Structural analysis of B-cell epitopes in antibody:protein complexes (original) (raw)
. Author manuscript; available in PMC: 2014 Jan 1.
Abstract
The binding of antigens to antibodies is one of the key events in an immune response against foreign molecules and is a critical element of several biomedical applications including vaccines and immunotherapeutics. For development of such applications, the identification of antibody binding sites (B-cell epitopes) is essential. However experimental epitope mapping is highly cost-intensive and computer-aided methods do in general have moderate performance. One major reason for this moderate performance is an incomplete understanding of what characterizes an epitope. To fill this gap, we here developed a novel framework for comparing and superimposing B-cell epitopes and applied it on a dataset of 107 non-similar antigen:antibody structures extracted from the PDB database. With the presented framework, we were able to describe the general B-cell epitope as a flat, oblong, oval shaped volume consisting of predominantly hydrophobic amino acids in the center flanked by charged residues. The average epitope was found to be made up of ~15 residues with one linear stretch of 5 or more residues constituting more than half of the epitope size. Furthermore, the epitope area is predominantly constrained to a plane above the antibody tip, in which the epitope is orientated in a −30 to 60 degree angle relative to the light to heavy chain antibody direction. Contrary to previously findings, we did not find a significant deviation between the amino acid composition in epitopes and the composition of equally exposed parts of the antigen surface. Our results, in combination with previously findings, give a detailed picture of the B-cell epitope that may be used in development of improved B-cell prediction methods.
Keywords: Antibody, Antigen, Epitope, Structure, Amino acid distribution
1. Introduction
One of the key events in the clearance of pathogens and foreign molecules by the immune system is the interaction between antibodies and antigens. Antibodies bind to antigens at sites known as antigenic determinant regions, which are also called B-cell epitopes since antibodies are produced by B-lymphocytes (B-cells). Identification of sites on the antigen surface capable of binding to antibodies is essential in several biomedical application such as; rational vaccine design, disease diagnostic and immune-therapeutics (Gershoni et al., 2007; Irving et al., 2001). Experimental identification of B-cell epitopes is costly and time consuming, and use of in silico screening methods is therefore an appealing alternative. The performance of methods for B-cell epitope prediction is however not optimal, with a significant proportion of the predicted epitopic sites being false positives and visa versa for the negative predictions. One important reason for this relative low predictive performance is our poor understanding of the properties that characterize a B cell epitope. Thus, a detailed description of the epitope area in terms of sequence composition and structural characteristics could potentially greatly contribute to development of improved methods for B cell epitope identification. Only in resent years has the number of publicly available structures of antigen:antibody complexes increased to a level where sound statistical characterization of B-cell epitopes can be accomplished and only a limited number of publications has focused entirely on B-cell epitope characterization. Studies on the broader field of protein-protein interactions either exclude antibody-antigen complexes (Bordner and Abagyan, 2005; Neuvirth et al., 2004) or fail to acknowledge antigen-antibody complexes as a special group of protein interactions (Bickerton et al., 2011; Bogan and Thorn, 1998; Chakrabarti and Janin, 2002; Keskin et al., 2005; Li et al., 2012; Lo Conte et al., 1999). This last point might be important as earlier work suggests that the physico-chemical and, to some extent, the structural composition of B-cell epitopes are different from the general composition of sites involved in protein-protein interactions (Ofran et al., 2008).
One of the most cited characteristics of the epitope is that they reside on the surface of the protein. This feature was first described in the work of Novotný et al. (1986) by calculating the solvent accessible surface area of residues involved in antigen-antibody binding from the 3-dimensional structures of lysozyme, myoglubin, myohemerythrin and cytochrome c. Furthermore, from the same set of structures, Thornton et al. (1986) demonstrated that antigenic areas protrude from the surface of the antigen. They approximated the shape of the proteins as an ellipsoid and observed that amino acids involved in antibody binding were predominantly located outside the ellipsoid surface. Recently, Lollier et al. (2011) challenged the general assumption that epitopes are confined to the protein surface. They were unable to establish a relationship between residues in continuous and discontinuous epitopes (data obtained from IEDB database, Vita et al., (2010)) and relative solvent accessibility (RSA), or the protrusion index (PI). However, the results might have a high degree of uncertainty, due to the fact that most epitopes in the data used were linear epitopes obtained by B-cell assays, which do not explicitly determine the residues in contact with the antibody (for a review of methods see Van Regenmortel, (2009)). Furthermore, other studies exclusively based on 3-dimensional structures conclude that epitope residues are more surface exposed compared to antigen residues in general (Andersen et al., 2006; Ofran et al., 2008; Rubinstein et al., 2008; Sun et al., 2011).
Another frequently investigated feature of the B-cell epitope is the amino acid composition (Andersen et al., 2006; Ofran et al., 2008; Rubinstein et al., 2008; Sun et al., 2011; Zhao and Li, 2010). It is generally agreed that epitopes are enriched in charged and polar amino acids and depleted of aliphatic hydrophobic amino acids, when comparing the epitope amino acid distribution to either the entire PDB database (Ofran et al., 2008) or amino acid composition of the antigen as a whole (Andersen et al., 2006; Zhao and Li, 2010) Furthermore, by recognizing that epitopes usually reside on the protein surface, Rubinstein et al. (2008) suggested that the amino acids Tyr and Trp are significantly over-represented in epitopes and that Val is significantly depleted. Besides individual amino acid preferences in epitopes, specific amino acid pairs have been observed more frequently in both linear (sequence based studies) (Chen et al., 2007) and conformational (structural based studies) (Rubinstein et al., 2008; Sun et al., 2011; Zhao and Li, 2010) epitopes than in non-epitope areas, suggesting that some amino acid pairs work cooperatively in mediating antibody binding. The concept of amino acid cooperatively has been adopted from studies on protein:protein interfaces, where pairs of hydrophobic and polar amino acids have been argued to play an important role in the binding formation (Lijnzaad and Argos, 1997; Ma et al., 2003; Neuvirth et al., 2004). This pattern is only to some extent supported in epitopes, where pairs of Tyr:Tyr, Cys:Pro, Asn:Tyr, Asp:Pro, Thr:Tyr and Arg:Tyr according to Rubinstein et al. (2008) and pairs of Asn:Try His:Tyr and His:Met according to Sun et al. (2011) appear more frequently compared to the antigen surface in general. Furthermore, expanding the definition of neighbor cooperatively to one position beyond the immediate neighbor reveals a different pattern, and especially pairs including charged amino acids, Asn and Gln, are found more often in epitopes (Zhao and Li, 2010).
The secondary structure of epitopes has been investigated by several authors (Liang et al., 2010; Ofran et al., 2008; Rubinstein et al., 2008), and epitopes are in general reported to have significantly less secondary structures (strands and helices) and significantly more loops compared to the remaining antigen. The over-representation of loops is small but significant and in agreement with the perception that protein-protein binding sites are flexible regions (Neuvirth et al., 2004). Furthermore, the overall secondary structure of epitopes has been reported to deviate from both that of protein-protein interfaces, and proteins in the PDB database in general (Ofran et al., 2008).
When comparing results from different publications, it is striking to observe, that authors to some degree reach different conclusions as to what defines the amino acid composition, amino acid cooperativeness and secondary structure of a B cell epitope. Besides the constant increase in the number of antigen-antibody complex structures in the PDB database, the inconsistencies in the conclusions from different studies most likely originate from deviations in three critical steps involved in defining the epitope data sets: 1) differences in data redundancy processing, e.g. removal of homologous entries, 2) different definitions of epitopes and epitope residues, and 3) different definitions of the non-epitope area and the non-epitope amino acid distribution, in particular the definition of surface (if defined at all). Given these observations, it remains clear that caution must be taken when comparing results and conclusions from different analyses.
Besides the multiple investigated epitope features presented above, Rubinstein et al. (2008) have established that the epitope area is significantly flatter and more convex (rugged), compared to equally sized patches on the antigen surface, and undergoes a small compression upon antibody binding.
In the later years, features of B-cell epitopes have extensively been utilized in computational mapping of B-cell epitopes (Andersen et al., 2006; Chen et al., 2007; Huang et al., 2007; Liang et al., 2010; Ponomarenko et al., 2008; Rubinstein et al., 2009; Sweredoski and Baldi, 2009, 2008; Zhao and Li, 2010). However, the performance of these methods has in general been moderate, thus stressing the importance of more detailed description of the B-cell epitope area. The objective of the work presented here was therefore to employ and develop methods for analyzing the antigen-antibody interface, especially the epitope area, with the purpose of identifying novel features that may be used in improvement of B-cell prediction methods. We present an analysis of B-cell epitope/paratope amino acid composition, the epitope size, shape and direction relative to the antibody and determine the epitope spatial amino acid distribution.
2. Methods
2.1 Data processing
Using antibody conserved residues as template (template residues are listed in Supplementary material Table S2), 801 antibody structures were identified from the PDB database (www.pdb.org) distributed with the MOE 2009.10 release (contains data from PDB up to August 24, 2009, http://www.chemcomp.com/software.htm). 376 of the 801 structures were protein antigen:antibody complexes. As some entries were duplicates or mutation of the same antigen, a subset of 224 structures with unique antibody sequences were retrieved and used for further data processing. Only the antigen chain interacting with the antibody was kept for further analysis. All other antigen chains were discarded. 26 of the 224 antigen structures (1BJ1, 1CZ8, 1I9R, 1KB5, 1KEN, 1NOX, 1OB1, 1OTS, 1QFW, 1QGC, 1RVF, 1TZH, 1W72, 1XIW, 1YNT, 2BC4, 2DQF, 2FJG, 2FX8, 2H2P, 2J6E, 2JIX, 2NR6, 2OTU, 2QR0, 3CSY) contained multiple chains in the vicinity of the antibody CDR loops. For these complexes, the chain with the main interaction with the antibody was identified and used in the study. If more than one asymmetric unit were present in the structure files, the antibody:antigen complex with the first occurrence was selected. Each residue in the 224 antigen:antibody structures was annotated as either epitope or non epitope (for the antigen) or paratope or non-paratope (for the antibody), based on a 4Å distance (between any pair of heavy atoms of two residues) threshold argued to give the best correlation to manual annotation (Andersen et al., 2006; Van Regenmortel, 2009). The annotation of epitope and paratope residues was used throughout this work in all analysis carried out. The dataset of 224 structures was further processed by first removing all complexes with antigen sizes below 20 amino acids resulting in 162 complexes, secondly by removing similar antigen-antibody interfaces. Similar entities were identified by defining a 400 dimensional “interaction vector” for each complex, holding the frequency of each contacting amino acid pair i.e. the first dimension was assigned the observed number of alanines in the epitope in contact with alanines in the paratope, the second dimension the observed number of alanine-valine contacts and so forth. Two entities were then defined as similar if the angle between the vectors was below 0.8 radians. The threshold was set based on the angle distribution in the data, which separated the data in two groups; below and above 0.8 radians (data not shown). Based on this similarity criterion, a hobohm2 algorithm (Hobohm et al., 1992) was employed to filter similar structures in the developed dataset of 162 structures, which resulted in 109 non-redundant antibody-antigen interactions. Additional two complexes: PDB ID: 1TZI and PDB ID: 1TZH (both from the same study (Fellouse et al., 2005)) were removed as very few (<4) amino acids were involved in complex formation and the entries were constantly identified as outliers in the analyses carried out. The remaining 107 complexes were structural superimposed to the antibody heavy chain. As the linker region between the two immunoglobulin folds of the antibody heavy chain is flexible, using the entire antibody FAB as template for superimposing structures is not feasible. Hence, only the antibody Fv region was used for superimposing. The PDB entry 1A2Y was used as template, as this structure only contains the antibody Fv region. From the superposition, a 3 dimensional visualization of the combined set of epitope amino acids was created which demonstrated that that the epitope residues form a homogeneous skewed disk-shaped distribution, with a few amino acids placed outside the disk (Supplementary materials Figure S1). As this study aims at finding common antigen-antibody features, amino acids outside the disk were treated as outliers and identified using a distance-based outlier algorithm (Knorr et al., 2000). In total, 70 residues (4.2%) were identified as outliers divided between 17 structures. The 107 PDB entries are listed and described in supplementary materials Table S1.
2.2 Surface measures
Surface accessibility for each residue was calculated using DSSP (Kabsch and Sander, 1983). Relative surface exposure values were obtained by normalizing the surface accessibility value against the maximum surface accessibility for the amino acid in question. The maximum exposure was computed by replacing X with the amino acid in question in a GGXGG sequence, where G is glycine. Upper half-sphere exposure (HSE) (Hamelryck, 2005) values were calculated using the biopython software package (Hamelryck and Manderick, 2003) and normalized according to the most buried (highest value) amino acid in question found in 1000 randomly extracted structures from the PDB database.
2.3 Comparison of amino acid distributions by log-odds scores
Two amino acid distributions were compared, by comparing the frequency of each amino acid. Log-odds scores (Sa) were calculated as the base 2 logarithm to the ratio between the frequencies of amino acid a in distribution A and B as described in (Lund et al., 2005). Negative log-odds scores indicate under-representation of a given amino acid and vice versa for positive log-odds scores.
2.4 Epitope and paratope amino acid composition
Epitope and paratope amino acid preferences were assessed by comparing the frequencies of amino acids in epitopes and paratopes to frequencies in non-epitope/paratope distributions. To eliminate the bias towards surface versus non-surface exposed residues, the following procedure was used to obtain the non-epitope and non-paratope amino acid distributions: 1) The epitope/paratope residues were divided into 12 bins of 0.1 intervals (0–0.1,0.1–0.2,…,1.1–1.2) based on their surface exposure (RSA or HSE) resulting in ni (i = 1,2,..,12) observations in each bin. 2) The pool of non-epitope and non-paratope residues were likewise divided into 12 bins based on their surface exposure and ni amino acids were randomly drawn from each corresponding bin, resulting in a distribution matching the epitope/paratope distribution in size and surface exposure profile. The epitope/paratope distribution was then compared to the ‘sampled’ non-epitope/paratope distribution by means of log-odds scores (Sa) as described in section 2.3. Statistical significance of log-odds scores being different from zero was obtained by bootstrapping using the following procedure: 1) 10,000 bootstrapped datasets were produced by sampling the epitope/paratope amino acid distribution with replacements N times (N being the number of residue in the distribution), 2) for each dataset the non-epitope/paratope distribution was obtained as described above and log-odds scores were calculated. In total 10,000 bootstrapped log-odds scores per amino acid (n = 10,000) were obtained, 3) a p-value indicating if a given amino acid is significant over or under-represented was calculated as
pval=min(n(Sa≤0),n(Sa≥0))n(Sa)
where n(statement) is the number of log-odds scores for which the alternative hypothesis was untrue. That is, the p-value for being overrepresented for an amino acid that in 9,900 of the 10,000 re-samplings has a positive log-odds score will be 100/10,000 = 0.01.
2.5 Fitting axis of inertia to epitopes
To get an accurate description of the antigen:antibody interface, the spatial representation of each epitope was defined as the antibody contacting points i.e. the coordinates of the subset of antigen heavy atoms in contact with the antibody (within 4Å of any antibody atom). Hence, each epitope was represented as a contact matrix of atom coordinates X c,j m_×_3 where m is the number of contacting atoms in the epitope and j = {1,2,…107} defines the individually epitopes. Axes of inertia were fitted to the each epitope individually, by centering Xc,j and subsequently compute the singular value decomposition (SVD) of Xc,j:
where the rows of U contain the left eigenvectors (eigenvectors to Xc,jXc,jt), V = [_v1;v2;v3_] the right eigenvectors (eigenvectors to Xc,jtXc,j) and D is a diagonal matrix holding the singular values; s1,s2,s3 (eigenvalues to the squared matrixes) to the corresponding eigenvectors. The axes of inertia (also referred to as principle components (PCs)) were then defined as the right eigenvectors and ranked according to the singular values (s1 > s2 > s3). As the singular values are related to the point variance on each axis (stdev =√ (s)), the first PC points in the direction of most point variation, the second, orthogonal to the first, points in the direction of second most point variance and the third PC is orthogonal to PC1 and PC2. To ensure that the rotated coordinate system spanned by the PCs is right-handed, PC1 were constrained to point into the positive space of the x-axis, PC2 to point into the positive space of the y-axis and PC3 to point into the positive space of the z-axis. The length of each PC was set to |PCi| = _2.5_√ (si), for i = 1,2,3 as the resulting ellipse plane spanned by PC1 and PC2 enclosed 90% of the epitope contact points when projected onto the plane, and PC3 enclosed 95% of the contacts when projected onto the axe. Approximating the epitope as an ellipsoid spanned by the PCs included on average 75% of the epitope contact points.
2.6 Statistical test of epitope shape
It was investigated if the data in the plane spanned by PC1 and PC2 formed an ellipse or was better described by a rectangle. For each epitope, the density of the epitope plane was calculated approximating the plane as an ellipse or a rectangle using the following procedure: 1) for each epitope in the dataset, the half-axis of an ellipse was defined by PC1 and PC2 respectively (see section 2.5) and a rectangle by ±PC1 and ±PC2, 2) epitope contact points were then projected onto the PC1-PC2 plane and the number of contacts enclosed by the ellipse and the number of contacts enclosed by the rectangle was calculated and divided by the area spanned by the ellipse and rectangle respectively, 3) a paired t-test was used to assess if the density of the ellipse was higher than the density of the square, hence testing if the data are best approximated by a square or an ellipse.
It was then investigated if the epitope plane was best described as an oblong ellipse or as a circle. The null-hypothesis of |PC2|/|PC1| = 1 was tested against the alternative hypothesis of |PC2|/|PC1| < 1. As the first PC is deliberately chosen to be larger than the second PC, and variation observed on the two axes is not strictly independent (data is drawn from the same population constrained to same biological patterns) statistics based on variance was not helpful (e.g. using a test of variance). Instead a random dataset was generated emulating the real dataset under the null-hypothesis and compared to the true dataset, using the following procedure: 1) For each epitope in the dataset a random “epitope”, comprising the same number of points in space, was drawn from a disk shaped uniform distribution. To emulate the epitope under the null-hypothesis the proportion of the disk shape distribution (determined by the halfaxis rx,ry,rz) was important. rx = ry were set to 1 to ensure a disk shape, and rz to the ratio between the length of PC3 and the average length of PC1 and PC2: rz = 2*|PC3|/(|PC1|+|PC2|). The definition of rz ensures that the ratio between rz and the ellipse area spanned by rx and ry equals the ratio between |PC3| and the ellipse area spanned by PC1 and PC2, thereby transferring the internal proportion of an ellipsoid fitted to the real epitope to the disk shape distribution, 2) axes of inertia were fitted to the each data point as described in section 2.5, 3) the |PC2|/|PC1| ratio was computed and compared to that of the true data point, counting the number of epitopes, for which the random ratio was lower than the real ratio as unsuccessful, and 4) a p-value was then obtained by using a binomial distribution with the assumption that, if the null hypothesis was true for all epitopes the frequency of unsuccessful events would be 0.5.
2.7 Statistical test for direction of epitopes
The direction of the common epitope was assessed by analyzing the direction of the primary axis of inertia (PC1) fitted to each epitope as described in section 2.5. As antibody heavy chains of the 107 antigen:antibody complexes were superimposed before axis of inertia were fitted to the epitopes, the direction of PC1 describes the epitope direction relative to the antibody.
The immunoglobulin fold of the antibody light and heavy chain gives the tip of antibody (the antigen binding site) a flat oblong shape, which constrains the directions of epitopes to lie in a narrow plane above the antibody tip. Hence, variation in PC1 directions perpendicular to the antibody tip were of less interest and to be able to describe the epitope directions accurately, the PC1s were projected onto a plane above antibody tip defined by the average plane spanned by the first and second principle components for all epitopes i.e. the normal vector to the plane was calculated as the mean of the normalized PC3s. As a measure of PC1 alignment we calculated the average angle between all 107 PC1s and tested the alternative hypothesis of a preferred direction by comparing this value to the average PC1 angle of a randomly obtained dataset. The random dataset were obtained as described in section 2.6 for statistical test of epitope shape, hence using disk shapes to emulate the epitopes under the null-hypothesis of no preferred direction. 1000 random datasets were obtained in this manner and the p-value for accepting the alternative hypothesis was calculated as number of random datasets with a lower average PC1 angle than the true dataset divided by 1000. Like the true data, PC1 of the random datasets were projected onto the average plane spanned by PC1 and PC2 of the random epitopes.
2.8 Spatial distribution of amino acids in epitopes
To assess the common spatial amino acids distribution in epitopes, the 107 epitopes in the dataset were first “structurally aligned” by superimposing the PCs fitted to each epitope (described in section 2.5). The PCs were used for epitope superposition rather than conventional structural alignment, as epitopes are highly diverse in sequence and structure, hence making conventional structural alignment unfeasible (e.g. using combinatorial extension (CE) alignment (Shindyalov and Bourne, 1998)). To avoid bias towards large amino acids, each residue was represented by only one coordinate; the centroid of the subset of residue heavy atoms in contact with the antibody. Hence each epitope were represented as the epitope residue matrix Xr,j w_×_3, where w is the number of residues in the epitope and j = {1,2,…107} defines the individually epitopes. The superposition of PCs was computed by centering the individually epitope residue coordinate matrixes Xr,j and subsequently transform the coordinates into the space spanned by the PCs fitted to the contact matrix Xc,j, (described in section 2.5) resulting in a new residue coordinate matrix Xr,j*. Each Xr,j* was then rescaled by dividing the transformed x*,y*,z*-coordinates by |PC1|, |PC2| and |PC3| respectively and all 107 Xr,j* were pooled to yield one epitope residue coordinate matrix: Xr*N_×_3 (N is the total number of epitope residues). Hence, the data was effectively transformed into a cubic orthogonal space. The spatial distribution of amino acids was assessed in 2-dimension by projecting the data onto the plane spanned by the primary and secondary PCs (effectively only using the first and second column of Xr*), dividing the plane in G ring-shaped bins, bx, determined by the radius; bx*sqrt(2)/G and calculating the log-odds scores (see section 2.3) for finding amino acid a in bx compared to the finding a in the epitope in general. Likewise, the spatial amino acid distribution in one-dimension were examined by projecting data onto PC3 (effectively using the third column of Xr*), dividing the axis in G bins and calculate the log-odds scores for each bin. G = 4 were used for both one and two-dimensional statistics. Statistical inference was accomplished by bootstrapping using the following procedure: 1) 10,000 bootstrapped XB* matrixes were produced by sampling the rows of Xr* with replacement, 2) Log-odds score for finding amino acid a in bx compared to finding a in XB* was calculated for each XB*. Log-odds values for amino acids absent from a bin were set to 0, 3) a p-value was calculated as described in section 2.4 for the epitope/paratope amino acid composition analysis. As data was relative scarce (only 1609 epitope residues), individually amino acids were often depleted from one or more bins when bootstrapping Xr*, thus complicating statistical inference. To overcome this shortcoming amino acids were grouped based on chemical properties as described in (Lund et al., 2005): i) Hydrophobic [ACFILMPVW], ii) hydrophilic [GNQSTY], iii) negatively charged [DE] and iv) positively charged [HKR]. Hence, the analysis was limited to investigate the spatial distribution of these 4 groups of amino acids.
2.9 Development of amino acid density heatmaps
Two dimensional heatmaps describing the density of a group of amino acids in a specific position in the plane were generated using a running average in 2-dimensions as follows; 1) The positions for each residue in the epitope were superimposed and rescaled as described above, 2) for each position (pixel) on a grid in the plane, the number of amino acids projected onto the plane within a radius of r from the position was computed, 3) each position was given a color on a scale from blue to yellow according to this number (density) of amino acids within r, normalized against the maximum density in data, 4) density heatmaps were plotted in Pymol using the Compiled Graphics Objects (CGO) file format. A value of r equal 0.3 was used corresponding to area of approximately 7% of the 2D projected and rescaled epitope area.
2.10 Identification of Complementary Determining Regions in antibodies
CDR regions were identified as described in (Ofran et al., 2008). In short, antibodies were structurally superimposed as described above and structurally aligned residues were identified as residing in the CDR region if above 10% were in contact with antigen residues.
3. Results
3.1 Data development
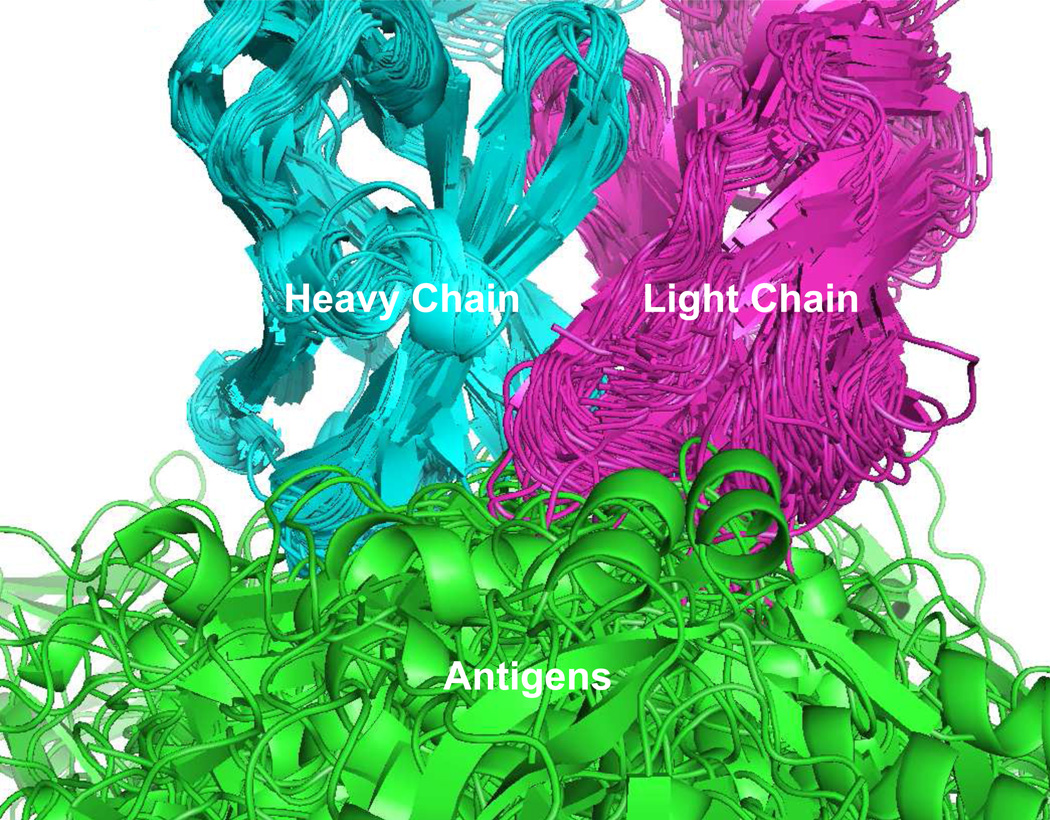
The basis for this study was a dataset of 107 unique non-similar antigen-antibody complexes available from the PDB database (www.pdb.org). Initially 224 unique antigen antibody complexes were identified, which by removing complexes with antigens shorter than 20 amino acids, resulted in a dataset of 162 complexes. The 162 complexes were subjected to a similarity analysis based on contacting amino acid pairs in the antigen-antibody interface (see methods), leading to the additional removal of 53 entries. The remaining complexes (107) were superimposed using the antibody heavy chain as template, as illustrated in Figure 1. The present study focuses on general features of the antigen-antibody binding side, hence interactions not mediated by the antibody variable regions, or in proximity of these, were of limited interest. An outlier algorithm was employed to identify such antigen amino acids, and 70 epitope-annotated residues (4.2%) were identified as outliers and removed from the epitope amino acid pool. PDB id, protein name, epitope length and antigen length are available in supplementary materials Table S1
Fig. 1. Alignment of data used in the study.
Antibody:Antigen complexes were structural superimposed using the antibody heavy chain as template. For illustrative purpose the number of structures displayed in the figure is limited to 60.
3.2 The antigen antibody interaction at a glance
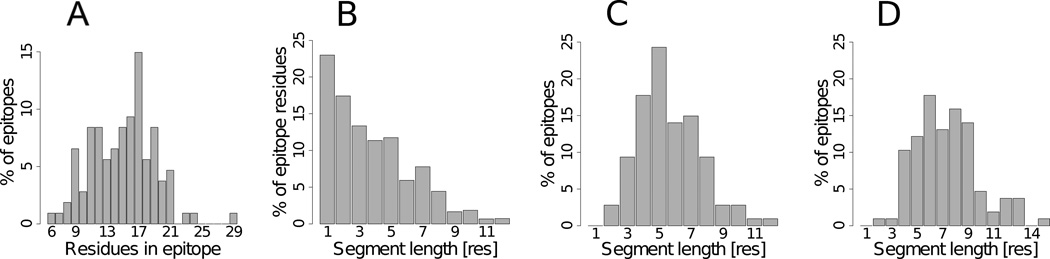
In total 1609 of 15797 (~10%) antigen residues in the data were identified as epitope residues, in contact with 1858 residues in the paratope. Of these, 985 interacted with the antibody heavy chain, 389 with the antibody light chain and 235 with both chains. The observed preference for heavy chain interactions was a general observation across antigens, and is a consequence of the heavy chain CDR regions being longer than the light chain CDR regions (~1.4 times longer in average in data, CDR regions defined as in Ofran et al., (2008), data not shown). These findings are in agreement with previous findings (Collis et al., 2003; MacCallum et al., 1996; Ofran et al., 2008). As a consequence of the high degree of diversity in the antibody CDR regions (Igawa et al., 2011), the size of epitopes were likewise highly diverse, with an average of ~15 residues and a standard deviation of ~4 residues (Figure 2A). However, neither the size of antibody CDR regions, nor the antigen size were found to correlate with the number of contacting amino acids in epitopes (data not shown). None of the epitopes included in the dataset were purely linear epitopes, however 60% of the epitope-annotated residues were found in a linear stretches of 3 amino acids or more (Figure 2B). The maximum linear stretch in each epitope varied from 2 to 12 residues (Figure 2C) and covered on average 40±16% of the residues in each epitope (where ± indicates the standard deviation). Expanding the definition of linear epitopes to allow for one non-epitope annotated residue between two linear stretches, demonstrated that more than 85% of the epitopes where characterized by a maximum linear stretch of 5 or more residues constituting on average 51±20% of the residues in each epitope (Figure 2D) and that 78% of the epitope residues reside in a linear stretch of 3 or more residues. Hence, the conformational epitope can in general be characterized as a sum of linear stretches with some amino acids not contributing in binding antibodies, and where a long linear stretch of 5 or more residues in general covers more than half the epitope residues. These findings are consistent with previously published results (Rubinstein et al., 2008; Sun et al., 2011).
Fig. 2. Size and segmentation of discontinues epitopes used in the study.
A) Distribution of epitope size. B) Distribution of epitope residues segmented by sequential stretches of residues. C) Distribution of maximum sequential stretches of residues in each epitope. D) Same as C, but allowing one non-binding residue in the sequential stretch.
3.3 Amino acid preferences in epitopes and paratopes
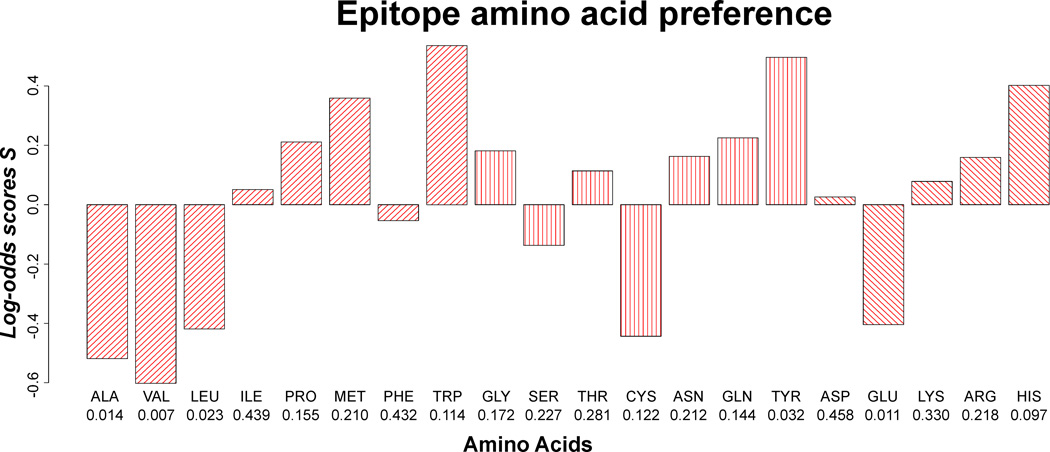
Previous work suggests that amino acid preference of epitopes and paratopes differs from the general composition of the antigen and antibody surface (Andersen et al., 2006; Ofran et al., 2008; Rubinstein et al., 2008; Sun et al., 2011; Zhao and Li, 2010). Epitopic (and paratopic) residues reside for the vast majority of cases exposed on the protein surface. Observed differences in amino acid composition between epitopic and non-epitopic residues then depend crucially on the explicit definition applied to define the antigen (and antibody) surface. Depending on the method used for defining surface residues, different conclusions in relation to epitopic amino acid prevalences have been reported. In the present work, the surface exposure parameter was eliminated by comparing the composition of the epitope amino acid population, to a population similar in size and relative surface accessibility (RSA) profile, drawn from the pool of non-epitope antigen residues. The frequency of each amino acid in epitopes was compared to the frequency of amino acids in non-epitopes, by means of log-odd scores (Figure 3), and statistical inference was obtained by bootstrapping (see Methods). When correcting for multiple testing in a strict Dunn-Bonferroni fashion none of the 20 amino acids were found to be significantly over- or under-represented in epitopes. However, there is a tendency for epitopes to be depleted of small hydrophobic amino acids (ALA, VAL, LEU), and enriched by tyrosines, which aligns with previous findings (Ofran et al., 2008; Rubinstein et al., 2008; Sun et al., 2011). Tryptophan is found to have a large, but insignificant, log-odds score, which is related to the fact that tryptophan is a rare amino acid in proteins. To test the influence of the surface exposure measure used, the analysis was repeated using half-sphere exposure. The result further validates the presented findings and suggests that alanine and valine are significantly under-represented in epitopes (p<0.001 in both cases, data not shown).
Fig. 3. Epitope amino acid composition.
P-values are stated beneath each amino acid.
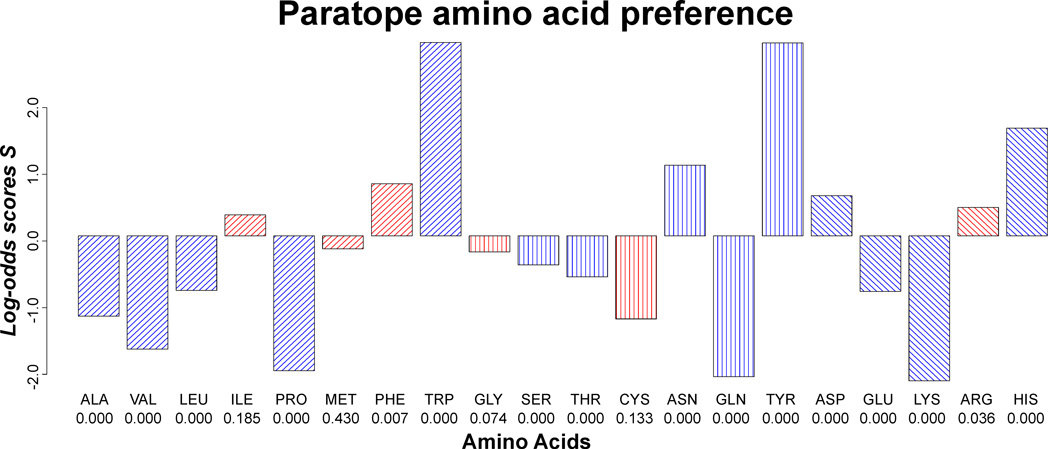
Similar to the epitope amino acid composition, the composition of amino acids in the paratope was compared to the composition of the antibody surface (Figure 4). 5 amino acids were significantly over-represented and 9 amino acids significantly under-represented, and the deviation from the overall antibody surface amino acid composition was in general more pronounced than what was found for epitopes. Interestingly, the 5 positively selected amino acids possess diverse chemical properties with at least one representative from each of the four chemical groups; hydrophobic (trp), hydrophilic (tyr and asn), negatively charged (asp) and positively charged (his).
Fig. 4. Paratope amino acid preferences.
Log-odds scores significantly different from zero are colered blue, and unsignificant red. P-values are stated beneath each amino acid.
3.4 The epitope size and shape
The shape and size of epitopes were quantified by fitting axes of inertia (also referred to as principle components (PCs)) to individual epitopes using atoms in contact with antibody as representative for the epitope shape. As illustrated in Figure 5B and supplementary materials Figure S2, the primary and secondary axis spanned a plane parallel to the antibody tip, describing the planer shape and size of the epitope, whereas the tertiary axis is perpendicular to the antibody and describes the depth (thickness) of the epitope. By projecting the epitope atoms in contact with antibody onto the epitope plane (spanned by PC1 and PC2), it was evident that the plane in general had an ellipse like shape (data not shown). This impression was supported by the findings that the density of contacting atoms in the plane (atom per Å2) was significantly higher when fitting an ellipse to data compared to a rectangular fit (p < 10−16, paired t-test). Approximating the epitope plane by an ellipse resulted in an average epitope plane of 401±133Å2 when the ellipse enclosed ~90% of the epitope atoms projected onto the plane. The general epitope thickness (PC3) was found to be 8.2±2.0Å when enclosing on average 95% of epitope atoms. The size of the epitope plane was strongly correlated to the number of contacting residues in the epitope (Pearson correlation coefficient: PCC = 0.775±0.047, 95% confidence interval found by bootstrapping), whereas the thickness of epitopes had a much lower correlation (PCC = 0.374±0.095, same method used). The tertiary axis was found to be significantly shorter than both the primary and secondary axis (p < 10−15 for both axes, one-tailed paired T-test), supporting previous findings, describing the epitope area as a flat rugged surface (Rubinstein et al., 2008). It was investigated if the shape of the plane was best described by a circle or an oblong ellipse, by comparing the |PC2|/|PC1| ratio for each epitope to the ratio of a random “epitope” generated from a uniform 3-dimensional disk-shaped distribution (see methods section 2.6). A binomial distribution was used to assess the number of times the true |PC2|/|PC1| ratio was lower than the randomly obtained ratio, and the results clearly demonstrated that the ratios of epitopes in general were lower than random (p < 10−11), thus suggesting that the epitope is best described as an flat oblong ellipse shape.
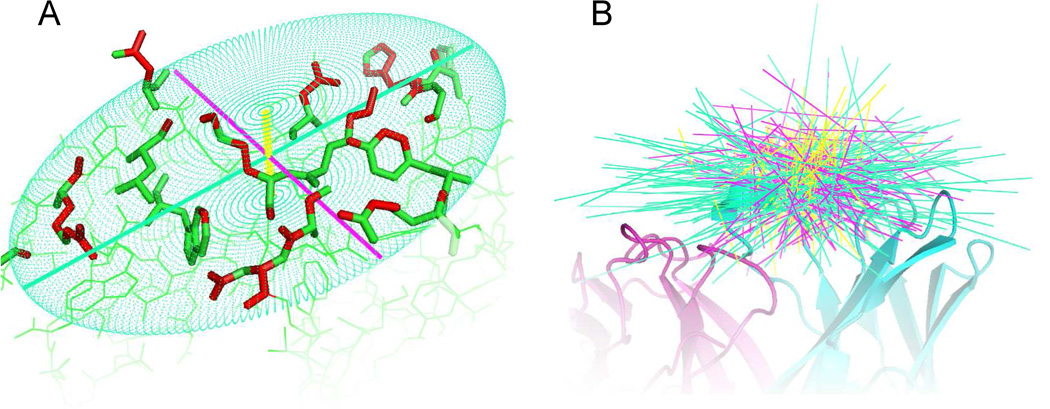
Fig. 5. Illustration of principle components fitted to epitopes.
A) The three principle components (axis of inertia) fitted to the antigen heavy atoms (marked red) in contact with the antibody. Primary axes are shown in green, secondary axes in purple and tertiary axes in yellow. B) Spatial orientation of principle components relative to the antibody. Refer to supplementary materials Figure S2 for an animation of B.
3.5 Spatial orientation of epitopes in relation to antibodies
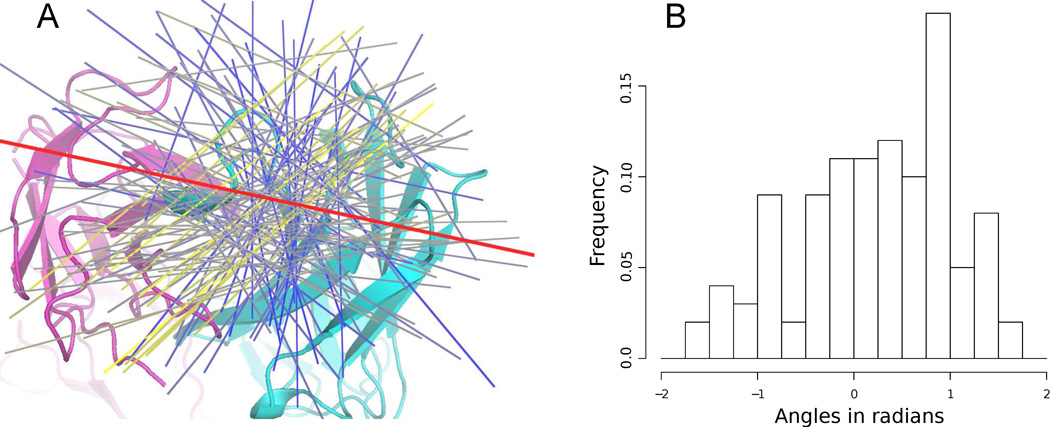
The immunoglobulin fold of the antibody light and heavy chain gives the tip of the antibody an oblong flat squircle-like shape (shape with properties between a circle and a square), which might force the epitope to bind in a specific direction. The oblong shape of the epitope described above, enabled us to represent the epitope direction by the direction of first principle components. The superposition of antibodies (Figure 1) prior to fitting axis of inertia ensures that PC1 described the epitope direction relative to the antibody. As a consequence of the flat shape of the antibody tip (antigen binding site), the spatial orientation of epitope relative to the antibody is constrained to a narrow plane above the antibody tip, which is also evident when inspecting the spatial distribution of PC1s (Figure 5B). Hence, assuming that the PC1 could have any direction in space would lead to wrong conclusions and the analysis of epitope spatial orientation was therefore limited to PC1 directions in the plane above the antibody tip, defined as the average epitope plane spanned by the first and second principle components (see methods section 2.7). The PC1s were projected onto the plane and the orientation of epitopes relative to the antibody was calculated as the angle between the PC1s and a vector pointing in the antibody light to heavy chain direction (see Figure 6A). Histograms of the angles are illustrated in Figure 6B, from which it is evident that no directions are disallowed. However, directions perpendicular to the antibody light to heavy chain direction are less preferred, and directions between −0.5 to 1 radians are overrepresented in the data, with a pronounced peak close to an angle of 0.8 radians. To test if the orientation of epitopes in the data indeed had a non-random preferred direction relative to the antibody, we calculated the average angle between all PC1s as a measure of PC1 alignment and compared it to the average PC1 angle obtained from a dataset of ‘random’ epitopes drawn from a disk shaped uniform distribution (same method as the test for epitope shape in section 3.4). The disk shape of the random epitope mimics the null-hypothesis of no preferred direction and the distribution of average PC1 angles under the null-hypothesis were estimated by generating 1000 random datasets and calculate the average PC1 angle for each set. Using this distribution the alternative hypothesis could be accepted with a p-value of 0.001, thus indicating that the epitope direction relative to the antibody indeed are not random.
Fig. 6. Directions of epitope relative to the antibody.
A) Cartoon drawing represents the antibody and the red line indicates the antibody light to heavy chain direction. Directions of epitopes are represented by the first principle component fitted to the epitope (see methods) and colored from blue to yellow based on the number of other epitopes pointing in roughly the same direction (within an angle of 0.2 radian). Blue indicates that the epitope points in a less preferred direction and yellow that the epitope points in a preferred direction. B) Histogram of angles between epitope direction and the antibody light to heavy chain vector.
3.6 Spatial distribution of amino acid in epitopes
The above mentioned epitope characteristics describe the epitope amino acid composition, shape and orientation but do not describe how amino acids are distributed spatially within the epitope. To address this, the spatial amino acid distribution in the epitope area was investigated by expanding the amino acid preference concept described in section 3.3 to 2-dimensions. The principle components fitted to each epitope were used as a point of reference for “aligning“ and rescaling epitopes (see methods section 2.8), thereby providing a framework for investigating common spatial patterns in amino acids across epitopes regardless of size and orientation in space.
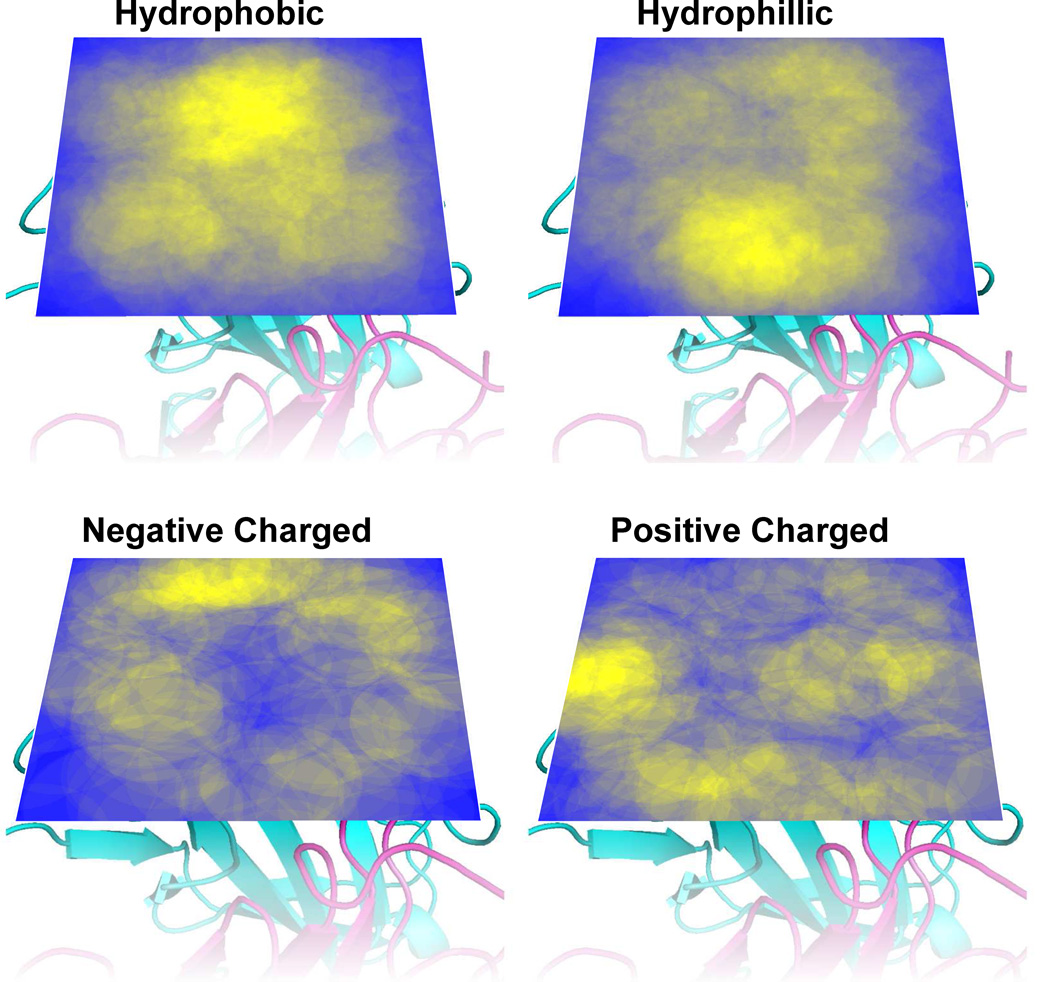
The amino acid dispersion in the epitope plane was examined by projecting positions (centroid of contacting atoms) of epitope-annotated amino acids, onto the plane spanned by the primary and secondary principle components. As data were scarce, amino acids were grouped based on chemical properties in order to enhance the signal to noise ratio in the analysis (see methods section 2.8). Heatmaps illustrating amino acid density for the 4 groups of amino acids are presented in Figure 7, from which it is evident that there is a tendency for hydrophobic amino acids to reside in the center of the epitope, flanked by charged amino acids. Furthermore, hydrophilic amino acids are dispersed in the entire plane, although less commonly in the center of the epitope.
Fig. 7. Density heatmaps of amino acid position in the epitopes plane above the antibody tip.
Areas are colored on a scale from yellow (high density) to blue (low density) (see methods). The antibody structures display are included to enhance visualization.
To further investigate the dispersion of amino acids, a statistical test was designed by expanding the concept of over- and under representation of amino acids presented in section 3.3 to 2 dimensions. The epitope plane was divided into rings (bins) from the center of the plane, and the over- or under-representation of amino acids in each ring, relative to the amino acid population in epitopes in general, were computed in means of log-odds scores. Note, that this test does not explicitly describe the dispersion of specific amino acids, but describes the environment in specific areas of the epitope plane (rings or bins). Statistic inference of over- or under-representation of amino acids was determined by bootstrapping as described in methods section 2.8.
The results presented in Table 1 clearly support the visual impression from the density heatmaps displayed in Figure 7, as hydrophobic amino acids are significantly over-represented in the center of the epitope plane and depleted further away, whereas charged residues have a significantly opposite distribution.
Table 1. Log-odd scores Sa and p-values for the 2-dimensional statistical description of the amino acid dispersion in the epitope plane.
b1 indicates the ring (bin) closest to the center of the plane, b2 the next ring (bin) and so forth. HyPho = group of hydrophobic amino acids [ACFILMPVW], HyPhi = group of Hydrophillic amino acids [GNQSTY], PosCha = group of positively charged amino acid [HKR], NegCha = group of negatively charged amino acids [DE]. Log-odds scores with a p-value below 0.05 are marked in bold.
| Ring | HyPho | HyPhi | NegCha | PosCha | ||||
|---|---|---|---|---|---|---|---|---|
| bx | Sa | Pval | Sa | Pval | Sa | Pval | Sa | Pval |
| 1 | 0.408 | 0.001 | −0.082 | 0.259 | −0.627 | 0.015 | −0.300 | 0.090 |
| 2 | 0.201 | 0.001 | 0.081 | 0.050 | −0.228 | 0.030 | −0.420 | 0.000 |
| 3 | −0.262 | 0.001 | −0.048 | 0.209 | 0.296 | 0.003 | 0.208 | 0.010 |
| 4 | −0.993 | 0.000 | −0.142 | 0.187 | 0.199 | 0.236 | 0.856 | 0.000 |
The spatial distribution of amino acids in the depth of the epitope, i.e. the direction pointing towards the antibody, was likewise analyzed by projecting amino acid position onto the tertiary PC and performing a statistically test for differences in amino acids dispersion similar to the one used for the epitope plane. The results are presented in Table 2. Positively charged amino acids are significantly under-represented close to the antibody and over-represented further away, hydrophilic amino acids are over-represented in the middle of the antibody depth and under-represented further away and hydrophobic amino acid are over-represented close to the antibody. Negatively charged amino acids seem to be equally dispersed along the length of the tertiary axis.
Table 2. Log-odd Sa scores and p-values for 1-dimensional statistical description of the amino acid dispersion in the epitope depth.
b1 indicates the bin closest to the antibody tip, b2 the second closest bin and so forth. HyPho = group of hydrophobic amino acids [ACFILMPVW], HyPhi = group of Hydrophillic amino acids [GNQSTY], PosCha = group of positively charged amino acids [HKR], NegCha = group of negatively charged amino acids [DE]. Log-odds scores with a p-value below 0.05 are marked in bold
| Bin | HyPho | HyPhi | NegCha | PosCha | ||||
|---|---|---|---|---|---|---|---|---|
| bx | Sa | Pval | Sa | Pval | Sa | Pval | Sa | Pval |
| 1 | 0.388 | 0.000 | −0.187 | 0.018 | −0.031 | 0.454 | −0.380 | 0.009 |
| 2 | −0.200 | 0.011 | 0.193 | 0.000 | 0.029 | 0.393 | −0.209 | 0.032 |
| 3 | −0.058 | 0.255 | −0.044 | 0.251 | 0.047 | 0.349 | 0.137 | 0.093 |
| 4 | −0.164 | 0.168 | −0.185 | 0.080 | −0.251 | 0.194 | 0.593 | 0.001 |
4. Discussion
The aim of this study was to develop a framework for describing the antigen:antibody interface, in particular the epitope area. With the presented framework, we were able to describe the shape, direction, and spatial amino acid distribution of epitopes and compare the amino acid composition of the epitopes and paratopes to that of the remaining antigen and antibody surface respectively. Combined with previous findings, our results give a detailed picture of the epitope area and constitute a general framework for analyzing structures that are neither sequentially nor structurally similar.
Table 3 summarize the findings presented here and compares them to earlier studies of B cell epitope properties.
Table 3. Characteristics of the epitope area.
The table lists both previously described characteristics and characteristics described in this study. Refer to results for detailed description of characteristics labeled ‘Present study’ in the reference list.
| Epitope characteristics | Reference | |
|---|---|---|
| Size | 10–25 residues is involved in binding 15±4 residues is involved in binding 22±8 residues is involved in binding 600–1000 Å2 is buried upon binding 847±279 Å2 accesible surface area The epitope plane (see results): 401±133Å2 when approximated by an ellipse Thickness (see results): 8.2±2.0Å | Van Regenmortal (2009) Present study Sun et al., (2011) Rubinstein et al. (2008) Sun et al., (2011) Present study Present study |
| Shape | Flat rugged area Flat oblong (ellipse) shaped area | Rubinstein et al. (2008) Present study |
| Segmentation | Above 60% epitope residues exists in linear stretches of 3 or more residues 85% of epitopes has a linear stretch of 5 or more residues | Rubinstein et al. (2008) and present study Sun et al., (2011) and present study |
| Secondary structure | Enriched by loops Depleted of strands and helixes | Rubinstein et al. (2008) and Ofran et al. (2008) |
| Epitope position on the antigen | Epitopes are more surface exposed than the remaining antigen Epitopes protrude from the antigen surface | Andersen et al. (2006) and Rubinstein et al. (2008) Thornton et al. (1986) |
| Orientation relative to the antibody | Epitopes bind predominantly in a −30 to 60 degrees angle relative to the light to heavy antibody chain direction | Present study |
| Amino acid composition | Enriched by polar and charged amino acids and depleted of hydrophobic amino acids compared to non-epitope antigen residues, surface exposed antigen residues or general protein composition No significant deviation from the non-epitope antigen surface, however a tendency for depletion of small hydrophobic amino acids is observed | Andersen et al. (2006); Ofran et al. (2008); Rubinstein et al. (2008); Zhao and Li (2010) and Sun et al., (2011) Present study |
| Amino acid coorporativeness | Pairs of Tyr:Tyr, Cys:Pro, Asn:Tyr, Gly:Tyr, Asp:Pro, Thr:Tyr and Arg:Tyr are more frequently observed in epitopes compared to the remaining antigen surface Pairs of Asn:Tyr, His:Tyr and His:Met are more frequently observed in epitopes | Rubinstein et al. (2008) Sun et al., (2011) |
| Spatial amino acid composition | Hydrophobic core flanked by charged amino acids Preferable; hydrophobic amino acids closes to the antibody, then hydrophilic and furthest away positive charged amino acids | Present study* Present study |
All through the analysis, the specific traits of the epitopes investigated were statistically tested against the hypothesis that the trait was obtained by random, and we able to establish that the epitope area is a flat, oblong, oval shaped area, that binds predominantly to the antibody tip in a −30 to 60 angle relative to the light to heavy chain antibody direction. Furthermore, the common epitope consists of hydrophobic amino acids in the center and charged residues on the edge and prefers hydrophobic amino acids close to the antibody, positively charged further away and hydrophilic amino acids in between hydrophobic and positively charged amino acids.
Contrary to previous findings, no particular amino acid was found to be significantly over- or underrepresented in epitopes compared to non-epitope surface residues. The difference between our results and previously studies originates from fundamental differences in the definition of non-epitope residues. Previously, non-epitope residues have been defined as either all non-interacting residues in antigens (Andersen et al., 2006), all residues in the PDB database (Ofran et al., 2008) or antigen residues with a relative surface accessibility above 5% (Rubinstein et al., 2008). These definitions do not fully reveal peculiarity of amino acids in epitope areas compared to remaining antigen surface but do, to some extent, describe surface/non-surface peculiarity. Here, the epitope amino acid composition was compared to a non-epitope antigen distribution with the same level of surface exposure, thus comparing epitope amino acid composition to sites at the antigen surface equally exposed to antibody binding. Hence, our findings demonstrate that the epitope amino acid composition deviates from the antigen composition as a whole, but not significantly from the surface of the antigen. However, more data might reveal significantly enrichment of tyrosine, depletion of valine and general depletion of small hydrophobic amino, as these amino acids in our study were in the vicinity of significantly deviating from the antigen surface.
The paratope was found to have a more distinct amino acid distribution than the epitope, as the paratope amino acid composition reflects the recombination and somatic hypermutations of the VDJ/VJ germline genes whereas the epitope originates from non-homologous genes. Predominately 5 amino acids; Trp, Asn, Tyr, Asp and His, were found to be over-represented in the paratope. CDR regions have previously been found to be generally enriched by Trp, Tyr and Ser (Collis et al., 2003; Martin, 2010; Ofran et al., 2008). Ofran et al., (2008) furthermore suggested that heavy chain CDRs are enriched in Asp and light chain CDRs are enriched in His and Asn and depleted of Asp. However, Collis et al., (2003) reported Asp to be depleted from CDRs and Asn and His to be non-significant over-represented. Note, that the work conducted by Ofran et al., (2008) and Collis et al., (2003) analyzed the CDR amino acid composition, whereas we here present the amino aced composition only of the paratope (residues interacting with the antigen). Interestingly, the five overrepresented amino acids found in this study posses diverse chemical properties with representatives from each of the four chemical groups: Hydrophobic (trp), hydrophilic (tyr and asn), positively (his) and negatively charged (Asp), suggesting that antibody diversity and binding to a very high degree can be accomplished by these 5 amino acids alone. Note, that the paratope amino acid pattern might be different for other classes of antigens than proteins utilized in this study, as the amino acid composition of CDRs in antibodies raised against lipids, sugars etc. deviates from CDRs raised against proteins (Collis et al., 2003).
The epitope area was found to have a significantly flat and oblong oval shape (ellipse), which adds to earlier findings, that the epitope area is a flat rugged area protruding from the antigen surface (Rubinstein et al., 2008; Thornton et al., 1986). The protrusion of epitopes was further supported by the observation that the size of the epitope (number of amino acids) was mainly determined by the size of the flat epitope plane, and to less degree by the depth of the epitope. Furthermore, the oblong tip of the antibody was not found to determine the oblong shape of the epitope, as the preferred epitope orientation relative to the antibody tip (−30 – 60 degrees) deviates from the light to heavy chain direction.
Neuvirth et al. (2004) observed a small, but significant non-random dispersion of hydrophobic amino acid in protein-protein interfaces, and suggested that patches of hydrophobic amino acids is important for protein binding. The present study supports this finding, and expands the concept of non-random amino acid disparity in antigen-antibody interfaces to specific patterns. In particular, epitopes are characterized by hydrophobic amino acids residing in the center of the antigen-antibody interface, flanked by charged amino acids. Results were obtained by normalizing the 3-dimensional amino acid distribution according to epitope shape and size. Hence, the observed patterns are generally applicable and independent of epitope shape and size, suggesting that antibodies bind specifically to areas comprising a hydrophobic core surrounded by charged amino acids. These findings aligns with the O-ring (Bogan and Thorn, 1998) theory, the recently proposed wet-rim-dry-core (Li et al., 2012) theory and the work of Chakrabarti and Janin, (2002) and Bickerton et al., (2011) for protein-protein interactions in general suggesting that the core of the interface predominantly consist of hydrophobic amino acids and the periphery of charged and hydrophilic amino acids. Hence, the spatial distribution of amino acids in the epitope to some extent mimics that of regular protein binding sites. Furthermore, the magnitude of the hydrophobic effect (the exclusion of water) at the center of the antigen:antibody interface has been experimentally shown to be twice that at the periphery (Li et al., 2005), thus suggesting that the role of hydrophobic residues in the epitope core is to mediate and stabilize complex formation, whereas the role of charged/hydrophilic residues residing on the edge is to exclude water from the interior of the antigen:antibody interface (Bogan and Thorn, 1998; Lo Conte et al., 1999). This perception of the antigen:antibody binding mechanism aligns with studies on antibody affinity maturation, which suggest that the increase in buried apolar surface at the expense of polar surface correlates with increased binding affinity (Li et al., 2003; Sundberg et al., 2003). However, exceptions to the wet-rim-dry-core epitope anatomy exist (Bhat et al., 1994).
Because the data used in this study was obtained from publicly available antigen-antibody complex structures, it bears some inherent limitations: 1) although all antigen-antigen complexes in the PDB database were extracted, data might be biased towards specific proteins of general interest, 2) 106 out of 107 complexes were solved by crystallization, which might induce an unnatural lattice like environment potentially introducing artificial contacts. However, contrary the biological patterns deduced from the data, it has been reported that such artificial contacts do not show any significantly patterns (Valdar and Thornton, 2001), 3) as non-antibody bound antigen structures were rarely available, the analyses were performed on epitopes obtained from complex bound antigens, which have been reported to slightly deviate from the nature of the unbound epitopes (Rubinstein et al., 2008).
Besides enhancing our understanding of antigen-antibody interaction, the position specific log-odds scores obtained in the epitope plane and depth and the deduced epitope shape, offers novel insights that potentially can aid our understanding of which antigen surface areas are more likely to host B cell epitopes. Integrating these features might therefore be very useful in a patch-searching algorithm that identifies patches on the protein surface, similar in shape and amino acid distribution, to the common epitope area. The B-cell epitope prediction server, Epitopia (Rubinstein et al., 2009) works much in this way, but does (naturally) not utilize the characteristics found here.
Supplementary Material
01
02
Highlights.
- The B-cell amino acid composition does not deviate from the antigen surface
- B-cell epitopes are flat oblong (ellipse) shaped areas
- B-cell epitopes consists of a hydrophobic core flanked by charged amino acids
- New method for superimposing protein structures unrelated in structure and sequence
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference list
- Andersen PH, Nielsen M, Lund O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006;15:2558–2567. doi: 10.1110/ps.062405906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat TN, Bentley GA, Boulot G, Greene MI, Tello D, Dall’Acqua W, Souchon H, Schwarz FP, Mariuzza RA, Poljak RJ. Bound water molecules and conformational stabilization help mediate an antigen-antibody association. Proc. Natl. Acad. Sci. U.S.A. 1994;91:1089–1093. doi: 10.1073/pnas.91.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickerton GR, Higueruelo AP, Blundell TL. Comprehensive, atomiclevel characterization of structurally characterized protein-protein interactions: the PICCOLO database. BMC Bioinformatics. 2011;12:313. doi: 10.1186/1471-2105-12-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. Journal of Molecular Biology. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- Bordner AJ, Abagyan R. Statistical analysis and prediction of proteinprotein interfaces. Proteins. 2005;60:353–366. doi: 10.1002/prot.20433. [DOI] [PubMed] [Google Scholar]
- Chakrabarti P, Janin J. Dissecting protein-protein recognition sites. Proteins. 2002;47:334–343. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- Chen J, Liu H, Yang J, Chou K-C. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids. 2007;33:423–428. doi: 10.1007/s00726-006-0485-9. [DOI] [PubMed] [Google Scholar]
- Collis AVJ, Brouwer AP, Martin ACR. Analysis of the antigen combining site: correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J. Mol. Biol. 2003;325:337–354. doi: 10.1016/s0022-2836(02)01222-6. [DOI] [PubMed] [Google Scholar]
- Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- Fellouse FA, Li B, Compaan DM, Peden AA, Hymowitz SG, Sidhu SS. Molecular Recognition by a Binary Code. Journal of Molecular Biology. 2005;348:1153–1162. doi: 10.1016/j.jmb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- Gershoni JM, Roitburd-Berman A, Siman-Tov DD, Tarnovitski Freund N, Weiss Y. Epitope mapping: the first step in developing epitopebased vaccines. BioDrugs. 2007;21:145–156. doi: 10.2165/00063030-200721030-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelryck T. An amino acid has two sides: a new 2D measure provides a different view of solvent exposure. Proteins. 2005;59:38–48. doi: 10.1002/prot.20379. [DOI] [PubMed] [Google Scholar]
- Hamelryck T, Manderick B. PDB file parser and structure class implemented in Python. Bioinformatics. 2003;19:2308–2310. doi: 10.1093/bioinformatics/btg299. [DOI] [PubMed] [Google Scholar]
- Hobohm U, Scharf M, Schneider R, Sander C. Selection of representative protein data sets. Protein Sci. 1992;1:409–417. doi: 10.1002/pro.5560010313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Honda W, Kanehisa M. Predicting B cell epitope residues with network topology based amino acid indices. Genome Inform. 2007;19:40–49. [PubMed] [Google Scholar]
- Igawa T, Tsunoda H, Kuramochi T, Sampei Z, Ishii S, Hattori K. Engineering the variable region of therapeutic IgG antibodies. MAbs. 2011;3:243–252. doi: 10.4161/mabs.3.3.15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving MB, Pan O, Scott JK. Random-peptide libraries and antigenfragment libraries for epitope mapping and the development of vaccines and diagnostics. Curr Opin Chem Biol. 2001;5:314–324. doi: 10.1016/S1367-5931(00)00208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Keskin O, Ma B, Nussinov R. Hot regions in protein--protein interactions: the organization and contribution of structurally conserved hot spot residues. J. Mol. Biol. 2005;345:1281–1294. doi: 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- Knorr EM, Ng RT, Tucakov V. Distance-based outliers: algorithms and applications. The VLDB Journal The International Journal on Very Large Data Bases. 2000;8:237–253. [Google Scholar]
- Li Y, Huang Y, Swaminathan CP, Smith-Gill SJ, Mariuzza RA. Magnitude of the hydrophobic effect at central versus peripheral sites in protein-protein interfaces. Structure. 2005;13:297–307. doi: 10.1016/j.str.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Li Y, Li H, Yang F, Smith-Gill SJ, Mariuzza RA. X-ray snapshots of the maturation of an antibody response to a protein antigen. Nature Structural & Molecular Biology. 2003;10:482–488. doi: 10.1038/nsb930. [DOI] [PubMed] [Google Scholar]
- Li Z, He Y, Wong L, Li J. Progressive dry-core-wet-rim hydration trend in a nested-ring topology of protein binding interfaces. BMC Bioinformatics. 2012;13:51. doi: 10.1186/1471-2105-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Zheng D, Standley DM, Yao B, Zacharias M, Zhang C. EPSVR and EPMeta: prediction of antigenic epitopes using support vector regression and multiple server results. BMC Bioinformatics. 2010;11:381. doi: 10.1186/1471-2105-11-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lijnzaad P, Argos P. Hydrophobic patches on protein subunit interfaces: characteristics and prediction. Proteins. 1997;28:333–343. [PubMed] [Google Scholar]
- Lollier V, Denery-Papini S, Larré C, Tessier D. A generic approach to evaluate how B-cell epitopes are surface-exposed on protein structures. Mol. Immunol. 2011;48:577–585. doi: 10.1016/j.molimm.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund O, Nielsen M, Lundegaard C, Kesmir C, Brunak S. Immunological bioinformatics. MIT Press; 2005. [Google Scholar]
- Ma B, Elkayam T, Wolfson H, Nussinov R. Protein-protein interactions: structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5772–5777. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum RM, Martin AC, Thornton JM. Antibody-antigen interactions: contact analysis and binding site topography. J. Mol. Biol. 1996;262:732–745. doi: 10.1006/jmbi.1996.0548. [DOI] [PubMed] [Google Scholar]
- Martin ACR. Protein Sequence and Structure Analysis of Antibody Variable Domains. In: Kontermann R, Dübel S, editors. Antibody Engineering. Berlin, Heidelberg: Springer Berlin Heidelberg; 2010. pp. 33–51. [Google Scholar]
- Neuvirth H, Raz R, Schreiber G. ProMate: a structure based prediction program to identify the location of protein-protein binding sites. J. Mol. Biol. 2004;338:181–199. doi: 10.1016/j.jmb.2004.02.040. [DOI] [PubMed] [Google Scholar]
- Novotný J, Handschumacher M, Haber E, Bruccoleri RE, Carlson WB, Fanning DW, Smith JA, Rose GD. Antigenic determinants in proteins coincide with surface regions accessible to large probes (antibody domains) Proc. Natl. Acad. Sci. U.S.A. 1986;83:226–230. doi: 10.1073/pnas.83.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofran Y, Schlessinger A, Rost B. Automated identification of complementarity determining regions (CDRs) reveals peculiar characteristics of CDRs and B cell epitopes. J. Immunol. 2008;181:6230–6235. doi: 10.4049/jimmunol.181.9.6230. [DOI] [PubMed] [Google Scholar]
- Ponomarenko J, Bui H-H, Li W, Fusseder N, Bourne PE, Sette A, Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Regenmortel MHV. What is a B-cell epitope? Methods. Mol. Biol. 2009;524:3–20. doi: 10.1007/978-1-59745-450-6_1. [DOI] [PubMed] [Google Scholar]
- Rubinstein ND, Mayrose I, Halperin D, Yekutieli D, Gershoni JM, Pupko T. Computational characterization of B-cell epitopes. Mol. Immunol. 2008;45:3477–3489. doi: 10.1016/j.molimm.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Rubinstein ND, Mayrose I, Martz E, Pupko T. Epitopia: a web-server for predicting B-cell epitopes. BMC Bioinformatics. 2009;10:287. doi: 10.1186/1471-2105-10-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- Sun J, Xu T, Wang S, Li G, Wu D, Cao Z. Does difference exist between epitope and non-epitope residues? Analysis of the physicochemical and structural properties on conformational epitopes from B-cell protein antigens. Immunome Res. 2011;7:1–11. [Google Scholar]
- Sundberg EJ, Andersen PS, Schlievert PM, Karjalainen K, Mariuzza RA. Structural, Energetic, and Functional Analysis of a Protein-Protein Interface at Distinct Stages of Affinity Maturation. Structure. 2003;11:1151–1161. doi: 10.1016/s0969-2126(03)00187-4. [DOI] [PubMed] [Google Scholar]
- Sweredoski MJ, Baldi P. PEPITO: improved discontinuous B-cell epitope prediction using multiple distance thresholds and half sphere exposure. Bioinformatics. 2008;24:1459–1460. doi: 10.1093/bioinformatics/btn199. [DOI] [PubMed] [Google Scholar]
- Sweredoski MJ, Baldi P. COBEpro: a novel system for predicting continuous B-cell epitopes. Protein Eng. Des. Sel. 2009;22:113–120. doi: 10.1093/protein/gzn075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JM, Edwards MS, Taylor WR, Barlow DJ. Location of “continuous” antigenic determinants in the protruding regions of proteins. EMBO J. 1986;5:409–413. doi: 10.1002/j.1460-2075.1986.tb04226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar WS, Thornton JM. Conservation helps to identify biologically relevant crystal contacts. J. Mol. Biol. 2001;313:399–416. doi: 10.1006/jmbi.2001.5034. [DOI] [PubMed] [Google Scholar]
- Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, Damle R, Sette A, Peters B. The immune epitope database 2.0. Nucleic Acids Res. 2010;38:D854–D862. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Li J. Mining for the antibody-antigen interacting associations that predict the B cell epitopes. BMC Struct. Biol. 2010;10(Suppl 1):S6. doi: 10.1186/1472-6807-10-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02