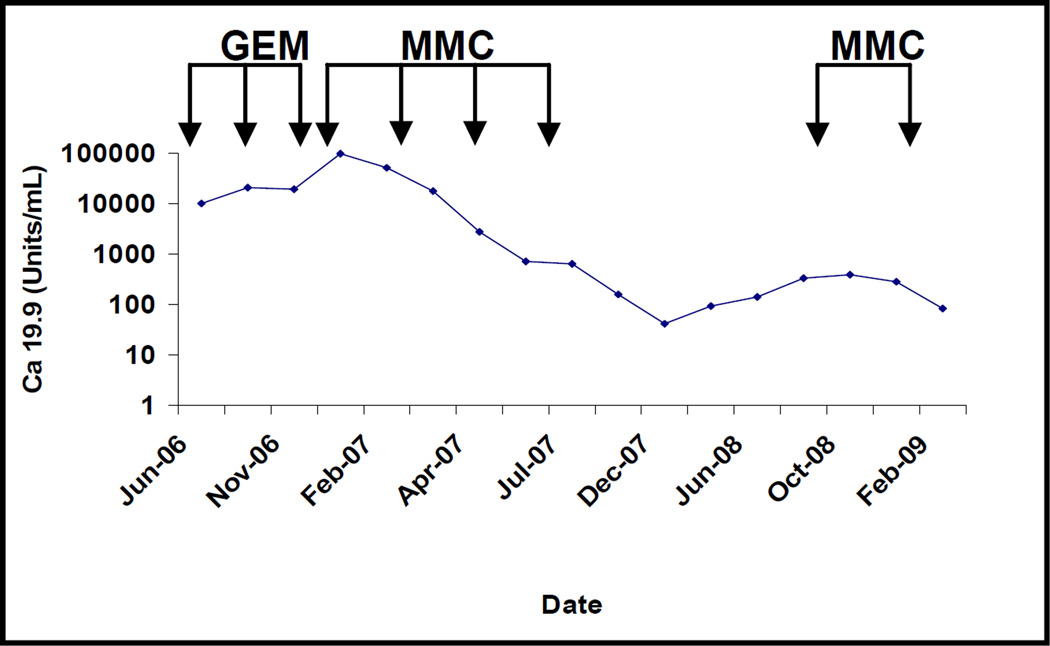
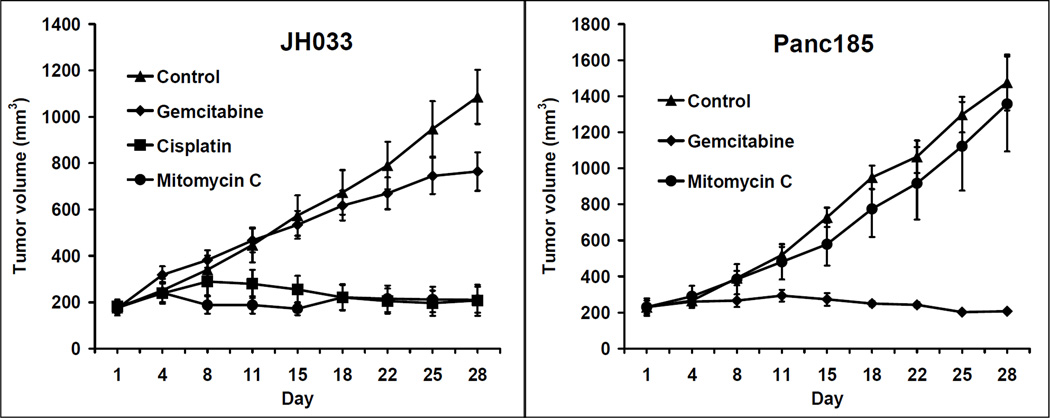
A 61 year-old male, with family history of pancreatic cancer, who had been previously tested and found to be wild-type for the BRCA2 gene, underwent a distal pancreatectomy and splenectomy for a pT3N1M0 infiltrating ductal adenocarcinoma of the pancreas. The patient had a 4 cm, poorly differentiated adenocarcinoma that had metastasized to 8 of 26 resected lymph nodes, with prominent extranodal extension. Venous and perineural invasion were identified and the carcinoma extended to involve the celiac artery margin of resection (R1). The patient was enrolled in the J0507 trial and a portion of the surgically resected tumor, coded as JH033, was xenografted in nude mice. Two months after surgery, prior to initiating adjuvant treatment, the patient was found to have a biopsy proven metastasis to a supraclavicular lymph node (Figure 1A), and his CA 19-9 rose to 10,132 U/ml (Figure 2). The patient was treated with single agent gemcitabine, but developed significant disease progression after 4 months with pleural effusion, locoregional progression in the abdominal cavity, and a CA 19-9 of 98,405 U/ml (Figure 1B and Figure 2). During this time, the results of the xenograft treatment studies became available (Figure 3), and based on the response of the patient’s xenografted cancer to MMC, the patient was treated with MMC 8 mg/m2/28 days for a total of five courses. After treatment with MMC the CT scan findings improved and the CA 19-9 level normalized (Figure 2). This response was maintained for 22 months, after which the CA 19-9 rose to 392 U/ml and a new lung nodule developed in the left upper lobe (Figure 2 and Figure 1 C). The patient was treated with 2 additional cycles of MMC, which resulted in a reduction in size of lung nodule (Figure 1 D), but the patient developed incipient renal failure. Because the xenograft was also sensitive to cisplatin, platinum-based chemotherapy was initiated, and the patient received three cycles of this agent. At his last follow up, three years after surgical resection, his CA 19-9 was 39 U/ml and the patient remains asymptomatic (data not shown).
Figure 1. Images of Clinical Outcome.
A) Pet-CT obtained at the first postoperative visit showing an enlarged left supraclavicular lymph node with increase FDG uptake (arrowhead). A biopsy of this lymph node showed metastatic adenocarcinoma; B) CT revealing extensive locoregional recurrent disease after four cycles of gemcitabine; C) Late pulmonary progression with a left upper lesion that developed 22 months of follow up after the initial treatment with MMC (arrowhead); D) Decrease in pulmonary lesion size after two additional courses of MMC (arrowhead).
Figure 2. CA 19-9 restores to normal levels with MMC treatment.
Time-course of CA 19-9 showing disease progression while on gemcitabine and complete normalization with MMC. Y-axis is log-scale of CA19-9 concentration presented in Units/mL;
Figure 3. MMC and cisplatin treatment remarkably suppressed the tumor growth of patient’s pancreatic carcinoma grown in nu/nu mice xenografts.
Tumor growth curves indicating resistance to gemcitabine and remarkable response to MMC and cisplatin in the patient’s own xenografts (JH033). Panc 185, a pancreatic cancer xenograft with wild-type PALB2 wass presented as a control. Mice were treated and tumor volumes were monitored over time (days) as indicated in the materials and methods. Tumor growth is expressed as mean tumor volume ± SEM.
Mechanism underlying unique sensitivity to DNA damaging agents
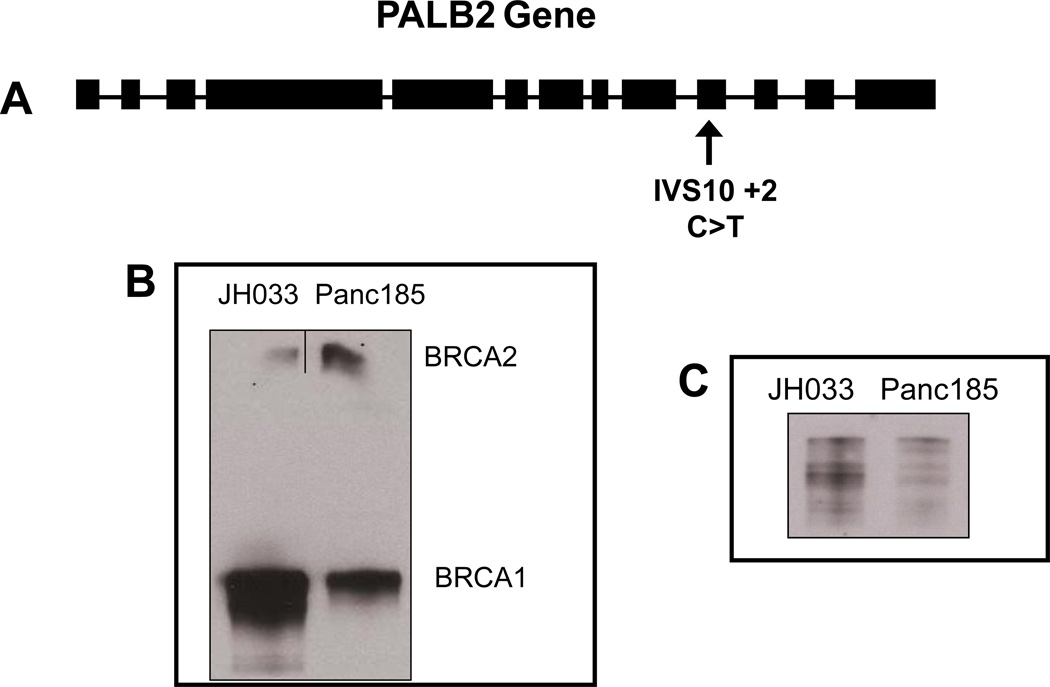
This patient’s carcinoma was recently sequenced as a part of an effort to sequence the pancreatic cancer genome (8). The results of this exomic sequencing allowed us to assess, in an unbiased fashion, potential genetic determinants of this patient’s remarkable response to MMC. The patient’s carcinoma was found to have a somatically acquired transition mutation (C to T) at a canonical splice site for exon 10 (IVS10+2) in the Partner and Localizer of BRCA2 (PALB2) gene (Figure 4A). A subsequent study identified a germline deletion of 4 base pairs (TTGT at ~172 to 175) that produced a frameshift mutation at codon 58 of the PALB2 gene (10). The PALB2 gene was therefore biallelically inactivated in this patient’s cancer. Functional analysis demonstrated that this tumor has an intact FA complex 1 system leading to successful mono-ubiquitination of the FANCD2 protein (Figure 4B, first lane), similar to the Panc 185 tumor used as a control, which has a wild-type PALB2 gene and is resistant to MMC. In contrast, the biallelic inactivation of the PALB2 gene in JH033 patient’s tumor disrupts the downstream interaction between the BRCA1/BRCA2 complex, an interaction essential for DNA double strand break repair (Figure 4C) (13).
Figure 4. Mechanistic Studies.
A) Location of somatic mutation in the PALB2 gene. The patient had somatically acquired a transition mutation (C to T) at a canonical splice site for exon 10 (IVS10+2). Exons are represented as black boxes and introns as black lines; B) Co-immunoprecipitation with a monoclonal antibody against BRCA1 of the BRCA1-BRCA2 complex. No complex is identified in the PALB2 mutant tumor JH033 as compared to the wild type Panc185 tumor used as a control; C) Western blot of FANCD2 ubiquitination. The upper band represents the ubiquitinated or long form (PFANCD2 Lys561) and the lower band represents the short, non ubiquitinated form. JH033 has competent proximal FA complex similar to the MMC resistant Panc185 control.
Discussion
In this report we highlight the remarkable clinical outcome of a patient with advanced, gemcitabine-resistant, pancreatic cancer who was treated with DNA damaging agents based on the observation of significant activity of this class of drugs against a personalized xenograft generated from the patient’s surgically resected tumor. Contrary to the expected median survival of 3 months for pancreatic cancer patients who progress on gemcitabine, this individual is virtually symptom-free for three years after progression to the first line chemotherapy. Nearly complete sequencing of all of the coding genes in this patient’s cancer revealed biallelic inactivation of the PALB2 gene, a DNA repair gene, loss of which mechanistically explains the observed sensitivity of the patient’s cancer to DNA damaging agents (8). Of note, in a conventional “protocol-based” regimen, MMC would not have been used in a second line setting for gemcitabine-refractory pancreatic cancer. Thus, this study highlights the considerable power of global genomic sequencing for the discovery of novel markers of drug activity, especially for cancers that demonstrate near uniform lethality. We have demonstrated that biallelic inactivation of the PALB2 gene in this patient’s cancer cells alters the interaction of the BRCA1 and 2 proteins, an interaction required for proper functioning of the DNA double strand break repair pathway (13).
Our study has at least two therapeutic implications in clinical oncology, one considerably more expansive in its scope than the other. First of all, response of pancreatic cancers to DNA damaging agents can now be predicted by sequencing PALB2 and BRCA2 genes. This situation is analogous to the EGFR gene mutations in lung cancer and response to EGFR inhibitors (6, 14). Based on somatic mutational rates, we anticipate that ~5–10% of pancreatic cancers will harbor such “synthetic lethal” interactions. Secondly, and more significantly, the process presented here can be generalized to other high mortality cancers, and systematically used to discover clinically relevant genetic defects that confer a vulnerability to therapeutic interventions. As the ability to obtain global genomic information from individual patient tumors becomes increasingly higher and inexpensive (15), live tumor xenografts with validated clinical response will become a viable platform to systematically explore “connections” between drug response and specific genetic alterations. In contrast to recurrent “driver” oncogenic mutations like EGFR, we anticipate that many therapeutic candidates identified by unbiased exomic or transcriptomic sequencing are likely to be rare (for example, PALB2 mutation was present in only 1 of 24 sequenced pancreatic cancers) (8), and might represent “passengers” acquired during clonal progression. Irrespective of the frequency or nature of these mutations, targeting genetic alterations identified by global sequencing represents a new paradigm in individualized cancer therapy.
In summary, we report a patient with advanced pancreatic cancer for whom a personalized xenograft model generated from the patient’s tumor, linked to unbiased exomic sequencing, led to the discovery of a highly effective treatment regimen, as well as to an understanding of the genetic defect underlying the observed sensitivity of this patient’s cancer to DNA damaging agents. This approach forms the basis for linking personalized xenografts with global genomic sequencing for the development of personalized treatment and biomarker discovery.
Acknowledgements
The authors are grateful to Bert Vogelstein, MD., Kenneth W. Kinzler, Ph.D., and Victor Velculescu, MD., Ph.D, Johns Hopkins University School of Medicine for their valuable input to this work.
Grant Support: NIH Grants CA116554 and CA129963 to Manuel Hidalgo
Footnotes
Disclosure of Potential Conflicts of Interest: Siân Jones, Ralph H. Hruban, James R. Eshleman and Alison Klein are co-inventors on PLAB2- related intellectual property managed by Johns Hopkins University and have the potential to receive royalty payments for the PALB2 invention. The other authors disclosed no potential conflicts of interest
References
- 1.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 2.Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363:1049–1057. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- 3.Burris HA, 3rd, Moore MJ, Andersen J, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15:2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 4.Moore MJ. Brief communication: a new combination in the treatment of advanced pancreatic cancer. Semin Oncol. 2005;32:5–6. doi: 10.1053/j.seminoncol.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Angulo AM, Hennessy BT, Mills GB. Future of personalized medicine in oncology: a systems biology approach. J Clin Oncol. 2010;28:2777–2783. doi: 10.1200/JCO.2009.27.0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 7.Bell DW, Lynch TJ, Haserlat SM, et al. Epidermal growth factor receptor mutations and gene amplification in non-small-cell lung cancer: molecular analysis of the IDEAL/INTACT gefitinib trials. J Clin Oncol. 2005;23:8081–8092. doi: 10.1200/JCO.2005.02.7078. [DOI] [PubMed] [Google Scholar]
- 8.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubio-Viqueira B, Jimeno A, Cusatis G, et al. An in vivo platform for translational drug development in pancreatic cancer. Clin Cancer Res. 2006;12:4652–4661. doi: 10.1158/1078-0432.CCR-06-0113. [DOI] [PubMed] [Google Scholar]
- 10.Jones S, Hruban RH, Kamiyama M, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajeshkumar NV, Tan AC, De Oliveira E, et al. Antitumor effects and biomarkers of activity of AZD0530, a Src inhibitor, in pancreatic cancer. Clin Cancer Res. 2009;15:4138–4146. doi: 10.1158/1078-0432.CCR-08-3021. [DOI] [PubMed] [Google Scholar]
- 12.Rajeshkumar NV, Rasheed ZA, Garcia-Garcia E, et al. A combination of DR5 agonistic monoclonal antibody with gemcitabine targets pancreatic cancer stem cells and results in long-term disease control in human pancreatic cancer model. Mol Cancer Ther. 2010;9:2582–2592. doi: 10.1158/1535-7163.MCT-10-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang F, Ma J, Wu J, et al. PALB2 links BRCA1 and BRCA2 in the DNAdamage response. Curr Biol. 2009;19:524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 15.Shendure J, Stewart CJ. Cancer genomes on a shoestring budget. N Engl J Med. 2009;360:2781–2783. doi: 10.1056/NEJMe0903433. [DOI] [PubMed] [Google Scholar]