The chromatin insulator CTCF and the emergence of metazoan diversity (original) (raw)
Abstract
The great majority of metazoans belong to bilaterian phyla. They diversified during a short interval in Earth’s history known as the Cambrian explosion, ∼540 million years ago. However, the genetic basis of these events is poorly understood. Here we argue that the vertebrate genome organizer CTCF (CCCTC-binding factor) played an important role for the evolution of bilaterian animals. We provide evidence that the CTCF protein and a genome-wide abundance of CTCF-specific binding motifs are unique to bilaterian phyla, but absent in other eukaryotes. We demonstrate that CTCF-binding sites within vertebrate and Drosophila Hox gene clusters have been maintained for several hundred million years, suggesting an ancient origin of the previously known interaction between Hox gene regulation and CTCF. In addition, a close correlation between the presence of CTCF and Hox gene clusters throughout the animal kingdom suggests conservation of the Hox-CTCF link across the Bilateria. On the basis of these findings, we propose the existence of a Hox-CTCF kernel as principal organizer of bilaterian body plans. Such a kernel could explain (i) the formation of Hox clusters in Bilateria, (ii) the diversity of bilaterian body plans, and (iii) the uniqueness and time of onset of the Cambrian explosion.
The stem groups of extant bilaterian phyla appeared with only few precursors and transitional forms during the Cambrian explosion, ∼540 million years ago (Mya). Abiotic, ecological, and genetic factors have been proposed to explain their sudden radiation. For example, an increase in deep-ocean oxygenation may have stimulated the evolution and fossilization of macroscopic animals (1) whereas complex ecological interactions could have favored the subsequent emergence and diversification of new body plans within a relatively short interval (2–4). However, genetic changes in body patterning must constitute the molecular basis of these events.
Studies in nonbilaterian animals revealed that many important developmental genes and signaling components are present in ancient phyla and thus evolved before the advent of the Bilateria (refs. 5–7 and references therein). Therefore, modification of existing gene regulatory networks rather than the invention of new genes is believed to drive the evolution of complex body plans. The term “kernel” has been introduced for a class of network components that control major aspects of body plan morphology (8). Kernels consist of inflexible regulatory subcircuits and are strictly conserved in evolution. According to Davidson and Erwin (8) a wave of morphological novelties at the phylum level, like in the Early Cambrium, must have been accompanied by the establishment of new kernels that underlie these innovations. However, kernels that could explain the emergence of the Bilateria more than 540 Mya have not been identified so far.
The appropriate expression of genes in time and space is crucial for cell identity and commitment. To control the transcriptional state of a locus at a given time, a network of interactions integrates developmental cues with chromatin organization. One particular factor implicated in the regulation of chromatin organization is CCCTC-binding factor (CTCF). CTCF is an 11-zinc-finger (ZF) protein that is functionally conserved in vertebrates and Drosophila melanogaster (9–11) and critically involved in diverse aspects of vertebrate biology (12, 13). It can act directly, as a positive or negative regulator of transcription, and indirectly as a mediator of long-range chromatin interactions and insulator protein (12, 14). Genome-wide ChIP experiments suggest that CTCF might exert its function on a global level (15, 16) as a master regulator of the genome (13). This view is reinforced by recent genome-wide ChIA-PET analysis that suggests a global function of CTCF in chromatin organization and transcriptional control through the establishment of distinct chromatin domains (17).
Despite the importance of CTCF for the biology of vertebrates and Drosophila, its phylogenetic distribution and role in other organisms have not been studied in detail so far.
Results and Discussion
CTCF Is Restricted to the Bilateria.
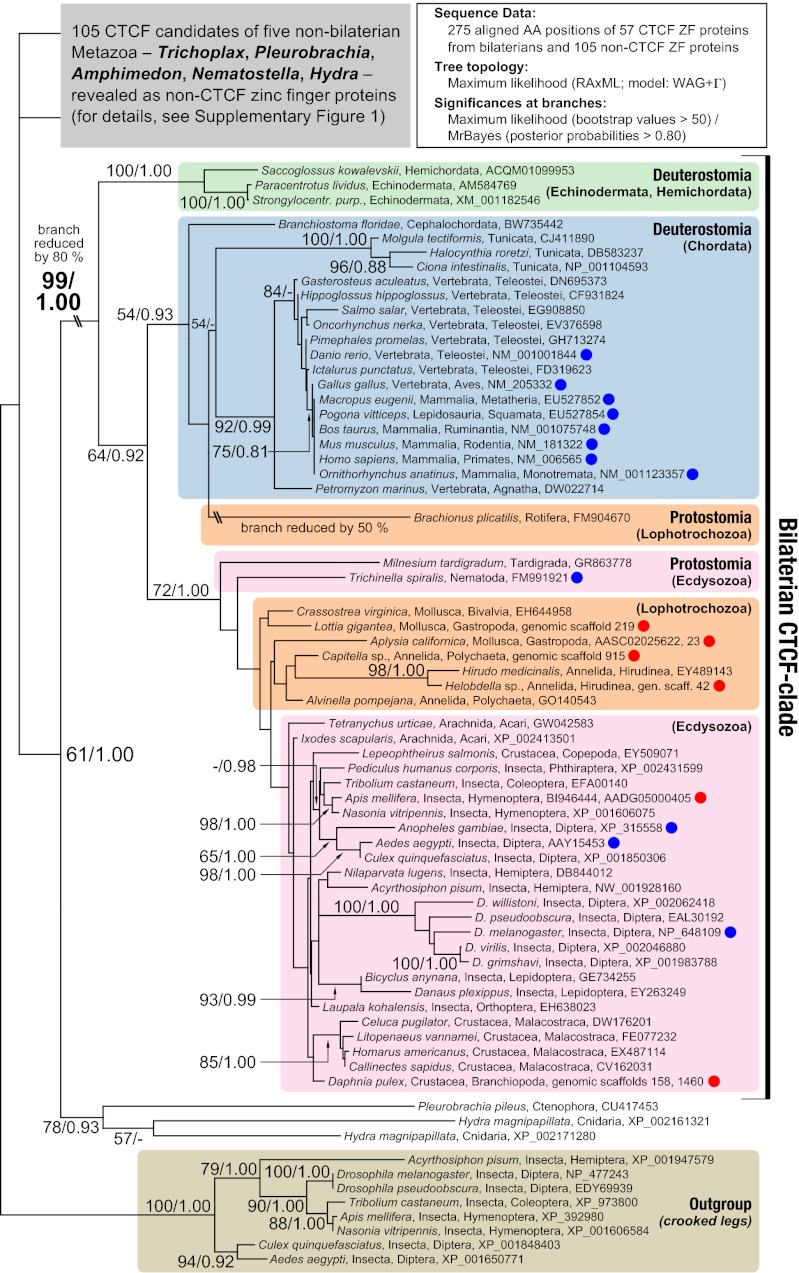
The presence of CTCF in vertebrates (11, 18–20), flies (10, 21), and some nematodes (22) suggests that all protostomes and deuterostomes might possess this 11-ZF protein. To investigate this possibility, we searched in public databases for CTCF candidates and determined their orthology to CTCF in a phylogenetic analysis. We found 24 previously undescribed CTCF orthologs in three of seven ecdysozoan phyla (tardigrades, nematodes, and arthropods). In the sister superphylum, the Lophotrochozoa, we detected eight orthologs, three from molluscs, four from annelids, and one from rotifers (Fig. 1 and SI Appendix, Fig. S1). We could not identify orthologs in other lophotrochozoan and ecdysozoan clades, most likely as a consequence of their low sequence coverage (SI Appendix, Fig. S2). Surprisingly, we were not able to identify CTCF in platyhelminthes despite abundant sequence resources (SI Appendix, Fig. S2). Absence of CTCF from this phylum is possibly due to a secondary loss, a phenomenon also reported for some nematodes (22).
Fig. 1.
Existence of a bilaterian CTCF clade. Phylogenetic analysis is shown of 162 CTCF candidates from Bilateria and early branching metazoans. CTCFs form a distinct, highly supported cluster. All major groups of Nephrozoa are represented within this cluster whereas candidates from early branching metazoans are not. Results of the latter are omitted for clarity (see SI Appendix, Fig. S1 for the complete tree). Blue dots indicate previously published CTCF orthologs, and red dots highlight orthologs we annotated from genomic contigs. Branch labels indicate origin and accession number of a sequence.
Next, we determined whether all deuterostomes (echinoderms, hemichordates, and chordates) (23) possess CTCF. We found many so far undetected orthologs in vertebrates and other chordates as well as orthologs in echinoderms and hemichordates (Fig. 1 and SI Appendix, Fig. S1), indicating that CTCF must have already been present in the gene repertoire of the last common ancestor of Nephrozoa (protostomes and deuterostomes). We were not able to identify CTCF in Acoelomorpha and Xenoturbella, two bilaterian sister groups of Nephrozoa (23), possibly as a consequence of scarce sequence data.
To test whether early branching metazoans also possess CTCF, we searched for candidate orthologs in the four available genomes of Trichoplax adhaerens (24) (Placozoa), Amphimedon queenslandica (25) (Porifera), Nematostella vectensis (5) (Cnidaria), and Hydra magnipapillata (26) (Cnidaria). All four lacked CTCF (Fig. 1 and SI Appendix, Fig. S1), as did a massive number of ESTs from early branching metazoans (SI Appendix, Fig. S2) and 40 complete or draft genomes of protozoa, fungi, and plants (SI Appendix, Table S1). These results demonstrate the absence of CTCF from diverse, possibly all, early branching metazoans and from all other eukaryotes. Three sequences from Cnidaria and Ctenophora clustered as a sister group to the CTCF clade (Fig. 1), but additional studies will be necessary to confirm this result.
Several conclusions can be drawn from our observations. First, CTCF originated in the last common ancestor of protostomes and deuterostomes or earlier (Fig. 1 and SI Appendix, Fig. S1). Second, a ZF protein clustering with CTCF is absent in protists, fungi, and plants (SI Appendix, Table S1). Third, all CTCFs are members of a highly supported clade (bootstrap value 99%, posterior probability 1.00; Fig. 1) with a distinct structure and conservation pattern (SI Appendix, Figs. S3 and S4). Fourth, branching pattern and lineage-specific synapomorphies of the CTCF clade point to a single origin and subsequent diversification of CTCF (SI Appendix, Fig. S4). Thus, CTCF must have appeared during the early evolution of Metazoa, most likely at the time of the protostome–deuterostome ancestor. It must have maintained a critical, albeit not necessarily identical, function in all descendants since.
Bilaterian Genomes Are Rich in CTCF Sites.
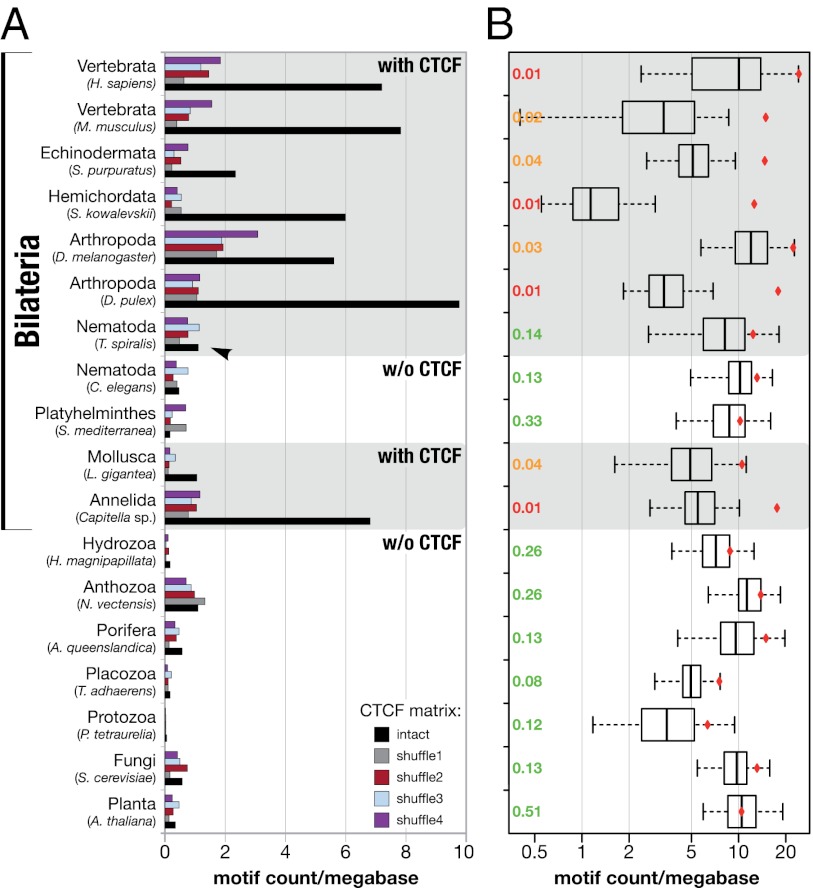
Studies in vertebrates and Drosophila have implicated CTCF in the coordination of chromatin organization via thousands of binding sites at a genome-wide scale (15–17, 27). To explore whether the presence of CTCF is associated with a similar enrichment of binding sites in other Bilateria, we scanned a broad range of bilaterian and control genomes (11 Bilateria; four early branching metazoans; one protist, fungus, and plant each) with known binding motifs for CTCF (16, 28) and slightly modified, but corrupt versions thereof (SI Appendix, Fig. S5). Two arguments justify this approach: a strong conservation in all bilaterian CTCFs of the amino acids responsible for target site recognition (SI Appendix, Fig. S3) (29) and the similarity of CTCF-binding motifs derived from Drosophila and diverse mammals (16, 28, 30) despite more than 500 My of divergence.
We found a significant overrepresentation of potential CTCF-binding sites in animals that contained CTCF (Fig. 2 A and B and SI Appendix, Fig. S6). In contrast, genomes from plants, fungi, protozoa, and early-branching metazoans had low motif counts, indicating that the enrichment of a CTCF-specific binding motif is restricted to bilaterian animals with CTCF (Fig. 2 A and B and SI Appendix, Fig. S6). In agreement with this view, such an enrichment was not detectable in the genomes of the platyhelminth Schmidtea mediterranea and the nematode Caenorhabditis elegans that lack CTCF (22). Another nematode, Trichinella spiralis, behaved similarly despite the presence of CTCF (22), illustrating that CTCF may have lost its genome-wide function in this lineage.
Fig. 2.
Enrichment of CTCF-binding sites in bilaterian genomes. (A) Relative abundance of predicted CTCF-binding sites in 18 different genomes (determined by PATSER). Black bars represent the number of binding sites resulting from the intact CTCF matrix. Colored bars indicate the results of four corrupted matrices that differ from the original matrix by a reciprocal nucleotide exchange at two conserved positions (SI Appendix, Fig. S5). The affiliation of an organism to the Bilateria (parentheses) and the distribution of CTCF (gray background) are indicated. Black arrowhead: no enrichment of CTCF sites in T. spiralis despite the presence of CTCF. Estimated specificity of binding-site prediction: >82% (SI Appendix, Table S2). (B) Significance of binding-site enrichment. Box plots show the relative distribution of motif counts in 18 genomes based on 100 randomized versions of the CTCF matrix. The number of hits based on the intact matrix is shown as a red diamond for each species. P values are indicated at the left (red, P ≤ 0.01; orange, P = 0.01–0.05; green, P ≥ 0.05). Whiskers extend to the most extreme data point, which is 1.5 times the interquartile range (indicated by the box) away from the box. Outliers are omitted for clarity. Differences in the motif count per megabase between Fig. 2_A_ and 2_B_ are caused by differences in the applied experimental procedures.
To quantify the reliability of these predictions, we analyzed the overlap between predicted and experimentally verified binding sites and found it to be greater than 82% for our parameter settings (SI Appendix, Table S2). Although these results do not prove actual binding of CTCF, they strongly support the idea that genome-wide binding is a conserved feature in animals that contain CTCF.
In conjunction with its phylogenetic distribution, these observations suggest that CTCF could play a role as an indispensable (31–33) genome organizer not only in D. melanogaster (27), humans (34), and mice (17), but also in protostomes and deuterostomes in general. Given its involvement in chromatin insulation, long-range DNA interactions, and higher-order chromosomal organization (13, 17, 34), it is conceivable that CTCF introduced gene regulatory mechanisms in Nephrozoa that are different or absent in other animals. Candidates for such mechanisms are, for example, developmentally regulated chromatin boundaries and long-range chromatin interactions. They facilitate the expression of complex chromosomal loci like the TCR, β-globin, MHC, Igh, protocadherin, or Hox loci in vertebrates in a CTCF-dependent manner (35–40) and might thus be instrumental to the formation of such loci. Although functional data are available so far only from mice and D. melanogaster, our findings are compatible with the idea that bilaterians have been equipped with a new layer of developmental possibilities through the invention of CTCF and its involvement in chromatin organization and transcriptional control.
Conservation of CTCF Sites in Vertebrate and Drosophila Hox Gene Clusters.
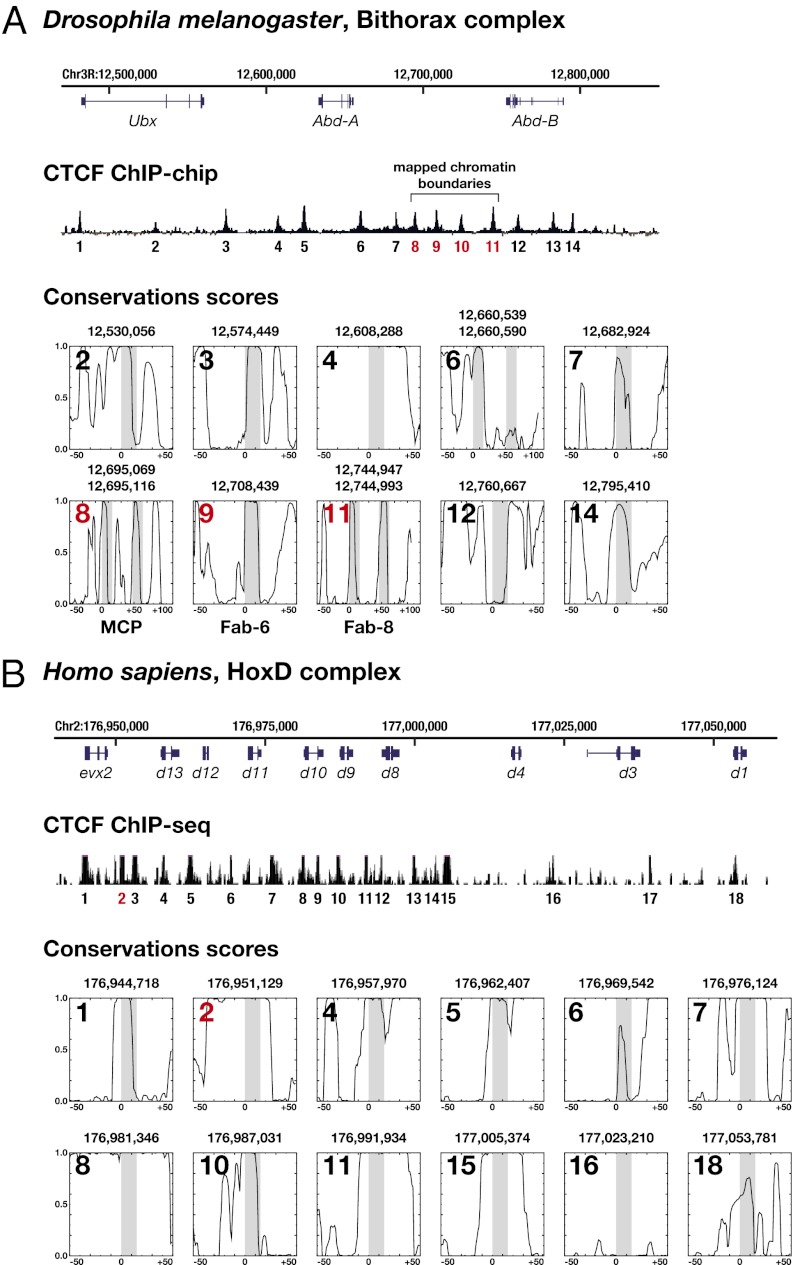
In D. melanogaster, CTCF confers insulator activity to the Fab-6 and Fab-8 chromatin boundaries within the Bithorax Hox complex (BX-C) (10, 41). With the exception of Fab-7, all known and postulated boundaries within BX-C bind CTCF in vivo (28). Both, CTCF null mutations and the deletion of CTCF-binding sites disrupt Hox gene expression and cause homeotic transformations (31, 42). Together, these findings illustrate the importance of CTCF for ordered Hox gene expression in D. melanogaster.
To investigate a possible link between CTCF and Hox genes in other Drosophilids, we determined the interspecies conservation of CTCF-binding sites in the Antennapedia (Antp-C) and Bithorax Hox complexes. Although we obtained similar results for Antp-C (SI Appendix, Fig. S7), we focus here on BX-C. ChIP-chip data from the Drosophila modENCODE project (43) revealed the presence of 14 CTCF peaks within this region (Fig. 3_A_), most of which (10/14) had exactly matching site predictions. In four cases, absence of a prediction suggested indirect binding of CTCF (sites 1, 5, 10, and 13). One of these sites (no. 10) was positioned within the Fab-7 boundary, in accordance with previous publications (28, 44). Next, we computed the conservation score of the remaining 10 sites, on the basis of genomic alignments of 12 Drosophila species with a sequenced genome (45). In 9/10 cases, the ChIP-positive, 15-bp CTCF target sequence precisely overlapped with a conservation peak, suggesting that the respective binding sequences are functionally important in most of the 12 Drosophila species (Fig. 3_A_). The ChIP signals of sites 6, 8, and 11, corresponding to the Fab-3, MCP, and Fab-8 chromatin boundaries, had two closely spaced predictions within a single ChIP-enriched region, and both were positioned exactly within separate conservation peaks (Fig. 3_A_). When we repeated the analysis with corrupted matrices (SI Appendix, Fig. S5), the association of predicted CTCF sites with ChIP signals and conservation disappeared (SI Appendix, Fig. S8_A_). Thus, sequences within the BX-C that resemble in vivo used CTCF sites and CTCF-dependent chromatin boundaries are subject to purifying selection throughout the Drosophila genus. These findings point at an emergence of the D. melanogaster Hox-CTCF interaction at least 60 Mya, in the common ancestor of Drosophilids (46).
Fig. 3.
Conservation of CTCF-binding sites in Hox clusters within Drosophilids and vertebrates. (A) (Top) Schematic view of the D. melanogaster Bithorax Hox complex with the Hox genes Ubx, Abd-A, and Abd-B (blue), drawn to scale. (Middle) Position of 14 CTCF ChIP-chip signals relative to the D. melanogaster BX-C (data from ref. 43). The known boundary elements MCP, Fab-6, Fab-7, and Fab-8 are highlighted in red (sites 8–11). (Bottom) Conservation of 10 ChIP-positive CTCF sites in 12 Drosophila species. Each plot shows the PHASTCONS score of the site(s) indicated on top, ±50 bp. It represents the conservation profile of a ChIP-positive target-site prediction. The 15-bp target sequence is highlighted as a gray box. All CTCF sites except no. 12 are positioned within a local conservation maximum. (B) (Top) Schematic view of the human HoxD complex with Hox genes d13_–_d1 (blue), drawn to scale. (Middle) Position of 18 CTCF ChIP-seq signals relative to the human HoxD cluster (as in ref. 55). Site 2 (red) is a verified binding site within a previously identified chromatin boundary (59). (Bottom) Conservation of 12 ChIP-positive CTCF sites in mammals. All sites except no. 16 are positioned within local conservation maxima.
Like D. melanogaster, vertebrate Hox clusters contain a large number of regulatory elements that direct expression of distinct Hox genes in multiple tissues at various times (e.g., refs. 47–54). Thus, one expects the existence of mechanisms that restrict regulatory crosstalk and allow a finely tuned expression of Hox genes. There is accumulating evidence that CTCF is involved in this regulation: (i) Similar to D. melanogaster, in vivo occupied CTCF binding sites are present between individual Hox genes in all human and murine Hox clusters (55, 56). (ii) Chromosome conformation capture data suggest that binding of CTCF to these sites influences Hox cluster architecture during development (57). (iii) Maternal depletion of CTCF in mice results in the misregulation of developmental genes, including several Hox genes (58). (iv) A chromatin boundary with a previously unrecognized CTCF site is required for the correct expression of HoxD genes in mice (59) and its loss is associated with altered Hox gene expression and body morphology in snakes (60). (v) CTCF’s barrier activity is responsible for higher-order organization and appropriate regulation of the HoxA locus in humans and mice (40).
To investigate whether vertebrates display a link between CTCF and Hox gene expression similar to that in Drosophila, we measured in their Hox complexes the interspecies conservation of CTCF-binding sites. As an example, we focused on the HoxD complex. ChIP-seq data from the ENCODE project (55) indicated the presence of at least 18 CTCF peaks within this cluster. We found exactly matching predictions for 16/18 signals and computed their conservation score in mammals. Again, the majority of sites (13/18) were conserved (Fig. 3_B_) and the association between ChIP-seq signals, computational prediction, and phylogenetic conservation was lost when we used corrupted matrices for binding-site prediction (SI Appendix, Fig. S8_B_ and Table S2).
Both our computational prediction and CTCF ChIP-seq data (55) revealed that a CTCF site is positioned within a chromatin boundary required for proper HoxD gene expression (59). This site (Fig. 3_B_, site 2) lies within a highly conserved 57-bp stretch and has been implicated in morphological alterations during squamate evolution (59, 60), suggesting that phenotypic variation can be a consequence of altered CTCF binding. Similarly, it has been proposed for humans that phenotypic variation can be caused by differences in transcription factor binding, including CTCF (61).
Thus, evolutionary constraints on the presence of CTCF-binding sequences have existed in the vertebrate lineage since more than 210 Mya, when the first mammals evolved (62). The conservation profile of site 2 indicates that such regulatory elements could have evolved even earlier, before the split between zebrafish and mammals ∼450 Mya (63). These findings are consistent with the existence of an ancient link that coupled Hox gene regulation to the CTCF protein. The presence of this link in Drosophila and vertebrates suggests that it dates back to the last common ancestor of protostomes and deuterostomes.
The Hox-CTCF Kernel.
On the basis of these observations, we propose that the Hox-CTCF link represents at least part of what Davidson and Erwin (8) called the kernel of a gene regulatory network. Once established in the ancestor of Nephrozoa, the Hox-CTCF kernel provides a mechanism for body patterning across the Bilateria due to CTCF’s ability to establish chromatin domains (17) and long-range interactions within Hox clusters in protostomes and deuterostomes (40, 44). These features are characteristic for CTCF and could have been a prerequisite for Hox cluster genesis and retention, permitting an individual spatiotemporal regulation of Hox genes.
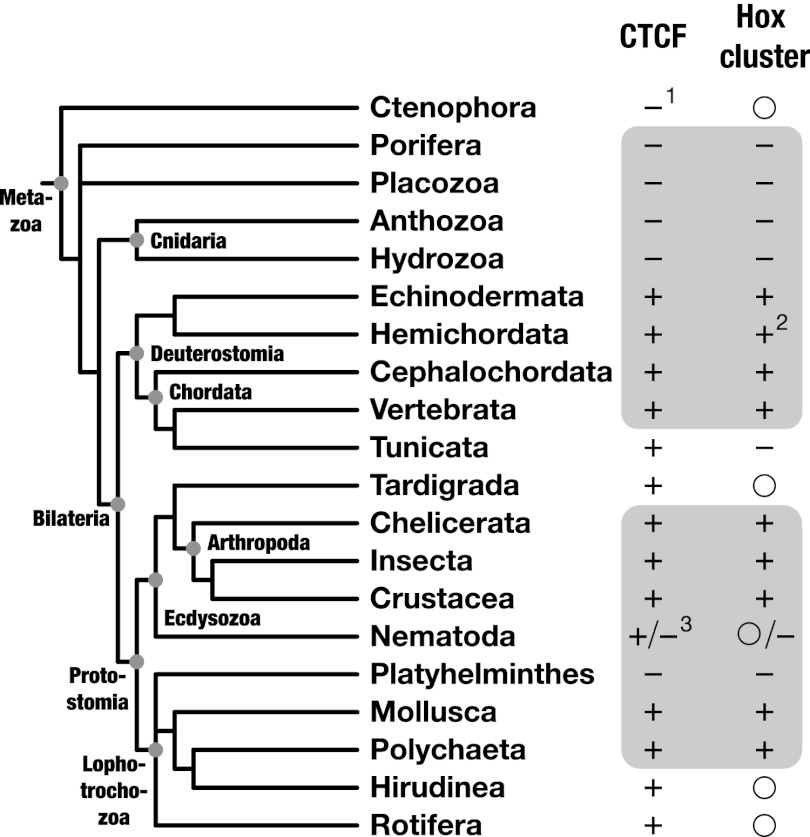
If this assumption is true, several requirements have to be met. First, CTCF-binding sites should exist between individual genes of a Hox cluster. This property is well documented in both vertebrates and Drosophila (10, 28, 40, 43, 55, 56), suggesting that it existed in the ancestor of protostomes and deuterostomes. Second, owing to the stability of the proposed kernel, one should find signs of conservation at least at some CTCF-binding sites. Our work demonstrates that the binding sequence of the majority of sites in Drosophila and vertebrate Hox clusters is evolutionarily conserved (Fig. 3 A and B and SI Appendix, Figs. S7 and S8). Although sites in Drosophila possess equivalent recognition profiles and high sequence similarity to those in vertebrates, they are not necessarily derived from a common ancestral sequence. High turnover rates of transcription factor binding sites and rearrangements observed in bilaterian Hox clusters (for references, see SI Appendix, Table S3) make an independent origin possible. Third, mutations in CTCF-binding sites should affect Hox gene regulation and body morphology. Although this has been shown at one site in D. melanogaster (42), a definite proof is missing in vertebrates. Work on the RXII regulatory element (59, 60) suggests that it might also be true in these organisms. Similarly, impairment of CTCF itself should induce Hox gene misexpression. Although confirmed in Drosophila (31), this has been questioned in vertebrates (56). These findings, however, refer to the mouse developing limb and do not rule out a role of CTCF during early embryogenesis when the proposed kernel is active. In line with this idea, a recent report demonstrated that CTCF knockout mice die after the blastocyst stage (33). Fourth, animals that display Hox gene clustering should possess CTCF, and vice versa. With the exception of tunicates, whose highly determinative development has been implicated in Hox cluster breakdown (64), this correlation is valid for all protostomian and deuterostomian phyla where information is available (Fig. 4 and SI Appendix, Table S3).
Fig. 4.
Correlation between CTCF and Hox gene clustering in Metazoa. Shown is the presence of CTCF and Hox gene clusters in animal phyla, mapped onto a phylogenetic tree. Only phyla for which information is available are shown. Plus and minus symbols indicate the presence or absence of CTCF and/or Hox clusters. Open circles indicate presence or absence is not known. Gray background: positive correlation between CTCF state and Hox gene clustering. 1, inferred from the absence in 51,000 ESTs; 2, SI Appendix, Fig. S10; 3, presence of CTCF in basal nematodes and absence in C. elegans and other derived nematodes (22). References for the state of Hox gene clustering are in SI Appendix, Table S3.
Although these pieces of evidence support the idea that a common Hox-CTCF kernel could be responsible for body patterning in bilaterians, we do not exclude that other genomic events in the bilaterian ancestor might have been important as well. Conservation of CTCF-binding sites, for example, is not restricted to Hox complexes, either in vertebrates (30) or in Drosophila (SI Appendix, Fig. S9) (65). Thus, other genomic loci might also have benefited from the emergence of CTCF. In addition, the Hox-CTCF link might have been acquired independently in Drosophila and vertebrates. Differences in CTCF’s interaction partners (CP190 vs. cohesin) (65–67) point at this possibility. However, an alternative explanation is that lineage-specific interaction partners have been acquired after establishment of the kernel.
It has been proposed that geological and ecological factors contributed to the Cambrian explosion (68, 69), but the ultimate explanation must lie in changes of the genetic program that regulates the development of body plans (8, 70). Candidates that would easily fit these requirements have not been identified so far. An exception is the acquisition of 32 new miRNA families at the base of bilaterian evolution (71) that may contribute to morphological innovations at a macroevolutionary scale. However, the assumption of a Hox-CTCF kernel is a more comprehensive explanation. As the Cambrian explosion affected mainly bilaterian animals (4), formation of the new kernel has to trace back to a time before this event when the common ancestor of protostomes and deuterostomes or of all Bilateria lived (∼600–680 Mya) (72). Our data indicate that CTCF originated not later than in the protostome–deuterostome ancestor and probably not before the last common ancestor of all Bilateria and thus provide a possible molecular explanation for the diversification of body plans during the Early Cambrian.
Materials and Methods
Search for CTCF Candidates.
BLASTX, BLASTP, and TBLASTN searches in public sequence databases with Drosophila CTCF as a query were performed and yielded CTCF candidates for many bilaterian phyla. In several cases, a CTCF coding region was constructed by spliced alignment to a genomic contig (e.g., for Apis mellifera). We included from Bilateria only sequences in the final dataset that suggested orthology to CTCF after reciprocal BLAST. A different strategy was used for early branching metazoans and other eukaryotes (protists, fungi, and plants) where CTCF seemed to be absent in initial tests. To minimize the chance of missing a potential ortholog, we downloaded the proteomes of completely sequenced representatives (SI Appendix, Table S4) and scanned them with a custom-made HMMer profile (73). The profile comprised ZF2–8 of an alignment of 23 CTCFs (SI Appendix, Fig. S3) and detects the conserved core of a potential CTCF with high specificity. A three-ZF–spanning fragment of human CTCF, spiked into the Hydra dataset, produced an _E_-value of 3 × 10−7 with this HMM. We set this value as a threshold for the detection of potential CTCFs and considered all sequences from nonbilaterian genomes with a lower _E_-value for phylogenetic analysis.
Multiple Sequence Alignment and Phylogenetic Analysis.
Sequence alignment of CTCF candidates was performed as described elsewhere (22). The sequence dataset for phylogenetic analysis consisted of 170 sequences. It included (i) eight vertebrate (11), three insect (10, 21), and one nematode (22) CTCFs as a positive control for identification of the CTCF clade; (ii) eight insect orthologs of the D. melanogaster ZF transcription factor crooked legs as outgroup; and (iii) 150 CTCF candidate sequences from early branching metazoans and bilaterians. We performed phylogenetic analysis with maximum likelihood and Bayesian methods according to the instructions given in SI Appendix.
Prediction of CTCF Binding Sites.
To predict CTCF-binding motifs, we constructed position weight matrices (PWMs) on the basis of the CTCF-binding profiles determined by ChIP-seq from human HeLa cells and ChIP-chip data from D. melanogaster S2 cells (67). Fastq files corresponding to CTCF ChIP-seq experiments were downloaded from the Encode repository at the University of California, Santa Cruz (UCSC), genome portal (http://hgdownload.cse.ucsc.edu/goldenPath/hg18/encodeDCC/wgEncodeHudsonalphaChipSeq/) and aligned to hg19, using BOWTIE (74). The MACS peak-finding algorithm (75) was used to identify binding regions. The top 500 regions with typical peak shape were picked for motif analysis. To identify CTCF-binding motifs within those regions, we used MEME (76) with default settings and derived a motif almost identical to that reported in multiple human cell lines (16, 77). Similarly, we used the top 500 binding regions derived from ChIP-chip experiments with Drosophila S2 cells to identify a Drosophila CTCF motif. The resulting multiple alignments were transformed into PWMs, using a custom Perl script. We constructed a mixed PWM by merging the two PWMs. Additionally, we cut from the 5′ and 3′ ends those positions with low or no conservation between Drosophila and humans, resulting in a core PWM with 15 nucleotide positions (SI Appendix, Fig. S5), slightly shorter than the 18- and 22-bp motifs identified for Drosophila and human CTCF, respectively. Repeat masked genomes were downloaded from the sources given in SI Appendix, Table S5. If a repeat masked genome was not available for a particular organism, we applied REPEATMASKER (http://www.repeatmasker.org/), using standard settings and repeat definitions from Repbase (http://www.girinst.org/). Motif prediction was performed with two alternative applications under stringent conditions to avoid detection of large numbers of false positives. To avoid inconsistencies caused by differing nucleotide frequencies between the analyzed genomes, we used default background frequencies for all searches (A:T, 0.3; G:C, 0.2). We used PATSER (78) with a cutoff score of 13 and STORM (79) with a cutoff of 17 as we found these settings to pick up similar numbers of hits with a high level of coincidence (calibration was done on the Drosophila dm3 genome). Validity of the predicted motif instances was verified by comparison of the motif locations with genome-wide binding data. In the case of hg19, we detected 15,929 instances of the mixed motif at the above settings with STORM, 9,929 of them (62%) being associated with in vivo binding (see SI Appendix, Table S2 for detailed statistical analysis). The total number of motif instances was normalized to the effective genome size of the respective organism. To judge the specificity of the identified motifs, we repeated the analysis with four corrupted matrices (SI Appendix, Fig. S5) that differed from the original matrix by the reciprocal exchange of two highly conserved nucleotide positions (67).
Statistical Evaluation of Observed vs. Expected CTCF Sites in Different Genomes.
To calculate the significance of the PWM analyses, we generated for each of the 18 genomes in Fig. 2 100 randomized versions of the PWM by a double-reshuffling procedure: First, columns were permutated; second, in each column the entries for nucleotides A and T and for C and G were exchanged with 50% probability. Every genome was first screened with the intact matrix, using the program FSS (http://jakob.genetik.uni-koeln.de/bioinformatik/software/fss/). Among all hits with a positive score we determined the upper 1% quantile to serve as score threshold (s*) for the subsequent screen with the shuffled matrices. The screen with the shuffled matrices yielded a distribution of the number of hits with a score larger than s* (Fig. 2_B_) and an estimated P value for the screen with the intact matrix. It was significant in all genomes that had CTCF (P < 0.05), except in T. spiralis. To produce Fig. 2_B_, PWMs with log2-transformed nucleotide frequencies, adjusted to the background distribution, and species-specific thresholds were used. This explains the slight discrepancies in motif counts shown in Fig. 2 A and B.
Computation of PHASTCONS Scores.
To calculate the conservation of in vivo relevant CTCF sites in Drosophila and vertebrates, we predicted CTCF sites in the D. melanogaster BX-C and Antp-C and in the human HoxD complex with PATSER and STORM (78, 79), using less stringent thresholds of 3 and 4, respectively, and determined their overlap with ChIP-seq/ChIP-chip signals. The majority of peaks exactly matched predicted sites (Drosophila BX-C, 10/14; Antp-C, 13/19; human HoxD, 11/12). From the resulting positive sites, we generated 100-bp windows of genomic alignments as input for the PHASTCONS program (80). The general conservation topology of these regions was similar with different parameter settings, indicating that the conservation of CTCF sites represents a robust signal. For the graphs shown in Fig. 3 A and B and SI Appendix, Fig. S7, we ran PHASTCONS with the settings used at the UCSC genome browser (ftp://hgdownload.cse.ucsc.edu/goldenPath/dm3/phastCons15way/ and ftp://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/).
Supplementary Material
Supporting Information
Acknowledgments
Preliminary sequence data were obtained from the Baylor College of Medicine Human Genome Sequencing Center website at http://www.hgsc.bcm.tmc.edu. The DNA sequence of Saccoglossus kowalevskii was supported by the National Human Genome Research Institute and National Institutes of Health. The genome sequences of Capitella teleta and Lottia gigantea were produced by the US Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) in collaboration with the user community. This research was supported by grants from the German Research Foundation (Sonderforschungsbereich 680).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.B.E. is a guest editor invited by the Editorial Board.
References
- 1.Canfield DE, Poulton SW, Narbonne GM. Late-Neoproterozoic deep-ocean oxygenation and the rise of animal life. Science. 2007;315:92–95. doi: 10.1126/science.1135013. [DOI] [PubMed] [Google Scholar]
- 2.Conway Morris S. Darwin’s dilemma: The realities of the Cambrian ‘explosion’. Philos Trans R Soc Lond B Biol Sci. 2006;361:1069–1083. doi: 10.1098/rstb.2006.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall CR. Explaining the Cambrian “explosion” of animals. Annu Rev Earth Planet Sci. 2006;34:355–384. [Google Scholar]
- 4.Budd GE. The earliest fossil record of the animals and its significance. Philos Trans R Soc Lond B Biol Sci. 2008;363:1425–1434. doi: 10.1098/rstb.2007.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 6.Erwin DH. Early origin of the bilaterian developmental toolkit. Philos Trans R Soc Lond B Biol Sci. 2009;364:2253–2261. doi: 10.1098/rstb.2009.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marshall CR, Valentine JW. The importance of preadapted genomes in the origin of the animal bodyplans and the Cambrian explosion. Evolution. 2010;64:1189–1201. doi: 10.1111/j.1558-5646.2009.00908.x. [DOI] [PubMed] [Google Scholar]
- 8.Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 9.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 10.Moon H, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hore TA, Deakin JE, Marshall Graves JA. The evolution of epigenetic regulators CTCF and BORIS/CTCFL in amniotes. PLoS Genet. 2008;4:e1000169. doi: 10.1371/journal.pgen.1000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filippova GN. Genetics and epigenetics of the multifunctional protein CTCF. Curr Top Dev Biol. 2008;80:337–360. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- 13.Phillips JE, Corces VG. CTCF: Master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zlatanova J, Caiafa P. CCCTC-binding factor: To loop or to bridge. Cell Mol Life Sci. 2009;66:1647–1660. doi: 10.1007/s00018-009-8647-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie X, et al. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci USA. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Handoko L, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lobanenkov VV, et al. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–1753. [PubMed] [Google Scholar]
- 19.Burke LJ, Hollemann T, Pieler T, Renkawitz R. Molecular cloning and expression of the chromatin insulator protein CTCF in Xenopus laevis. Mech Dev. 2002;113:95–98. doi: 10.1016/s0925-4773(02)00005-9. [DOI] [PubMed] [Google Scholar]
- 20.Pugacheva EM, et al. Cloning and characterization of zebrafish CTCF: Developmental expression patterns, regulation of the promoter region, and evolutionary aspects of gene organization. Gene. 2006;375:26–36. doi: 10.1016/j.gene.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 21.Gray CE, Coates CJ. Cloning and characterization of cDNAs encoding putative CTCFs in the mosquitoes, Aedes aegypti and Anopheles gambiae. BMC Mol Biol. 2005;6:16. doi: 10.1186/1471-2199-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heger P, Marin B, Schierenberg E. Loss of the insulator protein CTCF during nematode evolution. BMC Mol Biol. 2009;10:84. doi: 10.1186/1471-2199-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejnol A, et al. (2009) Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc Biol Sci 276(1677):4261–4270. [DOI] [PMC free article] [PubMed]
- 24.Srivastava M, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- 25.Srivastava M, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chapman JA, et al. The dynamic genome of Hydra. Nature. 2010;464:592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith ST, et al. Genome wide ChIP-chip analyses reveal important roles for CTCF in Drosophila genome organization. Dev Biol. 2009;328:518–528. doi: 10.1016/j.ydbio.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holohan EE, et al. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renda M, et al. Critical DNA binding interactions of the insulator protein CTCF: A small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J Biol Chem. 2007;282:33336–33345. doi: 10.1074/jbc.M706213200. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt D, et al. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012;148:335–348. doi: 10.1016/j.cell.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohan M, et al. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 2007;26:4203–4214. doi: 10.1038/sj.emboj.7601851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heath H, et al. CTCF regulates cell cycle progression of alphabeta T cells in the thymus. EMBO J. 2008;27:2839–2850. doi: 10.1038/emboj.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore JM, et al. Loss of maternal CTCF is associated with peri-implantation lethality of Ctcf null embryos. PLoS ONE. 2012;7:e34915. doi: 10.1371/journal.pone.0034915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Botta M, Haider S, Leung IX, Lio P, Mozziconacci J. Intra- and inter-chromosomal interactions correlate with CTCF binding genome wide. Mol Syst Biol. 2010;6:426. doi: 10.1038/msb.2010.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magdinier F, Yusufzai TM, Felsenfeld G. Both CTCF-dependent and -independent insulators are found between the mouse T cell receptor alpha and Dad1 genes. J Biol Chem. 2004;279:25381–25389. doi: 10.1074/jbc.M403121200. [DOI] [PubMed] [Google Scholar]
- 36.Splinter E, et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majumder P, Boss JM. CTCF controls expression and chromatin architecture of the human major histocompatibility complex class II locus. Mol Cell Biol. 2010;30:4211–4223. doi: 10.1128/MCB.00327-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Degner SC, et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci USA. 2011;108:9566–9571. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Golan-Mashiach M, et al. Identification of CTCF as a master regulator of the clustered protocadherin genes. Nucleic Acids Res. 2012;40:3378–3391. doi: 10.1093/nar/gkr1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim YJ, Cecchini KR, Kim TH. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proc Natl Acad Sci USA. 2011;108:7391–7396. doi: 10.1073/pnas.1018279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pérez-Lluch S, Cuartero S, Azorín F, Espinàs ML. Characterization of new regulatory elements within the Drosophila bithorax complex. Nucleic Acids Res. 2008;36:6926–6933. doi: 10.1093/nar/gkn818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iampietro C, Gummalla M, Mutero A, Karch F, Maeda RK. Initiator elements function to determine the activity state of BX-C enhancers. PLoS Genet. 2010;6:e1001260. doi: 10.1371/journal.pgen.1001260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy S, et al. modENCODE Consortium Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kyrchanova O, et al. Selective interactions of boundaries with upstream region of Abd-B promoter in Drosophila bithorax complex and role of dCTCF in this process. Nucleic Acids Res. 2011;39:3042–3052. doi: 10.1093/nar/gkq1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark AG, et al. Drosophila 12 Genomes Consortium Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 46.Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- 47.Whiting J, et al. Multiple spatially specific enhancers are required to reconstruct the pattern of Hox-2.6 gene expression. Genes Dev. 1991;5:2048–2059. doi: 10.1101/gad.5.11.2048. [DOI] [PubMed] [Google Scholar]
- 48.Knittel T, Kessel M, Kim MH, Gruss P. A conserved enhancer of the human and murine Hoxa-7 gene specifies the anterior boundary of expression during embryonal development. Development. 1995;121:1077–1088. doi: 10.1242/dev.121.4.1077. [DOI] [PubMed] [Google Scholar]
- 49.Frasch M, Chen X, Lufkin T. Evolutionary-conserved enhancers direct region-specific expression of the murine Hoxa-1 and Hoxa-2 loci in both mice and Drosophila. Development. 1995;121:957–974. doi: 10.1242/dev.121.4.957. [DOI] [PubMed] [Google Scholar]
- 50.Gérard M, et al. In vivo targeted mutagenesis of a regulatory element required for positioning the Hoxd-11 and Hoxd-10 expression boundaries. Genes Dev. 1996;10:2326–2334. doi: 10.1101/gad.10.18.2326. [DOI] [PubMed] [Google Scholar]
- 51.Morrison A, Moroni MC, Ariza-McNaughton L, Krumlauf R, Mavilio F. In vitro and transgenic analysis of a human HOXD4 retinoid-responsive enhancer. Development. 1996;122:1895–1907. doi: 10.1242/dev.122.6.1895. [DOI] [PubMed] [Google Scholar]
- 52.Zákány J, Gérard M, Favier B, Duboule D. Deletion of a HoxD enhancer induces transcriptional heterochrony leading to transposition of the sacrum. EMBO J. 1997;16:4393–4402. doi: 10.1093/emboj/16.14.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juan AH, Ruddle FH. Enhancer timing of Hox gene expression: Deletion of the endogenous Hoxc8 early enhancer. Development. 2003;130:4823–4834. doi: 10.1242/dev.00672. [DOI] [PubMed] [Google Scholar]
- 54.Wang WC, et al. Comparative cis-regulatory analyses identify new elements of the mouse Hoxc8 early enhancer. J Exp Zoolog B Mol Dev Evol. 2004;302:436–445. doi: 10.1002/jez.b.21009. [DOI] [PubMed] [Google Scholar]
- 55.Birney E, et al. ENCODE Project Consortium NISC Comparative Sequencing Program Baylor College of Medicine Human Genome Sequencing Center Washington University Genome Sequencing Center Broad Institute Children’s Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soshnikova N, Montavon T, Leleu M, Galjart N, Duboule D. Functional analysis of CTCF during mammalian limb development. Dev Cell. 2010;19:819–830. doi: 10.1016/j.devcel.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 57.Ferraiuolo MA, et al. 2010. The three-dimensional architecture of Hox cluster silencing. Nucleic Acids Res 38:7472–7484.
- 58.Wan LB, et al. Maternal depletion of CTCF reveals multiple functions during oocyte and preimplantation embryo development. Development. 2008;135:2729–2738. doi: 10.1242/dev.024539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kmita M, Tarchini B, Duboule D, Hérault Y. Evolutionary conserved sequences are required for the insulation of the vertebrate Hoxd complex in neural cells. Development. 2002;129:5521–5528. doi: 10.1242/dev.00151. [DOI] [PubMed] [Google Scholar]
- 60.Di-Poï N, et al. Changes in Hox genes’ structure and function during the evolution of the squamate body plan. Nature. 2010;464:99–103. doi: 10.1038/nature08789. [DOI] [PubMed] [Google Scholar]
- 61.McDaniell R, et al. Heritable individual-specific and allele-specific chromatin signatures in humans. Science. 2010;328:235–239. doi: 10.1126/science.1184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rougier GW, Novacek MJ. Early mammals: Teeth, jaws and finally a skeleton! Curr Biol. 1998;8:R284–R287. doi: 10.1016/s0960-9822(98)70174-5. [DOI] [PubMed] [Google Scholar]
- 63.Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 64.Seo HC, et al. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature. 2004;431:67–71. doi: 10.1038/nature02709. [DOI] [PubMed] [Google Scholar]
- 65.Nègre N, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 67.Bartkuhn M, et al. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009;28:877–888. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Erwin DH. Colloquium paper: Extinction as the loss of evolutionary history. Proc Natl Acad Sci USA. 2008;105(Suppl 1):11520–11527. doi: 10.1073/pnas.0801913105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dahl TW, et al. Devonian rise in atmospheric oxygen correlated to the radiations of terrestrial plants and large predatory fish. Proc Natl Acad Sci USA. 2010;107:17911–17915. doi: 10.1073/pnas.1011287107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Valentine JW. Late Precambrian bilaterians: Grades and clades. Proc Natl Acad Sci USA. 1994;91:6751–6757. doi: 10.1073/pnas.91.15.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peterson KJ, Dietrich MR, McPeek MA. MicroRNAs and metazoan macroevolution: Insights into canalization, complexity, and the Cambrian explosion. Bioessays. 2009;31:736–747. doi: 10.1002/bies.200900033. [DOI] [PubMed] [Google Scholar]
- 72.Peterson KJ, Cotton JA, Gehling JG, Pisani D. The Ediacaran emergence of bilaterians: Congruence between the genetic and the geological fossil records. Philos Trans R Soc Lond B Biol Sci. 2008;363:1435–1443. doi: 10.1098/rstb.2007.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 74.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- 77.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 78.Hertz GZ, Hartzell GW, 3rd, Stormo GD. Identification of consensus patterns in unaligned DNA sequences known to be functionally related. Comput Appl Biosci. 1990;6:81–92. doi: 10.1093/bioinformatics/6.2.81. [DOI] [PubMed] [Google Scholar]
- 79.Smith AD, Sumazin P, Xuan Z, Zhang MQ. DNA motifs in human and mouse proximal promoters predict tissue-specific expression. Proc Natl Acad Sci USA. 2006;103:6275–6280. doi: 10.1073/pnas.0508169103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information