Evaluation of the Compact High-Field Orbitrap for Top-Down Proteomics of Human Cells (original) (raw)
. Author manuscript; available in PMC: 2013 Aug 3.
Published in final edited form as: J Proteome Res. 2012 Jul 13;11(8):4308–4314. doi: 10.1021/pr3004216
Abstract
Mass spectrometry based proteomics generally seeks to identify and fully characterize protein species with high accuracy and throughput. Recent improvements in protein separation have greatly expanded the capacity of Top Down Proteomics (TDP) to identify a large number of intact proteins. To date, TDP has been most tightly associated with Fourier Transform Ion Cyclotron Resonance (FT-ICR) mass spectrometry. Here, we couple the improved separations to a Fourier-Transform instrument based not on ICR but using the Orbitrap Elite mass analyzer. Application of this platform to H1299 human lung cancer cells resulted in the unambiguous identification of 690 unique proteins and over 2000 proteoforms identified from proteins with intact masses <50 kDa. This is an early demonstration of high throughput TDP (>500 identifications) in an Orbitrap mass spectrometer and exemplifies an accessible platform for whole protein mass spectrometry.
Keywords: Top-down proteomics, Orbitrap Elite, high-resolution mass spectrometry
Introduction
In recent years, a variety of proteomic work flows have evolved to provide increasingly complete descriptions of pleiotropic protein molecules in human cells. Mass spectrometry-based proteomics now plays an important role in modern research of complex systems. Digestion-based methods may characterize thousands of peptides in a single project, providing significant insights into cellular dynamics at the level of protein molecules.1 The complementary technique of top-down proteomics (TDP) has also made great strides to increase the number of proteins identified. Recent publications show twenty-fold increases over past studies for large scale proteome mapping rather than characterization of single proteins.2 The complete characterization of protein species remains an enduring goal of proteomics, and the top-down method excels in this regard.3 Full characterization of proteins with alternate splice forms, diverse modifications and endogenous protein cleavages is very challenging for peptide-based proteomics.4 With improvements in sample handling, front-end separations, chromatography and informatics, many elements are now in place to routinely characterize intact proteins in a high-throughput manner. Despite the significant aforementioned improvements, TDP will benefit from further mass spectrometry improvements in mass resolution, scan speed, and dynamic range to obtain more useful information for systems biology.5
The instrumentation selected for TDP has often been Fourier Transform Ion Cyclotron Resonance (FT-ICR) mass spectrometers, which provide the high resolution and mass accuracy required for such analyses.6,7 While current FT-ICR instruments afford the highest available resolving power for intact proteins, other factors such as speed and ease of use become important for TDP to expand. Since resolution and scan speed are linearly related to the magnetic field strength, practical limitations prohibit large improvements in these metrics for ICR-based instruments. The development of electrostatic based Orbitrap instruments proves a route to higher resolution, allowing FTMS techniques to be applied to an increasing diversity of applications.8 Early uses of Orbitrap mass spectrometers were largely limited to peptide-based proteomics.9,10 While work by Michalski, et al., did show the potential for using the Orbitrap Elite for complex samples, they limited top down analysis to known proteins standards.10 The implementation of complex high-throughput TDP on these systems was limited by two main factors: a) Lower resolving power in a standard Orbitrap as compared to available FTICR mass analyzers (e.g. 60,000 vs 170,000 at 768 msec transient at 400 m/z for 12 Tesla FTICR) and b) A more rapid collision-mediated transient decay rate in the Orbitrap mass analyzer versus the FTICR cell.
The recent introduction of the compact high field Orbitrap hybrid mass spectrometer ameliorates these limitations and provides new opportunities to advance high-throughput intact protein analysis.10 This newest variety of Orbitrap mass analyzer (Orbitrap Elite) has outer electrodes with a reduced inner diameter (from previous generation Orbitraps: 30 mm to 20 mm), which provides a 1.78-fold increase in either resolution or speed. The reduced spacing between the central electrode and outer electrodes results in a higher electric field. The increased electric field offsets the reduced volume and enables a similar space charge capacity compared to previous Orbitrap mass analyzers.11 Additionally, the device has been capacitively balanced which prevents the preamplifier from entering saturation near the injection event and thus enables the start of detection within hundreds of microseconds of the injection event. With detection beginning almost immediately and the knowledge that all ions begin in phase, advanced signal processing methods have been applied to extract the absorption-mode spectrum from the transformed data, resulting in an additional factor of two increase in resolving power for a fixed transient length over traditional magnitude-mode spectra.12 Combined, these changes to the Orbitrap mass analyzer and the advanced signal processing should provide a 3.6 fold increase in resolution (or speed) compared to previous instruments. For comparison, at 400 m/z and a 768 ms transient, the new Orbitrap affords a resolution of 240,000 on compared to 170,000 with a 12 Tesla FTICR instrument utilizing magnitude-mode spectra. In addition, the effective resolution at higher mass to charge ratios decreases at a slower rate in the Orbitrap while the resolution of FTICR decreases as 1/(m/z)11,12
The resolution and speed enhancement of Orbitraps synergize well with recent improvements in protein separation.10 As the separation of intact proteins improves, faster scan speeds are essential for the efficient identification. In addition to faster scan speeds, alternative fragmentation methods can enhance protein characterization by potentially providing more diverse peptide backbone cleavage and retention of labile PTMs.13 Most TDP studies, to date, have used some form of collisionally activated dissociation (CAD). Electron transfer dissociation (ETD) is well suited for TDP because fragmentation of the peptide backbone occurs on a shorter time scale when compared to internal energy redistribution, is indifferent to peptide sequence, and preserves labile PTMs.14,15 Reaction rates for electron-based fragmentation have has been shown to scale inversely with protein mass, making it faster for intact proteins than small peptides.16 Combining ETD with other fragmentation methods used previously for top down can provide complementary information desirable for large scale intact protein analysis. For proteins in a known mass range, the most appropriate fragmentation and conditions can be chosen to provide the highest characterization possible in a dynamic LC-MS/MS experiment.
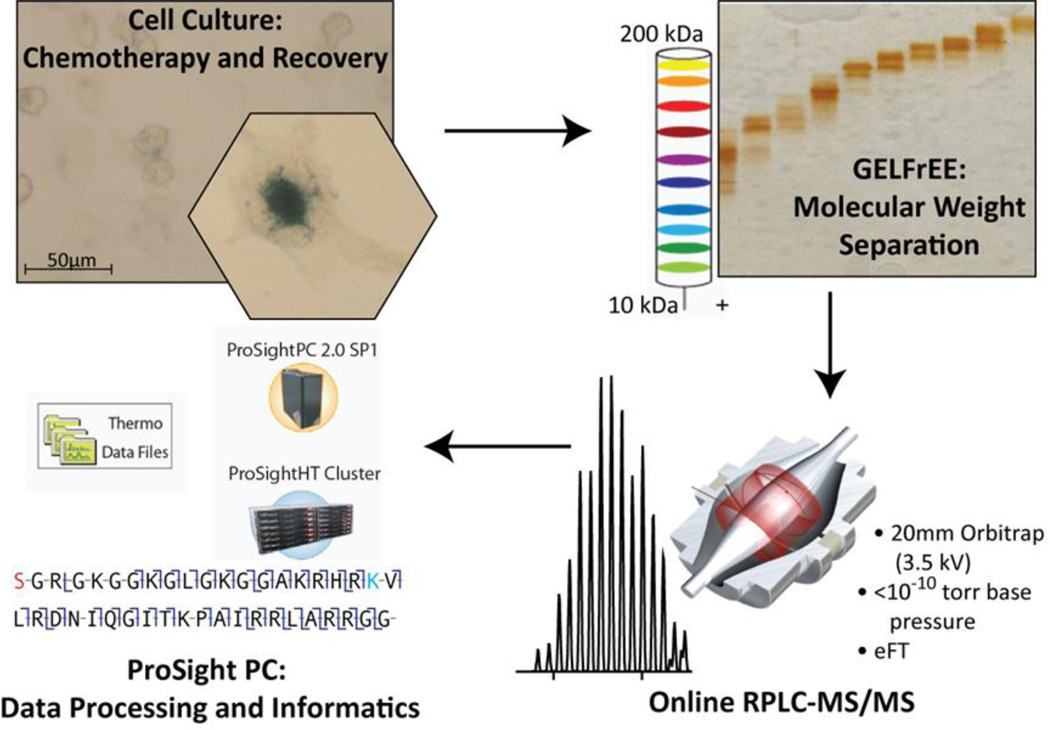
To evaluate coupling the Orbitrap Elite with our previously published high-throughput TDP platform,2 we chose the H1299 cancer cell line established from a patient with non-small cell lung cancer. These cells can be challenged with the DNA topoisomerase I inhibitor, camptothecin, to induce a cell cycle arrest and an accelerated senescence phenotype within several days post-treatment.17,18,19 Studies performed with H1299 cells provide a benchmark for future development of TDP and provide specific examples of post-translational dynamics in cells undergoing stress-induced senescence.17 These cells also provide an opportunity to test the current level of robustness of the high throughput top down platform (Figure 1), shown here to readily provide >500 identifications with an Orbitrap mass spectrometer.
Figure 1.
The overall platform used in this study of proteins from H1299 cells. Cell cultures treated and untreated were harvested in pellets from ~107 cells, and lysed. Protein aliquots of 200–400 µg were separated by GELFREE and SDS was removed. Analysis by nano-capillary RPLC-MS/MS with CID, ETD, HCD, or SID fragmentation followed. Data analysis and informatics was performed using ProSight running on a cluster or single PC with data processing routines detailed in the main text.
Experimental Section
Intact Protein Standards
Intact proteins were obtained from Sigma-Aldrich and mixed such that 1 pmol of myoglobin, 0.5 pmol of trypsinogen, 0.5 pmol carbonic anhydrase, and 0.1 pmol of ubiquitin were loaded on-column. Superoxide dismutase, a contaminant present in the carbonic anhydrase standard, was also analyzed. Proteins were separated as described below for intact proteins.
H1299 Cell Culture
H1299 cells were obtained from American Type Culture Collection (CRL-5803), and grown in Dulbecco’s Modified Eagle Medium (Sigma) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin solution. Stress-induced accelerated senescence was induced by 24 hour treatment with 25 nM camptothecin followed by 4 days of recovery in normal medium. The senescence phenotype was monitored by cell morphology changes, senescence-associated-β-galactosidase staining, and fluorescence imaging for senescence-associated heterochromatin foci as described previously.2 Cells (approximately 107) were harvested using 5% Trypsin-EDTA solution (Invitrogen) and centrifuged. After rinsing with phosphate buffered saline, cells were lysed in 150 mM Tris-HCl, 10 mM DTT, 4% sodium dodecyl sulfate buffer, pH 7.5, by boiling, then centrifuged. Aliquots (300 µL) were assayed for protein concentration by BCA protein assay (ThermoFisher, Rockford, IL) and then frozen.
Gelfree analysis and SDS removal
Gelfree analysis and SDS removal were performed as described previously.6,20 Briefly, approximately 400 µg of protein were precipitated with acetone to remove salts, and resuspended in 4% SDS solution prior to the addition of Gelfree loading buffer. Separation was performed using a commercial Gelfree 8100 Fractionation System, a technology based on the GELFrEE system published by Tran and Doucette,20,21 (Expedeon, Knoxville, TN) with either 10% or 12% cartridges to isolate proteins in ~5 kDa mass bins from 5 kDa to approximately 45 kDa. SDS was removed using the method of Wessel and Flügge.22 Resuspension in 30 µL Buffer A (95% water, 5% acetonitrile, 0.2% formic acid) allowed for multiple (3–5) technical replicates.
Reversed-phase Nano-capillary Liquid Chromatography
Resuspended fractions were loaded onto a 150 µm inner diameter, 2 cm bed length, (Phenomenex, Torrance, CA) trap column using a polymeric reversed phase media, PLRP-S, 1000Å pore size, 5 µm particle size. This media has Proteins were then transferred to a 75 µm inner diameter self-packed analytical column PLRP-S 1000 Å pore size, 10 cm, 5 µm particle size (Phenomenex) column for online separation. The nano-HPLC system (Dionex, Thermo Fisher, Sunnyvale, CA) was operated at 2.5 µL/min for loading onto the trap for 10 min. Afterwards, proteins were eluted at a constant flow rate of 300 nL/min using the following gradient: 5% B (95% acetonitrile, 5% water, 0.2% formic acid) at start, 25% at 5 min, 60% B at 45 min, 80% B at 50 min, 5% B at 60 min.
Mass Spectrometry Data Acquisition
All data were obtained on an Orbitrap Elite (Thermo Fisher Scientific, San Jose, CA), using the Orbitrap mass analyzer with a range of 500–2000 m/z in the precursor measurement and a target value of 1 million charges. Fragmentation was performed in a data-dependent fashion (top 3) with higher-energy collisional dissociation (HCD), CID, ETD, or source-induced dissociation (SID). In discovery-mode for proteins <25 kDa, each fragmentation method hit the same target before moving onto the next most intense, so a total of 9 fragmentation scans were acquired in this type of a top 3 experiment. For fractions containing >25 kDa proteins, fragmentation was performed with either HCD or SID (fragmentation of all incoming species). ETD activation time varied between 5 –25 ms depending on the mass range analyzed. For HCD, the normalized collision energy setting used was 30, while CID used 41. SID fragmentation scans were performed with an offset voltage of 100 V. Dynamic exclusion was enabled with a repeat count of 2, exclusion duration of 600 s, and a repeat duration of 240 s. Intact protein standards were analyzed with 240,000 resolving power (at 400 m/z) but routine intact protein analysis was performed using 120,000 resolving power for MS1 scans and either 120,000 or 60,000 for MS/MS scans.
Software and Data Analysis
Software solutions used for intact mass determination and species differentiation have been described recently.2 Briefly, RAW files collected were processed inside ProSightPC 2.0 SP1 using the CRAWLER algorithm average related scans to assign precursor and fragment masses using the Xtract algorithm. All fragmentation data were filtered (minimum fragment intensity at 100, retaining only the top 3 most intense neutral fragment masses within a 100 Da window below 2000 Da, and the top 5 most intense neutral fragment masses within a 100 Da window above 2000 Da). CRAWLER grouped one or more precursor mass(es) and observed fragment masses from each summed unit to create individual ProSightPC Experiments which were concatenated into an XML file in ProSight Upload Format (PUF).
PUF files were searched against a human proteome database using an implementation of ProSightPC 2.0 with iterative search logic on a 168-core Rocks Linux cluster. Iterative search trees were designed to begin with specific searches to take advantage of high mass accuracy (i.e., both 10 ppm at the intact and fragment ion level). Subsequent searches used larger intact mass tolerances (100 or 2000 Da) if a result of sufficient quality could not be determined initially. If the top result matched with a p-score of <1 × 10−8 the search engine accepted this result as valid and analyzed the next experiment in the PUF file, otherwise the next search in the tree was executed. As all fragmentation data were obtained in the Orbitrap mass analyzer at high resolving power, searches were performed with a 10 ppm fragment ion tolerance against a database made via shotgun annotation from the Swiss-Prot Homo sapiens release from the UniProt Knowledgebase release 2011_04.23 Data were searched against both forward and sequence scrambled databases (for FDR estimation purposes) separately, but with identical search parameters. Upon completion, all search results were loaded into a ProSight data repository and a report was produced for the top result of each experiment. The result with the best q-value (calculated as previously described)2 was chosen for each gene product. The q-value is based on first calculating a raw ProSight p-score based on the Poisson model.24,25 Next, the ProSight p-scores from scrambled sequence searches are fit to a Gamma distribution. Posterior probabilities are calculated for each result from the forward database to determine the probability that the score obtained could have been explained by the scrambled data. Data are reported at an estimated 5% FDR at the protein level.
Results and Discussion
Analysis of intact protein standards
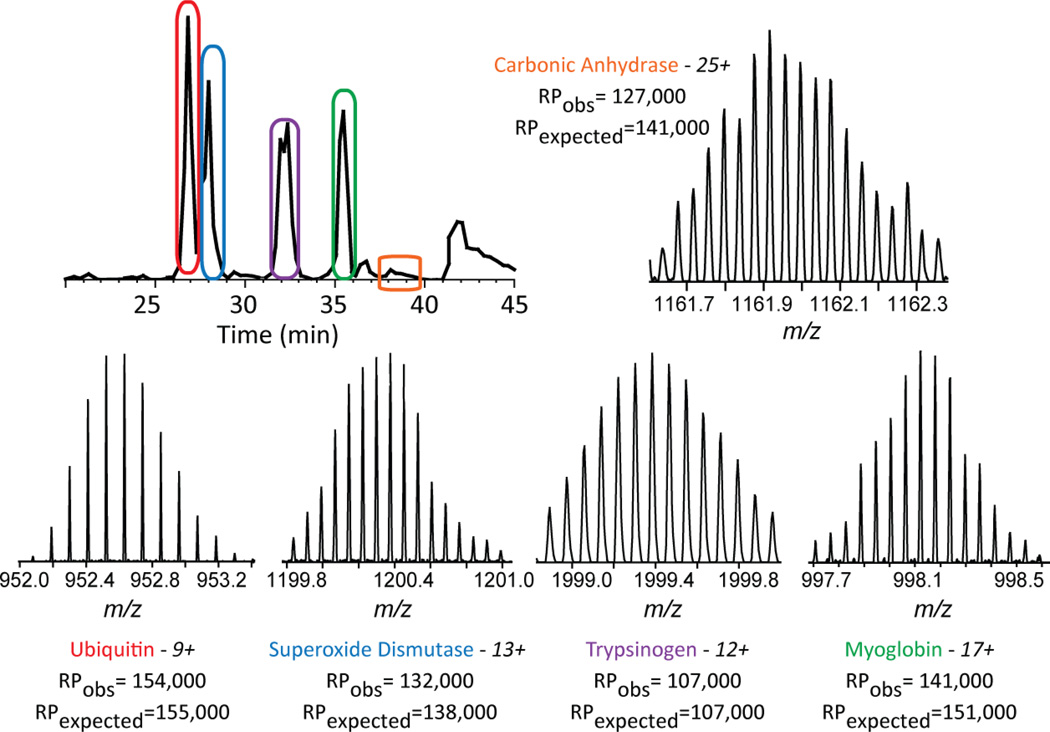
To assess the analytical figures of merit for the new Orbitrap, single LC-MS/MS analyses with a mix of standard proteins encompassing a range of masses from 8 kDa to 29 kDa were performed using multiple fragmentation techniques. Intact masses were readily observed with signal-to-noise values above 200:1 and resolved isotopic distributions (Figure 2) using a resolving power setting of 240,000 at 400 m/z. For superoxide dismutase, the observed resolving power at m/z 1114.57 was 138,500, agreeing with the resolving power expected at this m/z, 143,000. This represents an effective resolving power of 231,200 at m/z 400. For ubiquitin, online fragmentation with ETD resulted in a raw p-score of 9 × 10−118, CID gave a p-score of 1 × 10−75, and HCD resulted in a p-score of 8 × 10−53, all exceptionally confident identifications. The other protein standards were also fragmented with all three methods, producing highly confident identifications with the exception of trypsinogen. For future analyses within the mass range of these standards, a lower resolution setting (120,000) was used for MS2 spectra. This allows an increase in the rate of spectral acquisition without sacrificing the ability to resolve isotopic distributions or mass accuracy. Resolving power settings for fractions containing complex mixtures were chosen as described in the experimental section. These metrics indicate that this instrument should perform well for TDP of low and medium sized proteins using LC-MS/MS.
Figure 2.
Intact protein standards. Five protein standards were separated in a linear gradient. All proteins were isotopically resolved, and with the exception of trypsinogen, were completely characterized by MS/MS; RP= resolving power.
Multiple fragmentation methods for TDP
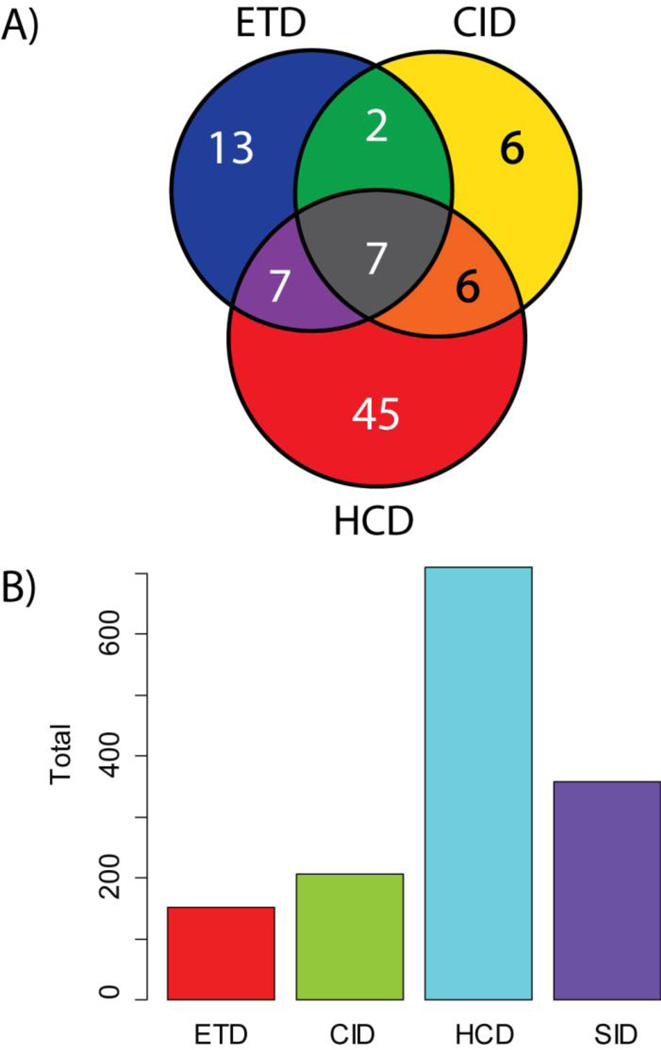
Previously, studies achieving high throughput TDP have taken advantage of mass-based separation using GELFREE to tailor the methods of fragmentation for LC-MS/MS runs based on the expected mass of the precursors present in particular GELFREE fractions (Figure 1). Adding to previously published studies utilizing CID and SID, the Orbitrap Elite allowed the use of HCD and ETD fragmentation in a robust and high-throughput fashion. To compare ETD, HCD and CID in the 5–20 kDa range, the fractions that elute earliest off the GELFREE device were analyzed with all three methods. These LC-MS/MS runs resulted in a total of 96 identifications distributed as shown in Figure 3a. HCD fragmentation had the highest number of unique identifications by accession number, but many of those identified with ETD had greater sequence coverage (Figure 4). For this study as a whole, the observed distribution of proteins with unique accession numbers identified with each fragmentation method is shown in Figure 3b. In these analyses, a greater number of LC-MS/MS runs were subjected to HCD fragmentation providing the observed skew of the data. Despite this bias, many of these proteins were identified by two or more fragmentation methods.
Figure 3.
A) Comparison of identifications across the three main fragmentation methods used in this study. For 6 LC-MS/MS runs encompassing the mass range of 5–20 kDa, a top 2 experiment, rotating between the three main fragmentation methods, produced a total of 96 identifications of unique Swiss-Prot accession numbers, separated as shown. ETD, HCD, and CID fragmented each target for a total of 6 fragmentation scans in a top 2 data-dependent experiment. B) Histogram of unique identifications separated by method of fragmentation. 45% of LC-MS/MS runs were fragmented using HCD, partially explaining the observed bias. Many proteins were also identified using multiple fragmentation methods.
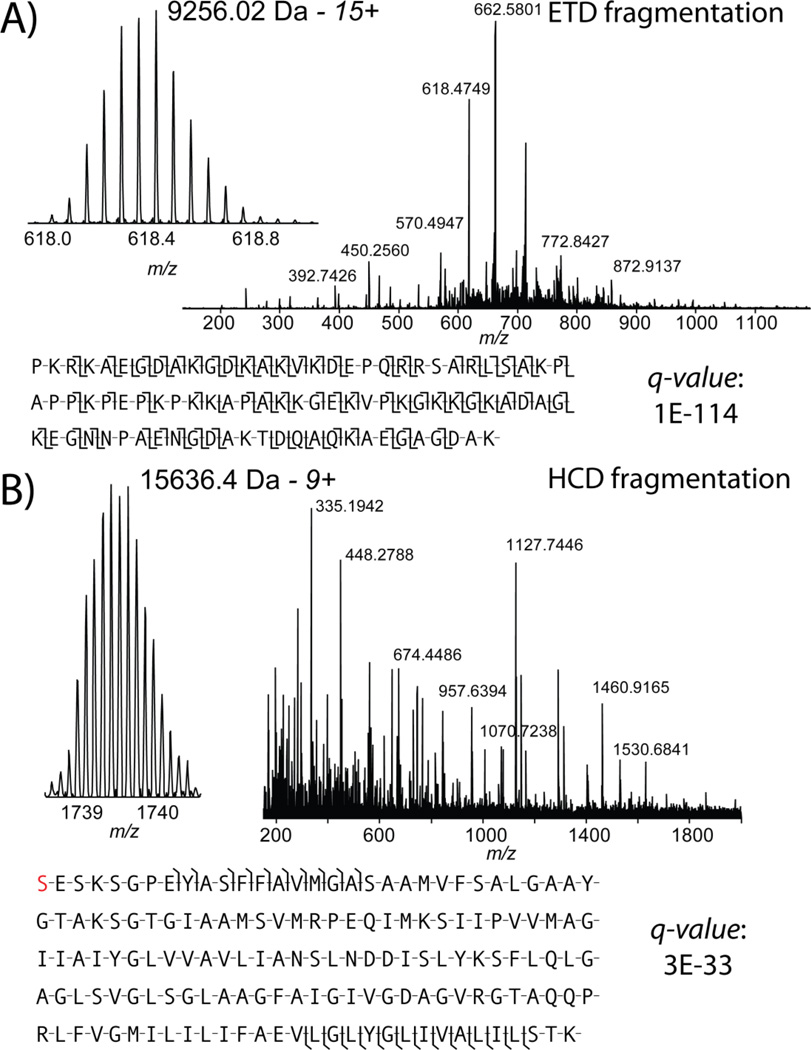
Figure 4.
Two example identifications obtained in discovery mode. A) Online electron transfer dissociation provided complete characterization of HMG-17 with a q-value of 1 × 10−114. B) Online HCD also provided a confident identification of the 15.6 kDa proteolipid subunit from the V-type ATPase with a q-value of 3 × 10−33.
Identifications from H1299 proteins below 50 kDa
Overall, 690 unique gene products were identified from 9 full GELFREE-LC-MS/MS experiments of H1299 cell proteins (abbreviated list in Table 1, for complete list see Supplementary Table 1). When considering posttranslational modifications, isoforms, alternative splices and peptide cleavages, evidence for up to 2366 proteoforms was accumulated. Proteoforms containing 337 phosphorylations, 75 monomethylations, 58 dimethylations, 31 trimethylations, and 892 acetylations were detected, and extensively or partially characterized. Overall, 50% of the identifications were consistent with precursor mass values determined with high mass accuracy. Of the 690 unique gene products identified, 105 automatically matched exactly to a protein form in the database within 1 or 2 Da. Another 94 gene products were validated manually to within 2 Da as a result of multiple precursors in the isolation window. The others were identified with mass shifts corresponding to cleavage events, multiple post-translational modifications, or artificial adducts. Many of the intact mass spectra for the 690 unique gene products were observed in a multiplicity of forms and/or at a low signal-to-noise in the full MS scan.
Table 1.
Selected proteins identified in this study. For a complete table, please see Supplementary Table 1
| ID | Protein | TheoreticalMass (Da) | ObservedMass(Da) | ΔMass(ppm) | q-value | PTMsObserved | Frag.Type |
|---|---|---|---|---|---|---|---|
| P05204 | HMG-17 | 9256.0 | 9256.03 | 1.6 | 1E-114 | N/A | ETD |
| P20962 | Parathymosin | 11434.2 | 11434.2 | 0.2 | 2E-71 | N-acetyl | ETD |
| P06454-2 | Prothymosinalpha | 11977.9 | 11977.9 | 0.6 | 2E-62 | N-acetyl, O-phospho-L-serine | ETD |
| P07195 | L-Lactatedehydrogenase Bchain | 36834.7 | 36834.9 | 5.9 | 2E-46 | N-acetyl, N6-acetyl-L-lysine | SID |
| Q07021 | Complementcomponent 1 Qsubcomponent-binding protein | 23768.6 | 23768.7 | 7.5 | 5E-46 | N6-acetyl-L-lysine, O-phospho-L-tyrosine/serine, | SID |
| P84090 | Enhancer ofrudimentaryhomolog | 12162.0 | 12161.9 | −6.6 | 3E-34 | N-acetyl, N6-acetyl-L-lysine | CID |
| P27449 | V-type protonATPase 16 kDaproteolipidsubunit | 15636.4 | 15636.5 | 5.9 | 3E-33 | N-acetyl | HCD |
| P60174 | Triosephosphateisomerase | 26521.7 | 26521.6 | −3.0 | 7E-31 | N-acetyl, O-phospho-L-serine | SID |
| P22392 | Nucleosidediphosphatekinase B | 17197.9 | 17197.9 | −0.6 | 5E-30 | N-acetyl, N6-acetyl-L-lysine,O-phospho-L-threonine | ETD |
| Q9H299 | SH3 domain-binding glutamicacid-rich-likeprotein 3 | 10342.2 | 10342.3 | 3.4 | 3E-25 | N-acetyl | HCD |
The identification power of this platform is exemplified by Figure 4a. In a single scan (four microscans), a 9.2 kDa protein, HMG-17 was completely characterized. Unambiguously identified with an exceptional q-value of 1 × 10−144, the fragmentation map shows that 68% of all backbone bonds were cleaved in this chromatin-related protein. The q-value is calculated (as above) to obtain a FDR by correcting for multiple hypotheses in large scale LC-MS runs, while p-score is a probability score reflecting the quality of match between a single spectrum and a single sequence. This protein has two known phosphorylations (though only one low-level phosphorylation was observed) and an acetylation that can be monitored via top-down mass spectrometry in the future if present at a stoichiometry greater than ~1%. With the fragmentation displayed in Figure 4a, localization of any post-translational modification would be unequivocal. Online HCD also provided confident identification of the 15.6 kDa protein V-type proton ATPase proteolipid subunit with a q-value of 3 × 10−33(Figure 4b).
Only one high-throughput TDP study has been published with more identifications (1,043 gene products), but this analysis added an additional dimension of isoelectric focusing prior to the mass-based GELFREE. Our study presents a sizable increase in identifications in this mass range over previous high-throughput TDP studies without this additional fractionation.2,26 Also, all data for this study from 9 GELFREE-LC-MS/MS analyses were collected in approximately one month, and limited to the mass range below 50 kDa. This compares quite favorably to the amount of time required to gather previous TDP data sets.2
The typical cycle time for a successful top-down experiment on a high resolution FT-ICR instrument was 4–8 s when performing a top 3 experiment for proteins below 30 kDa.2 Performing the same experiment on the Orbitrap Elite, this cycle time is reduced to a total cycle time of 1.5 s. The reduction in cycle time is because of the increased resolution per unit time provided by both enhanced signal processing and the compact HF Orbitrap mass analyzer described above. Additional sensitivity and speed gains are provided by predictive AGC and the brighter source (i.e. higher number of ions per time the source is able to transmit) provided by the LTQ Velos Pro.
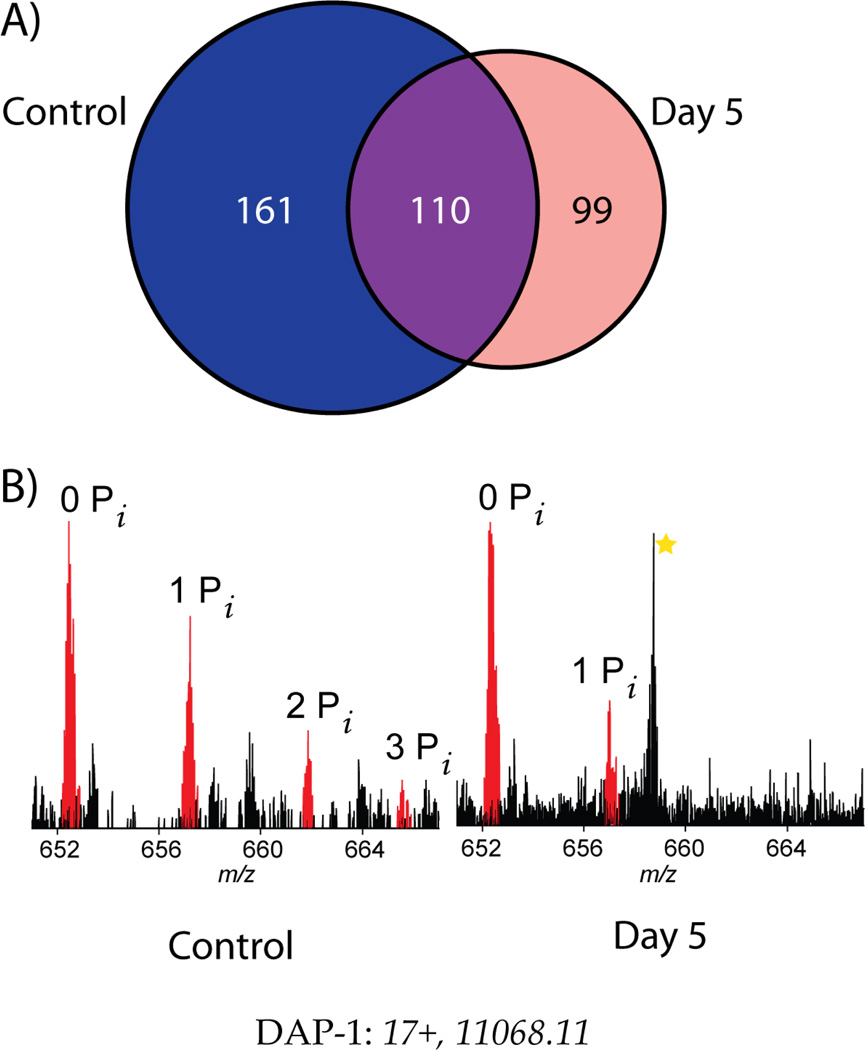
In addition to large-scale analysis of H1299 cells in an asynchronous state, a comparison was performed between H1299 control cells and those harvested in an accelerated senescence phenotype. Many proteins were identified in identical analyses of 2 biological replicates (24 LCMS/MS runs) for each state (Figure 5a). Within this initial set of identifications, several different proteins indicated relative PTM and proteoform abundance changes. For example, Figure 5b shows a clear change in PTM dynamics between treated and untreated cells for Death Associated Protein-1 (Uniprot ID P51397). This protein is known to be involved in apoptosis and autophagy, and our evidence shows reduced phosphorylation occurring during stress-induced senescence. Soon, this target and several others will be investigated to detect and characterize full phosphorylation hierarchies linked to decisions of cellular fate.
Figure 4.
Initial scan for proteins dynamic with the onset of cellular senescence 5 days after treatment with a DNA-damaging agent. A) A Venn diagram detailing identifications from two biological replicate injections of whole cell lysate separated on a 12% GELFREE cartridge (masses up to 35 kDa). B) An example of PTM changes across senescence with Death Associated Protein-1 (DAP-1). Prior to the onset of senescence, two phosphorylations are clearly visible with a third possible. After the onset of senescence, only one phosphorylation is observed, and at a lower stoichiometry than originally observed. The absolute signal intensities are comparable, strongly supporting the assertion that a hypophosphorylated state is adopted on DAP-1 during senescence.
Conclusion
Utilizing new Orbitrap instrumentation to provide increased speed and options for fragmentation has allowed more TDP identifications per unit time than has been previously achieved.6,26 This foray into TDP has focused on a traditionally amenable mass range (<50 kDa), but both the mass range and number of identifications and characterizations will expand rapidly in the near future. Improvements to previously developed software allow for continued confidence in protein identification, streamlined database searching, automatic characterization of modifications, and detection of yet-unexplained mass shifts in endogenous human proteins. This proof-of-concept for high throughput top down proteomics on Orbitrap instrumentation also lays the groundwork for biological studies into the post-translational dynamics of accelerated senescence. We project an increase in confident characterization of proteins using the integrated approach described here.
Supplementary Material
1_si_001
2_si_002
Acknowledgements
The authors thank all of the members of the Top Down Development Team and the Kelleher Group for their assistance, as well as NIH grant GM067193 and DA018310. This work was partially funded by the Chicago Biomedical Consortium with support from The Searle Funds at The Chicago Community Trust. For graduate student support we thank the NSF-GK12 fellowship Reach For The Stars, Northwestern University to D.A. Image of the Orbitrap in Figure 1 reproduced with permission of Thermo Fisher Scientific.
Footnotes
Supporting Information Available
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
Full list of identifications (.XLSX format)
References
- 1.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nature Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 2.Tran JC, Zamdborg L, Ahlf DR, Lee JE, Catherman AD, Durbin KR, Tipton JD, Vellaichamy A, Kellie JF, Li M, Wu C, Sweet SMM, Early BP, Siuti N, LeDuc RD, Compton PD, Thomas PM, Kelleher NL. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Service RF. Proteomics ponders prime time. Science. 2008;321:1758–1761. doi: 10.1126/science.321.5897.1758. [DOI] [PubMed] [Google Scholar]
- 4.Nesvizhskii A, Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Molecular & Cellular Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Compton PD, Zamdborg L, Thomas PM, Kelleher NL. On the scalability and requirements of whole protein mass spectrometry. Analytical Chemistry. 2011;83:6868–6874. doi: 10.1021/ac2010795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JE, Kellie JF, Tran JC, Tipton JD, Catherman AD, Thomas HM, Ahlf DR, Durbin KR, Vellaichamy A, Ntai I, Marshall AG, Kelleher NL. A Robust Two-Dimenensional Separation for Top-Down Tandem Mass Spectrometry of the Low- Mass Proteome. Journal of the American Society for Mass Spectrometry. 2009;20:2183–2191. doi: 10.1016/j.jasms.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roth MJ, Parks BA, Ferguson JT, Boyne II MT, Kelleher NL. “Proteotyping”: population proteomics of human leukocytes using top down mass spectrometry. Analytical Chemistry. 2008;80:2857–2866. doi: 10.1021/ac800141g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scigelova M, Makarov A. Orbitrap Mass Analyzer -Overview and Applications in Proteomics. Proteomics. 2006;6:16–21. doi: 10.1002/pmic.200600528. [DOI] [PubMed] [Google Scholar]
- 9.Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, Wouters ER, Senko M, Makarov A, Mann M, Horning S. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Molecular & Cellular Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michalski A, Damoc E, Lange O, Denisov E, Nolting D, Mueller M, Viner R, Schwartz J, Remes P, Belford M, Dunyach J-J, Cox J, Horning S, Mann M, Makarov A. Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Molecular & Cellular Proteomics. 2012;11:1–11. doi: 10.1074/mcp.O111.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarov A, Denisov E, Lange O. Performance evaluation of a high-field Orbitrap mass analyzer. Journal of the American Society for Mass Spectrometry. 2009;20:1391–1396. doi: 10.1016/j.jasms.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Lange O, Damoc E, Wieghaus A, Makarov A. Enhanced FT for Orbitrap Mass Spectrometry; Presented at the 59th ASMS Conference on Mass Spectrometry; 2011. [Google Scholar]
- 13.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annual Review of Biomedical Engineering. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 14.Compton PD, Strukl JV, Bai DL, Shabanowitz J, Hunt DF. Optimization of electron transfer dissociation via informed selection of reagents and operating parameters. Analytical Chemistry. 2012;84:1781–1785. doi: 10.1021/ac202807h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Syka J, Coon J, Schroeder M. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Electron Capture Dissociation for Structural Characterization of Multiply Charged Protein Cations. Analytical Chemistry. 2000;72:563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 17.Roberson RS, Kussick SJ, Vallieres E, Chen S-YJ, Wu DY. Escape from Therapy-Induced Accelerated Cellular Senescence in p53-Null Lung Cancer Cells and in Human Lung Cancers. Cancer Research. 2005;65:2795–2803. doi: 10.1158/0008-5472.CAN-04-1270. [DOI] [PubMed] [Google Scholar]
- 18.Zhang R, Chen W, Adams PD. Molecular dissection of formation of senescenceassociated heterochromatin foci. Molecular and Cellular Biology. 2007;27:2343–2358. doi: 10.1128/MCB.02019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collado M, Blasco M, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Tran JC, Doucette A. Gel-eluted liquid fraction entrapment electrophoresis: an electrophoretic method for broad molecular weight range proteome separation. Analytical Chemistry. 2008;80:1568–1573. doi: 10.1021/ac702197w. [DOI] [PubMed] [Google Scholar]
- 21.Tran JC, Doucette A. Multiplexed size separation of intact proteins in solution phase for mass spectrometry. Analytical Chemistry. 2009;81:6201–6209. doi: 10.1021/ac900729r. [DOI] [PubMed] [Google Scholar]
- 22.Wessel D, Flügge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Analytical Biochemistry. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 23.Roth MJ, Forbes AJ, Boyne MT, Kim Y-B, Robinson DE, Kelleher NL. Precise and parallel characterization of coding polymorphisms, alternative splicing, and modifications in human proteins by mass spectrometry. Molecular & Cellular Proteomics. 2005;4:1002–1008. doi: 10.1074/mcp.M500064-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng F, Cargile BJ, Miller LM, Forbes AJ, Johnson JR, Kelleher NL. Informatics and multiplexing of intact protein identification in bacteria and the archaea. Nature Biotechnology. 2001;19:952–957. doi: 10.1038/nbt1001-952. [DOI] [PubMed] [Google Scholar]
- 25.Taylor GK, Kim Y-bin, Forbes AJ, Meng F, Mccarthy R, Kelleher NL. Web and Database Software for Identification of Intact Proteins Using" Top Down" Mass Spectrometry harboring posttranslational modifications ( PTMs ), a" top. Analytical Chemistry. 2003;75:4081–4086. doi: 10.1021/ac0341721. [DOI] [PubMed] [Google Scholar]
- 26.Vellaichamy A, Tran JC, Catherman AD, Lee JE, Kellie JF, Sweet SMM, Zamdborg L, Thomas PM, Ahlf DR, Durbin KR. Size-Sorting Combined with Improved Nanocapillary-LC-MS for Identification of Intact Proteins up to 80 kDa. Analytical Chemistry. 2010;82:1234–1244. doi: 10.1021/ac9021083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1_si_001
2_si_002