Structure and assembly of a paramyxovirus matrix protein (original) (raw)
Abstract
Many pleomorphic, lipid-enveloped viruses encode matrix proteins that direct their assembly and budding, but the mechanism of this process is unclear. We have combined X-ray crystallography and cryoelectron tomography to show that the matrix protein of Newcastle disease virus, a paramyxovirus and relative of measles virus, forms dimers that assemble into pseudotetrameric arrays that generate the membrane curvature necessary for virus budding. We show that the glycoproteins are anchored in the gaps between the matrix proteins and that the helical nucleocapsids are associated in register with the matrix arrays. About 90% of virions lack matrix arrays, suggesting that, in agreement with previous biological observations, the matrix protein needs to dissociate from the viral membrane during maturation, as is required for fusion and release of the nucleocapsid into the host’s cytoplasm. Structure and sequence conservation imply that other paramyxovirus matrix proteins function similarly.
The structure of icosahedral viruses has been extensively studied by X-ray crystallography and by a combination of X-ray crystallography and cryoelectron microscopy, but less is known about the atomic structure of viral proteins in the context of pleomorphic viruses. Paramyxoviridae, Rhabdoviridae, Filoviridae, and Bornaviridae are pleomorphic viruses belonging to the order Mononegavirales. These enveloped viruses all have a linear, nonsegmented, negative-sense, 10- to 20-kb–long RNA genome that is encapsidated by a nucleocapsid protein into a helical assembly (1). Paramyxoviruses include human pathogens that infect the respiratory system. Newcastle disease virus (NDV) is an avian paramyxovirus that is the prototypical species of the genus Avulavirus, belonging to the Paramyxovirinae subfamily. Poultry are especially susceptible to NDV infections, and some virulent strains have been classified by the US Department of Agriculture as select agents (2, 3).
Paramyxoviruses (Fig. 1) are pleomorphic and encode at least six proteins (1). They all have two transmembrane glycoproteins—the attachment protein termed HN (approximately 75 kDa), G, or H (depending upon the virus) and the F protein (approximately 60 kDa)—that form spikes protruding from the lipid bilayer. HN is a dual-function hemagglutinin/neuraminidase, capable of binding to cell surface sialic acids (4). F is required for fusion with the host cell plasma membrane (5–7). The nucleocapsid protein, NP (approximately 50 kDa), together with the genomic RNA forms a helical structure that encapsidates and protects the viral RNA genome. The helical parameters of the nucleocapsid structure exhibit variability among paramyxoviruses (8, 9). The matrix (M) protein of many pleomorphic, membrane-enveloped viruses directs assembly and budding (10–14). The structures of three matrix proteins of viruses belonging to the order Mononegavirales [respiratory syncytial virus (RSV) (14), Ebola virus (12), and Borna virus (13)] have been determined. These structures are built of one or two domains that have similar β-sandwich folds, suggesting gene duplication during evolution. The dimers of paramyxovirus M protein (approximately 40 kDa) can form a grid-like array on the inner surface of the viral membrane (15–18), and probably interact with both the cytoplasmic tails of the HN and F glycoproteins (19, 20) as well as the nucleocapsid (21–24) to initiate virus assembly and budding (20). The NDV M protein has greater than 20% sequence identity with other paramyxovirus M proteins including measles, mumps, and the parainfluenza viruses. However, the NDV M protein has no sequence homology to any proteins with known structure.
Fig. 1.
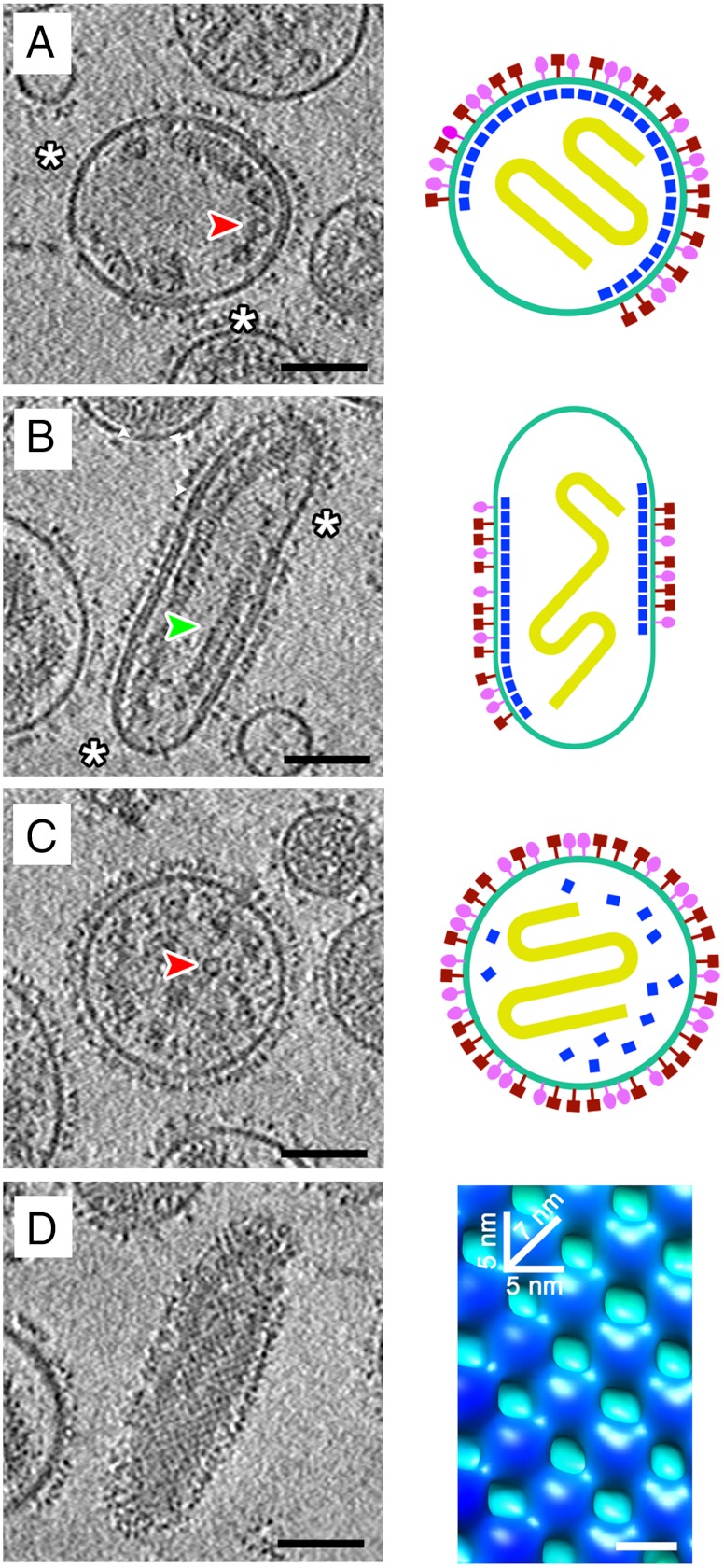
Tomographic and diagrammatic representations of NDV virions. (A) A tomographic section through the center of a nearly spherical virion, which has an approximately 4–5 nm thick matrix layer lining the inner surface of the viral membrane (Left). The white asterisks indicate the absence of glycoproteins and nucleocapsids where the virus lacks a matrix layer. The red arrowhead indicates a portion of the nucleocapsid as viewed end-on. The virion is also depicted schematically (Right). The HN and F glycoproteins are colored in red and magenta, respectively. The nucleocapsid is depicted in yellow and the matrix protein and membrane are colored dark blue and light blue, respectively. (B) A tomographic section through the center of an elongated virion (Left) and the corresponding schematic representation (Right). The white asterisks indicate the absence of glycoproteins and nucleocapsids where the virus lacks a matrix layer. The green arrowhead indicates a portion of the nucleocapsid that lies in plane. (C) A tomographic section through the center of a spherical virion that lacks an organized matrix protein layer (Left). The red arrowhead indicates the nucleocapsid as viewed end-on. Presumably, the matrix layer has dissociated from the viral membrane (Right). (D) The array of matrix proteins is evident in tomographic sections tangential to the viral membrane for some virions (Left). Subtomographic averaging generated an improved structure for a portion of the matrix protein layer (Right). The averaged map, as viewed from the inside of the virion, is colored according to the radius of a cylinder such that the density attributed to the membrane is light blue and the matrix protein subunits are dark blue. The on-edge and diagonal dimensions of a matrix protein subunit are indicated. For all the tomographic sections, strong density is black and the scale bar represents 75 nm. The tomographic images (A_–_C, Left) represent the average of three layers of voxels over a thickness of approximately 4.5 nm. The tangential section (D, Left) represents the average of seven layers of voxels over a thickness of 10.5 nm and is from the same virion represented in B.
Using X-ray crystallography in combination with electron tomography we determined the pseudoatomic structure of assembled M protein arrays in NDV and showed how the matrix protein arrays organize the glycoproteins and nucleocapsid and suggest the sequence of events during budding and fusion. Here, we report one of the few structural investigations of whole pleomorphic virions and a unique description of a matrix array in atomic detail.
Results and Discussion
Morphology of Newcastle Disease Virions.
Consistent with previous observations on paramyxoviruses (1), the morphology and size of NDV, as seen here in tomographic reconstructions (Fig. 1A_–_C, Left), varied from approximately spherical to ellipsoidal. The diameters of the spherical virions ranged from about 100 to 250 nm, whereas the ellipsoidal virions were sometimes as long as 350 nm and as narrow as 125 nm. On most virions, a layer of glycoprotein spikes extended about 12 to 18 nm beyond the surface of the 4–5 nm thick membrane. The electron tomographic results reported here show more detail of the paramyxovirus matrix proteins and nucleocapsid proteins in the virus than reported previously (17).
Within the virions, the nucleocapsids have both linear and bend segments. When viewed end-on, they have a ring-like cross-section with an outer diameter of about 20 nm and an inner diameter of 4–5 nm (Fig. 1A and C). For straight segments, the nucleocapsid has an approximately 7 nm repeating structure along its long axis, representing a helical arrangement of the nucleocapsid (Fig. 1B and Fig. S1).
Some virions have an additional layer of density on the inner surface of the viral membrane (Fig. 1A and B, Left). This layer is about 4 to 5 nm thick and represents the matrix protein. Although this layer was visible in only about 10% of the virions, SDS-PAGE showed that the matrix protein was in approximately the same abundance as the other structural proteins (Fig. S2). In those NDV particles where there is at least a partial matrix layer, the surface glycoproteins and internal nucleocapsids associate with the matrix array (Fig. 1A and B).
The Averaged Matrix Protein Array and Intercalating Glycoproteins.
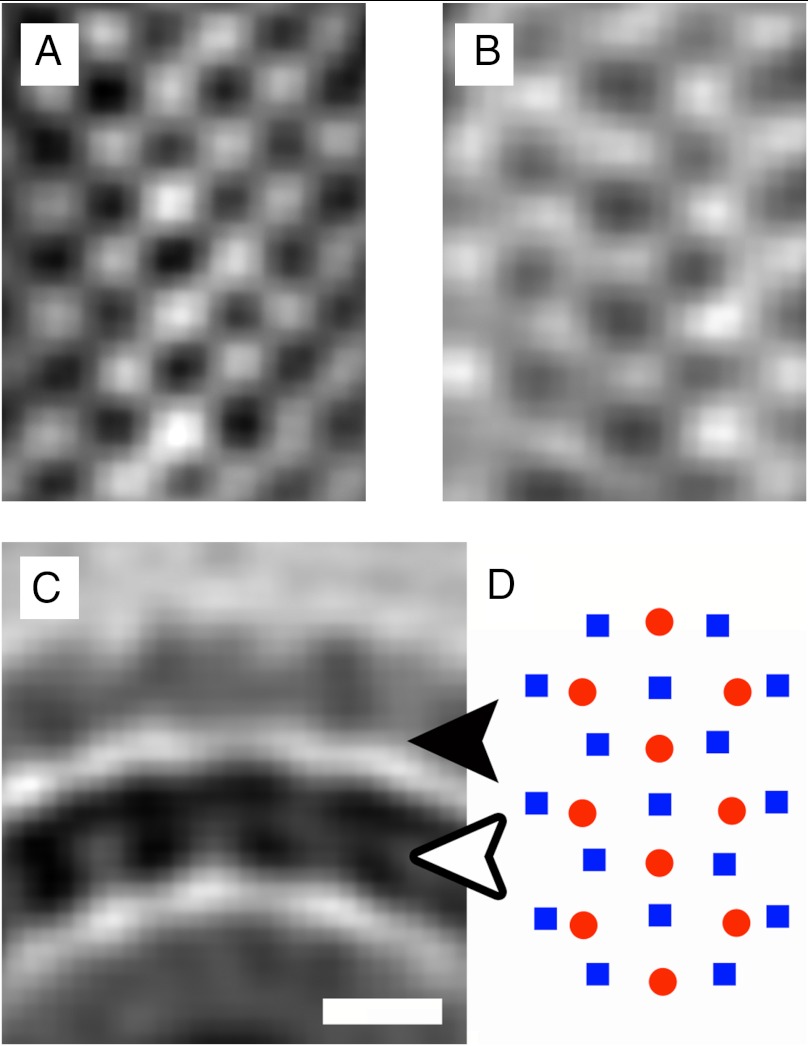
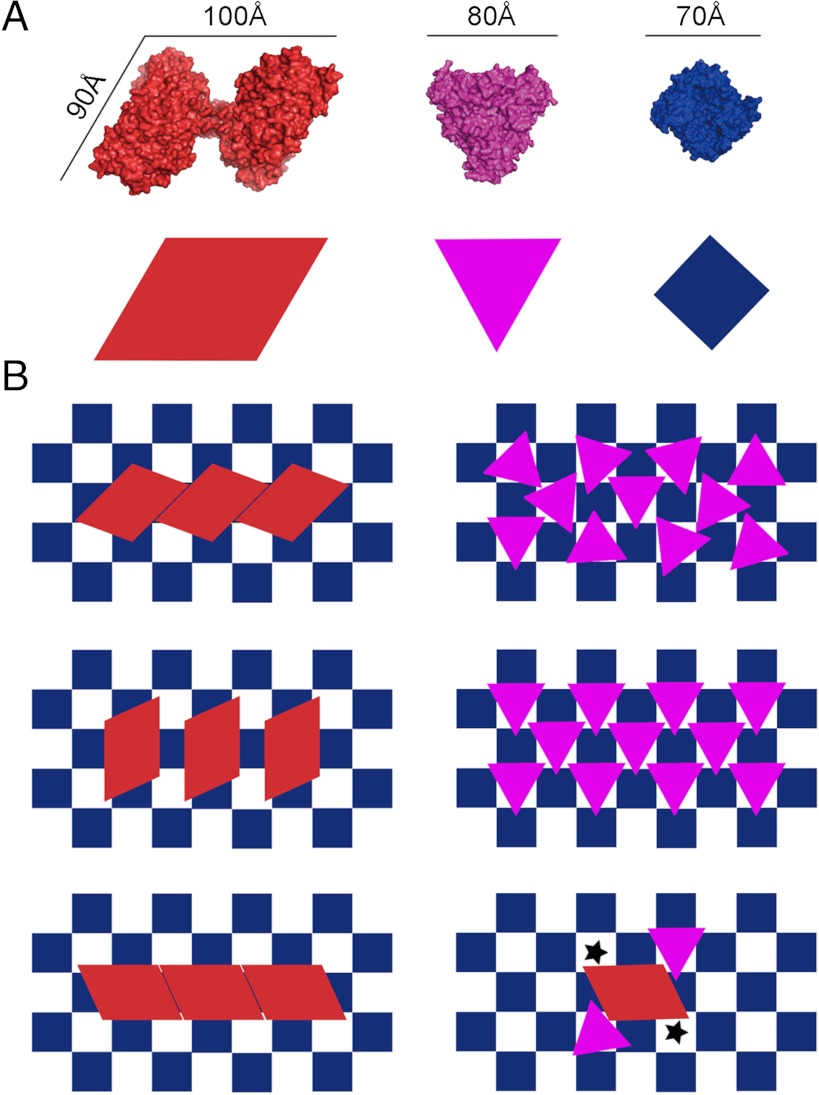
Tomographic sections tangential to the viral membrane show that the matrix layer forms a grid-like array (Fig. 1D, Left). An improved map representing the matrix protein and membrane was generated by averaging over repeating matrix subunits (SI Materials and Methods). This showed that the repeating unit of matrix protein has a nearly square shape, 5 nm on edge, which forms an array with a 7 nm repeat in orthogonal directions along the diagonals of the square (Fig. 1D, Right). Although visual inspection of the tomographic sections tangential to the virion surface did not show any obvious organization for the HN and F glycoprotein ectodomains, averaging the glycoprotein layer using the periodicity of the matrix protein showed the existence of an extracellular array with high densities in register with the gaps between repeating units in the matrix array on the inner side of the membrane (Fig. 2). The volume of the gaps between the matrix protein units is about equal to the volume of the repeating matrix protein unit itself, equivalent to about 70 kDa (SI Materials and Methods). The volume of this gap would be ample to accommodate the cytoplasmic tails of the tetrameric HN or trimeric F glycoproteins. Modeling of the NDV HN (25) or parainfluenza virus F (6) structures into the glycoprotein array shows that the glycoprotein ectodomains have the correct size to fit into neighboring sites consistent with the matrix protein periodicity. Although the head of the HN protein can be accommodated in arrays along one direction, the spaces in the orthogonal direction would only be able to accommodate the F protein (Fig. 3).
Fig. 2.
Periodic glycoprotein densities. (A) A section through the matrix protein array, which lines the inside of the viral membrane in the averaged tomographic map. (B) A section through the averaged map indicating an array on the extracellular side of the membrane. The extracellular densities were averaged assuming the same periodicities that were determined for the matrix protein array. The high densities in B oppose the low-density regions in A, indicating that the HN and F glycoproteins are anchored in the spaces between adjacent proteins in the matrix layer. (C) A cross-section of the averaged map, orthogonal to the ordered arrays. The white and black arrows indicate the planes from which A and B are derived, respectively. (D) A schematic diagram showing the relative locations of the matrix protein dimers (blue squares) and intercalating extracellular densities (red circles). For A_–_C, black represents high density.
Fig. 3.
The relationship between matrix and glycoproteins. (A) The crystal structures of NDV HN (red) (25), prefusion parainfluenza virus F (magenta) (6), and NDV M (navy blue) (present results) are shown (Top) and represented schematically (Bottom). The structures are shown such that the viral membrane would be in the plane of the page. (B) Three possible packing arrangements (Top, Middle, and Bottom) of the HN protein (Left) and F protein (Right) relative to the spaces between the matrix proteins. The size of the HN globular head would restrict the placement of other HN proteins in some neighboring spaces. Because the globular head of F is smaller than that of HN, the placement of the F glycoprotein within the array would not be restricted. (Lower Right) The HN and F glycoproteins as they might be situated relative to the matrix layer. Black stars indicate spaces between matrix proteins that could not be occupied by a glycoprotein because of steric hindrance.
The Tomographic Nucleocapsid Density.
The tomograms of NDV showed long regions of nucleocapsids that have a repeating pattern (Fig. S1). This pattern was shown to correspond to a helix with a pitch (the axial translation for one turn of the helix) of 7 nm, consistent with that of other paramyxovirus nucleocapsids (8, 9, 26). The twist (or number of NP repeats per turn of the helix) could not be determined because of the limited resolution of the tomographic maps (SI Materials and Methods and Fig. S1). The nucleocapsids observed in the tomograms were often in register with the matrix protein dimers, such that the 7 nm pitch along the long axis of the nucleocapsid was placed along the same axis as the 7 nm repeat along the diagonal of the matrix array (Fig. S3). However, a helically organized matrix protein enveloping the nucleocapsid structure as described for measles virus (24) was not observed.
Structure of the Dimeric Matrix Protein Determined by X-Ray Crystallography.
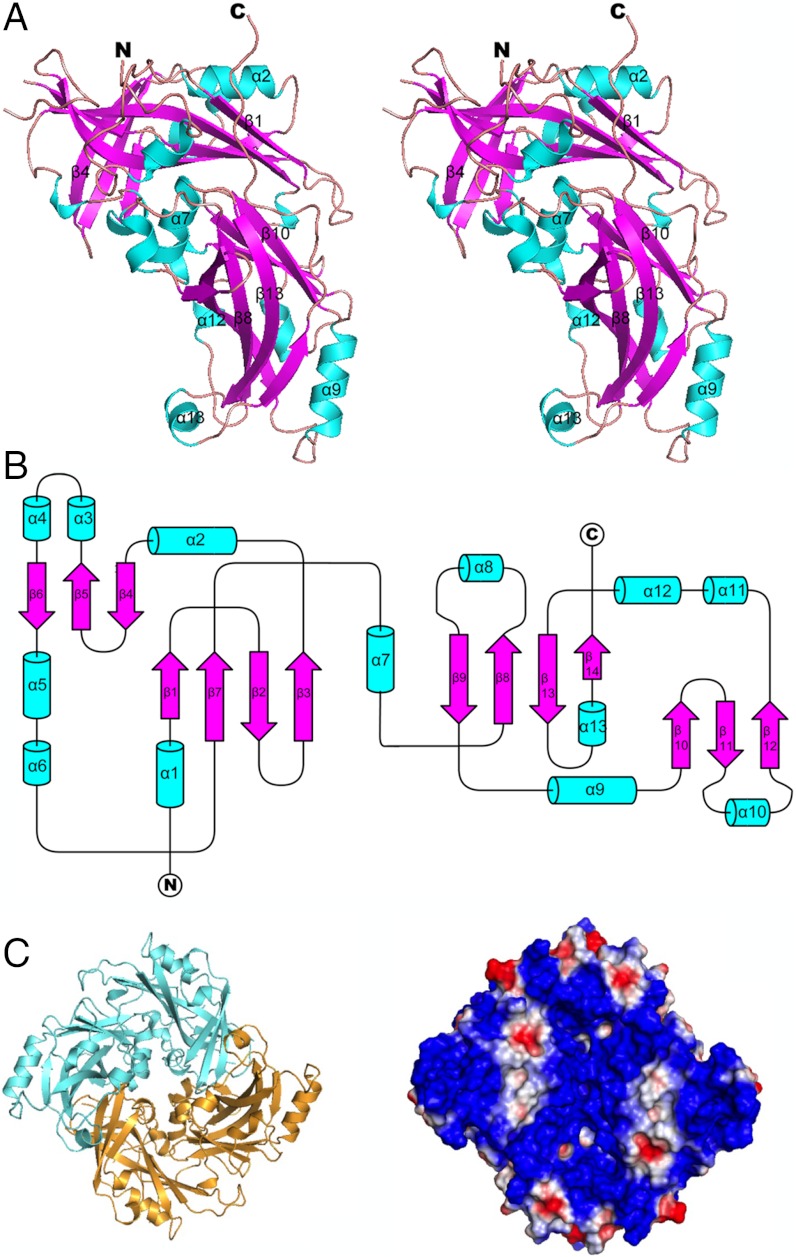
The full-length NDV M protein, containing 364 amino acids, was recombinantly expressed, purified, and crystallized. Elution peaks from size-exclusion chromatography indicated a dimeric matrix protein (Fig. S4). The structure was determined in three different crystal forms to at least 2.2 Å resolution (Table S1). Each crystal form had two monomers in the asymmetric unit related by the same dimer axis. The M protein monomer has two similarly folded domains (Fig. 4 A and B) joined by a 16 amino acid linker and related by a roughly fourfold axis coincident with the dimer twofold axis (Fig. 4C and SI Materials and Methods). The N-terminal domain extends from residues 1 to 181 and the C-terminal domain stretches from residues 198 to 364. Each domain consists of a β-sandwich in which the β-strands in the opposing β-sheets are approximately orthogonal to each other. Various α-helices are situated mostly on the surface of these β-sandwich domains (Fig. 4A). The domains can be superimposed with a rmsd of 3.8 Å between 74 equivalent Cα atoms of 167 residues.
Fig. 4.
The matrix protein structure from X-ray crystallography. (A) The ribbon drawing of the monomeric structure shown as a stereo diagram. The NDV matrix protein monomer consists of two similarly folded β-sandwich domains (magenta) that are flanked by a number of α-helices (cyan). (B) A schematic diagram representing the secondary structure elements of the matrix protein monomer. (C) The surface of the matrix protein dimer that faces the viral membrane is positively charged and has a nearly square envelope. The two monomers (blue and gold) are shown as a ribbon diagram (Left). The surface charge is represented on a space-filling model (Right), with the positively charged regions shown in blue and the negatively charged regions in red.
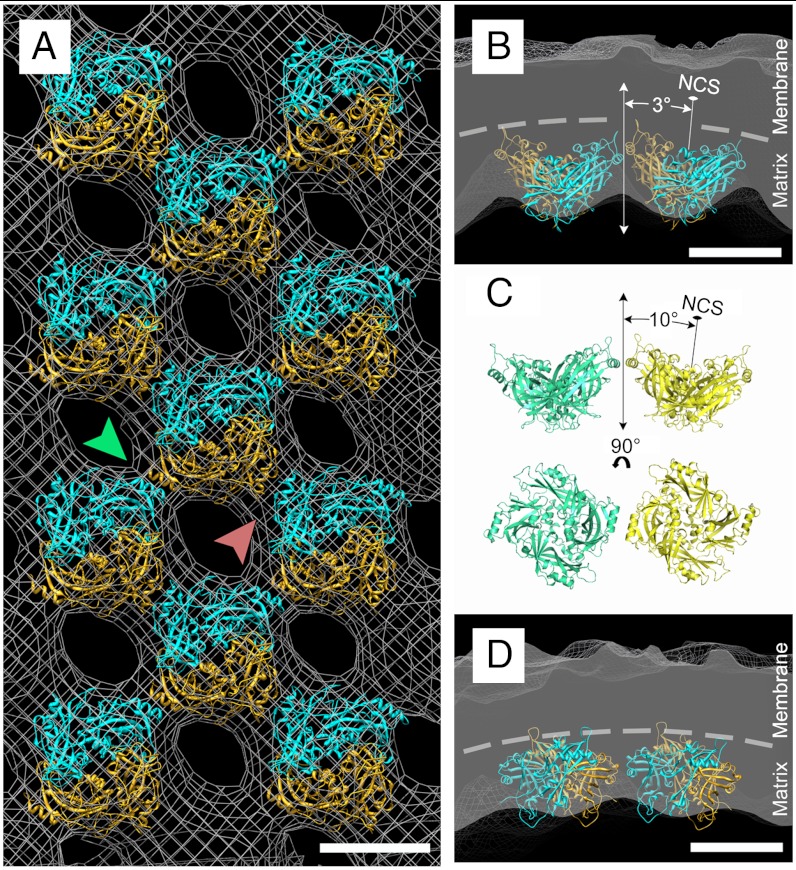
The dimeric M protein crystal structure (Fig. 4C) was fitted into the subunit density in the matrix protein layer of the tomograms (Fig. 5 and Table S2) using the program EMfit (27). The volume of the tomographic density was consistent with the volume of the crystallographic dimer structure (SI Materials and Methods and Table S2). The good quality of the fit verified that the crystallographic dimer is the physiologically relevant unit. Furthermore, the side of the matrix protein facing the membrane is highly positively charged as is required to associate with the negatively charged membrane surface (Fig. 4C, Right).
Fig. 5.
The matrix protein environment in the virus. (A) Fit of the dimeric matrix protein crystal structure (cyan and gold ribbon diagram) into the matrix protein density (gray mesh), which was generated by subtomographic averaging. The matrix protein array lies in the plane of the page. The green arrow indicates the contacts between helix α9 of adjacent dimers. The red arrow indicates the contacts between helix α2 of adjacent dimers. The scale bar represents 5 nm. (B and D) Cross-sections of the viral membrane and matrix layer. B shows the contacts between adjacent dimers generated by helix α9. The dashed line demarcates the membrane and matrix layer interface and illustrates the membrane curvature generated by the matrix protein array. D shows the contacts between adjacent monomers generated by helix α2. The scale bars represent 5 nm. (C) Contacts between matrix dimers similar to those shown in B are made between neighboring dimers in the monoclinic crystal structure. The NCS dimer axes related by a crystallographic twofold subtend an angle of 20° between them. Similar interdimer contacts are made between neighboring dimers in the virus (B), which subtend an angle of about 6° between them.
Structural Comparison with Other Mononegavirales Matrix Proteins.
A Dali search (28) gave a significant match between the NDV and the RSV matrix proteins as indicated by a Z score of 18 and a rmsd of 3.0 Å for 242 out of 254 residues. However, the crystal structure of RSV is a monomer (14), not a dimer as in NDV. This may be because the crystallized RSV matrix protein is missing the 25 carboxyl-terminal residues, including what would be helix α13 in NDV that is involved in the monomer-to-monomer contact within the dimer. The greater conservation of residues in the dimer contact surface as compared to elsewhere in the protein (Fig. S5) provides further evidence that the biologically relevant assembly unit is dimeric for paramyxoviruses. The Dali search also showed that the matrix protein of Borna virus consists of a single domain similar in fold to each β-sandwich domain of the NDV matrix protein. These domains are related by an exact fourfold axis in Borna virus (13) corresponding to the approximate fourfold axis in the NDV matrix protein. Ebola virus matrix protein, VP40, also consists of two β-sandwich domains (12), but their rotational relationship to each other is different than that in the NDV matrix protein. Although the sequence divergence between these different Mononegavirales matrix proteins is great, their function and tertiary structure are well-conserved (Fig. S6 and Table S3), as has frequently been observed in other proteins (29–32).
The Matrix Protein Environment.
The result of fitting the crystal structure into the tomographic density shows that there are two separate contact areas (Fig. 5) related by quasi-fourfold symmetry between neighboring matrix protein dimers in the virus. The first contact region consists of helix α9 interacting with the antiparallel helix α9 of a neighboring dimer. The other contact region consists of helix α2 interacting with the antiparallel helix α2 of a different neighboring dimer. The first contact region also occurs between dimers related by a crystallographic twofold axis in the monoclinic crystal structure. This contact involves hydrogen bonding and electrostatic interactions between Asp 255 and Arg 263 as well as Arg 262 and Glu 258. The angle between the noncrystallographic symmetry (NCS) dimer axes of the abutting dimers in the crystal is 20° (Fig. 5C), giving a similar curvature as is produced by the 6° angle between neighboring dimers in virions (Fig. 5B). Previously it was reported that the assembly of the matrix protein array is essential for virus budding (21). As is shown here, the contact between dimers controls the approximate curvature of the assembled matrix array and thus the virion’s shape while budding.
Implications for the Virus Life Cycle.
The contacts between the dimeric matrix proteins will determine the curvature of the matrix arrays as shown above and, hence, that of the budding virus membrane. The matrix protein is also required for recruiting the nucleocapsid and glycoproteins into the virus (21). The relatively few particles that were observed to have at least a partial matrix protein layer presumably represent the virus immediately after budding, because this event requires the matrix protein arrays for the generation of membrane curvature (33). A similar observation has been hinted for the distribution of viral proteins in the organization of Sendai virus (17). Therefore, other particles that did not show distinct matrix protein arrays probably represent the virus structure sometime after budding. These particles still contain the matrix protein (Fig. S2) but the arrays have disassembled, either as a result of the normal virus life cycle or the purification procedure. The signal for the dissociation of the matrix protein arrays in vivo might be, for instance, recognition of a receptor molecule by the glycoproteins. This disassembly is necessary for infection when the glycoproteins must be freed from the matrix array, allowing the F glycoprotein to undergo the conformational change required to transform from a pre- to postfusion state (34). In addition, the matrix protein has to be released from the membrane for the formation of fusion pores and the release of the nucleocapsid into the host’s cytoplasm. Thus, the matrix protein orders the maturation and infection process in time and space.
Materials and Methods
NDV was purified and vitrified for cryotomography (SI Materials and Methods). Tilt-series images were acquired and used to reconstruct 3D tomographic maps. Using the periodicity of the matrix protein seen in the reconstructed virions, an improved structure of the matrix protein array was generated by averaging subtomographic densities. The pitch of the helical nucleocapsid was determined by measuring the repeat distance along the length of the helix in the tomographic data. In addition, the full-length NDV matrix protein was recombinantly expressed, purified, and crystallized (SI Materials and Methods). The structure was determined by X-ray crystallography using single-wavelength anomalous dispersion data. The dimeric crystallographic structure was then fitted into the tomographic density and compared to the previously determined structures of homologous matrix proteins.
Supplementary Material
Supporting Information
ACKNOWLEDGMENTS.
We thank Anastasia Aksyuk and Giovanni Cardone for helpful comments and suggestions regarding image processing; Ken Hibler and Zhiheng Yu for assistance with the FEI Titan Krios Microscope; Paul Chipman, Siyang Sun, and Ye Xiang for helpful comments and suggestions throughout the course of this project; Xinzheng Zhang for assistance with electron microscopy; and Sheryl Kelly for help in the preparation of this manuscript. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-06CH11357. We thank National Institutes of Health for both an individual research award (AI11219 to M.G.R.) and support via the intramural research program of National Institute of Arthritis and Musculoskeletal and Skin Diseases (to A.C.S.). Support for A.J.B. was provided by a Biophysics Training Grant (T32 GM008296-20) from the National Institutes of Health/General Medicine (C.V. Stauffacher, principal investigator) and a Bilsland Dissertation Fellowship from Purdue University Graduate School.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates of the Newcastle disease virus M crystal structures have been deposited with the Protein Data Bank, www.pdb.org (PDB ID codes 4G1G, 4G1L, and 4G1O); the averaged cryoelectron tomography map of the matrix layer has been deposited with the Electron Microscopy Data Bank, www.emdatabank.org (EMDB ID code 5448).
References
- 1.Lamb RA, Parks GD. Paramyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Philadelphia: Lippincott Williams and Wilkins; 2007. pp. 1449–1496. [Google Scholar]
- 2.McGinnes LW, Pantua H, Reitter J, Morrison TG. Newcastle disease virus: Propagation, quantification, and storage. Curr Protoc Microbiol. 2006 doi: 10.1002/9780471729259.mc15f02s01. Chapter 15:Unit 15F.2. [DOI] [PubMed] [Google Scholar]
- 3.National Select Agent Registry. http://www.selectagents.gov. Accessed August 2012.
- 4.Scheid A, Caliguiri LA, Compans RW, Choppin PW. Isolation of paramyxovirus glycoproteins. Association of both hemagglutinating and neuraminidase activities with the larger SV5 glycoprotein. Virology. 1972;50:640–652. doi: 10.1016/0042-6822(72)90418-7. [DOI] [PubMed] [Google Scholar]
- 5.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci USA. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagai Y, Klenk H-D, Rott R. Proteolytic cleavage of the viral glycoproteins and its significance for the virulence of Newcastle disease virus. Virology. 1976;72:494–508. doi: 10.1016/0042-6822(76)90178-1. [DOI] [PubMed] [Google Scholar]
- 8.Bhella D, Ralph A, Yeo RP. Conformational flexibility in recombinant measles virus nucleocapsids visualised by cryo-negative stain electron microscopy and real-space helical reconstruction. J Mol Biol. 2004;340:319–331. doi: 10.1016/j.jmb.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 9.Egelman EH, Wu SS, Amrein M, Portner A, Murti G. The Sendai virus nucleocapsid exists in at least four different helical states. J Virol. 1989;63:2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris A, Forouhar F, Qiu S, Sha B, Luo M. The crystal structure of the influenza matrix protein M1 at neutral pH: M1–M1 protein interfaces can rotate in the oligomeric structures of M1. Virology. 2001;289:34–44. doi: 10.1006/viro.2001.1119. [DOI] [PubMed] [Google Scholar]
- 11.Rao Z, et al. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature. 1995;378:743–747. doi: 10.1038/378743a0. [DOI] [PubMed] [Google Scholar]
- 12.Dessen A, Volchkov V, Dolnik O, Klenk H-D, Weissenhorn W. Crystal structure of the matrix protein VP40 from Ebola virus. EMBO J. 2000;19:4228–4236. doi: 10.1093/emboj/19.16.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neumann P, et al. Crystal structure of the Borna disease virus matrix protein (BDV-M) reveals ssRNA binding properties. Proc Natl Acad Sci USA. 2009;106:3710–3715. doi: 10.1073/pnas.0808101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Money VA, McPhee HK, Mosely JA, Sanderson JM, Yeo RP. Surface features of a Mononegavirales matrix protein indicate sites of membrane interaction. Proc Natl Acad Sci USA. 2009;106:4441–4446. doi: 10.1073/pnas.0805740106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell PH, Almeida JD. A regular subunit pattern seen on non-infectious Newcastle disease virus particles. J Gen Virol. 1984;65:1023–1031. doi: 10.1099/0022-1317-65-6-1023. [DOI] [PubMed] [Google Scholar]
- 16.Heggeness MH, Smith PR, Choppin PW. In vitro assembly of the nonglycosylated membrane protein (M) of Sendai virus. Proc Natl Acad Sci USA. 1982;79:6232–6236. doi: 10.1073/pnas.79.20.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bachi T. Intramembrane structural differentiation in Sendai virus maturation. Virology. 1980;106:41–49. doi: 10.1016/0042-6822(80)90219-6. [DOI] [PubMed] [Google Scholar]
- 18.Hewitt JA. Studies on the subunit composition of the M-protein of Sendai virus. FEBS Lett. 1977;81:395–397. doi: 10.1016/0014-5793(77)80562-0. [DOI] [PubMed] [Google Scholar]
- 19.Waning DL, Schmitt AP, Leser GP, Lamb RA. Roles for the cytoplasmic tails of the fusion and hemagglutinin-neuraminidase proteins in budding of the paramyxovirus simian virus 5. J Virol. 2002;76:9284–9297. doi: 10.1128/JVI.76.18.9284-9297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pantua HD, McGinnes LW, Peeples ME, Morrison TG. Requirements for the assembly and release of Newcastle disease virus-like particles. J Virol. 2006;80:11062–11073. doi: 10.1128/JVI.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison MS, Sakaguchi T, Schmitt AP. Paramyxovirus assembly and budding: Building particles that transmit infections. Int J Biochem Cell Biol. 2010;42:1416–1429. doi: 10.1016/j.biocel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghildyal R, Mills J, Murray M, Vardaxis N, Meanger J. Respiratory syncytial virus matrix protein associates with nucleocapsids in infection cells. J Gen Virol. 2002;83:753–757. doi: 10.1099/0022-1317-83-4-753. [DOI] [PubMed] [Google Scholar]
- 23.Schmitt PT, Ray G, Schmitt AP. The C-terminal end of parainfluence virus 5 NP protein is important for virus-like particle production and M–NP protein interaction. J Virol. 2010;84:12810–12823. doi: 10.1128/JVI.01885-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liljeroos L, Huiskonen JT, Ora A, Susi P, Butcher SJ. Electron cryotomography of measles virus reveals how matrix protein coats the ribonucleocapsid within intact virions. Proc Natl Acad Sci USA. 2011;108:18085–18090. doi: 10.1073/pnas.1105770108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan P, et al. Structure of the Newcastle disease virus hemagglutinin–neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc Natl Acad Sci USA. 2011;108:14920–14925. doi: 10.1073/pnas.1111691108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tawar RG, et al. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science. 2009;326:1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- 27.Rossmann MG, Bernal R, Pletnev SV. Combining electron microscopic with x-ray crystallographic structures. J Struct Biol. 2001;136:190–200. doi: 10.1006/jsbi.2002.4435. [DOI] [PubMed] [Google Scholar]
- 28.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perutz MF, et al. Structure of haemoglobin. A three-dimensional Fourier synthesis at 5.5-Å resolution, obtained by X-ray analysis. Nature. 1960;185:416–422. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- 30.Rossmann MG, Moras D, Olsen KW. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974;250:194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- 31.Rossmann MG, et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 32.Nandhagopal N, et al. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc Natl Acad Sci USA. 2002;99:14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shnyrova AV, et al. Vesicle formation by self-assembly of membrane-bound matrix proteins into a fluidlike budding domain. J Cell Biol. 2007;179:627–633. doi: 10.1083/jcb.200705062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamb RA, Jardetzky TS. Structural basis of viral invasion: Lessons from paramyxovirus F. Curr Opin Struct Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information