Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling (original) (raw)
. Author manuscript; available in PMC: 2013 Dec 1.
Published in final edited form as: Curr Opin Chem Biol. 2012 Nov 3;16(5-6):498–506. doi: 10.1016/j.cbpa.2012.10.016
Abstract
Although it is widely accepted that _S_-nitrosation occurs in vivo, questions remain regarding _S_-nitrosation as a signaling mechanism. The chemistry of _S_-nitrosation includes NO oxidation to N2O3 followed by reaction with thiolates, radical recombination of NO and thiyl radicals, and transition metal catalyzed pathways. Once formed, nitrosothiols can be transferred between small molecule or protein thiols through transnitrosation reactions. The pathways that lead to selective _S_-nitrosation of only a subset of cellular cysteines remain largely unknown. Selectivity may be conferred through colocalization with NOS isoforms, protein-protein interaction driven transnitrosation reactions, regulation of _S_-nitrosoglutathione levels, or directed denitrosation of protein nitrosothiols.
Introduction
Nitric oxide (NO) is synthesized by three mammalian nitric oxide synthase isoforms: endothelial (eNOS), inducible (iNOS), and neuronal (nNOS). The primary NO receptor is the soluble isoform of guanylate cyclase (sGC), and NO captured by the heme cofactor of sGC activates sGC to produce the secondary messenger cyclic GMP (cGMP) [1]. However, cGMP-independent NO signaling pathways have been reported and attributed to _S_-nitrosation of structurally and functionally important cysteine residues. Dysregulation of _S_-nitrosation is implicated in the pathology of a variety of disorders including asthma, hypertension, cancer, and neurodegeneration [2]. Although it is widely accepted that _S_-nitrosation occurs in vivo, the mechanism(s) by which nitrosothiols are formed and removed are unclear. This review focuses on potential mechanisms by which cellular nitrosothiols are formed and how selectivity for individual cysteine residues might be conferred.
Nomenclature
_S_-nitrosation is a post-translational modification in which a thiol is converted to a nitrosothiol. Although nitrosation and nitrosylation are often used interchangeably, nitrosation is the proper term to describe formation of a nitroso group (R-NO), which involves a one-electron oxidation from the NO radical. Although “nitrosylation” was originally coined to suggest an analogy to phosphorylation (the transfer of the phosphoryl group), nitrosylation is chemically misleading. The nitrosyl group is formally a NO radical, and the nitroso group (not the nitrosyl group) is transferred during nitrosation reactions. Nitrosylation instead refers to the coordination of the nitrosyl group with a transition metal. Therefore, we utilize _S_-nitrosation to describe the conversion of a thiol to a nitrosothiol.
Mechanisms of nitrosothiol formation
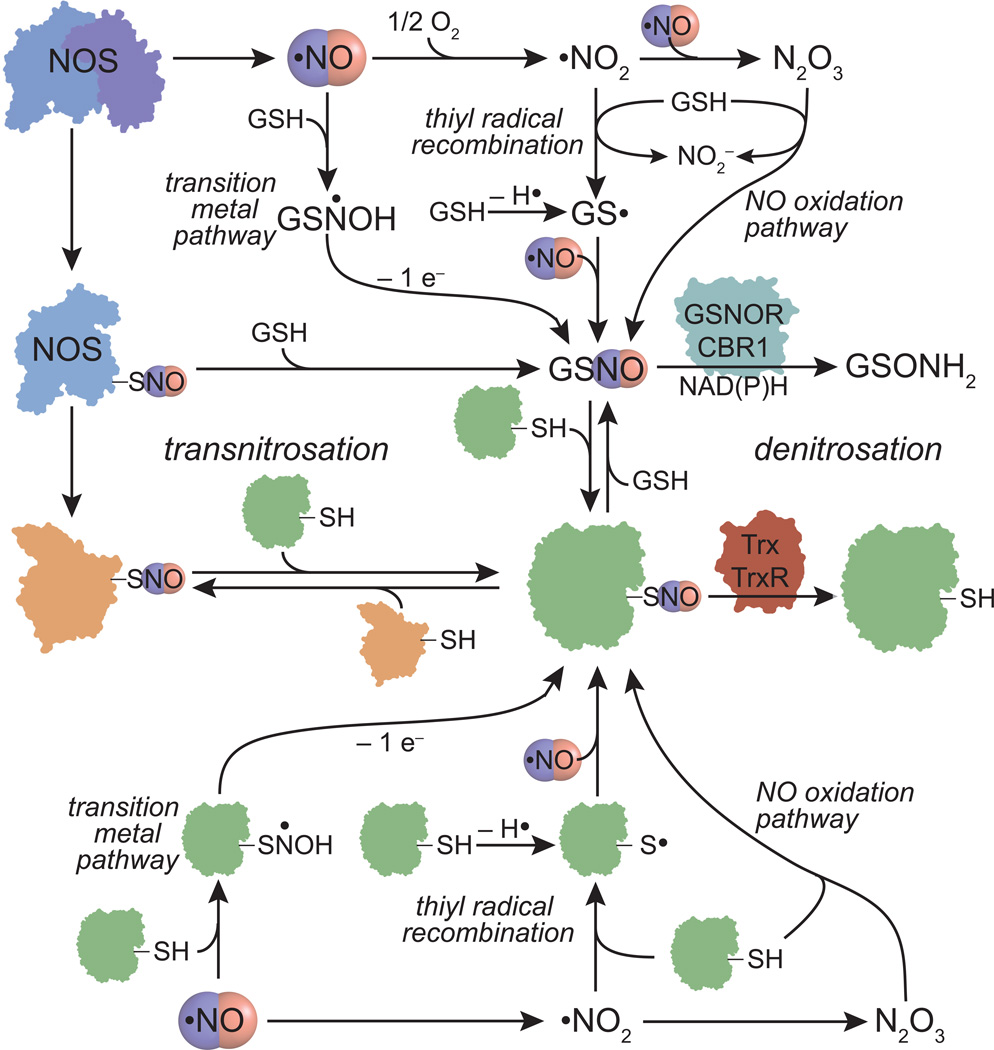
_S_-nitrosation requires a one-electron oxidation of the initial addition complex between NO and a thiol. In the absence of an electron sink (e.g. under anaerobic conditions and in absence of transition metals), a nitrosothiol will not form. Nitrosothiol formation can take place through NO autooxidation to N2O3, radical recombination between NO and a thiyl radical, and transition metal catalyzed pathways (Figure 1). Once formed, nitrosothiols can be transferred to small molecule or protein thiols through transnitrosation reactions.
Figure 1.
Potential mechanisms of protein nitrosothiol formation and degradation. Activation of nitric oxide synthases (NOS) results in NO production and auto-_S_-nitrosation. _S_-nitrosoglutathione (GSNO) or protein nitrosothiols can be generated from NO through either a transition metal catalyzed pathway, thiyl radical recombination, NO oxidation to N2O3, or transnitrosation with protein nitrosothiols such as NOS. Once formed, GSNO or _S_-nitrosated NOS can transfer nitrosothiols to other proteins through transnitrosation reactions. Degradation of nitrosothiols can occur through direct denitrosation of GSNO by the NAD(P)H-dependent enzymes GSNO reductase (GSNOR) or carbonyl reductase 1 (CBR1). Thioredoxin (Trx) can denitrosate protein nitrosothiols, a process that becomes catalytic through the activity of thioredoxin reductase (Trx).
NO autooxidation to N2O3
Aerobic solution decomposition of NO generates N2O3 in a reaction that is second order in NO and first order in O2 (Figure 2). N2O3 is highly reactive with water (forming NO2−) or with thiolates to produce nitrosothiols and NO2− [3]. However, kinetic modeling has predicted that physiological NO concentrations (≤1 µM) result in femtomolar N2O3 concentrations [4]. As N2O3 is formed from radical recombination of NO and NO2, NO must compete with the rapid reaction of NO2 radical with cellular thiols and other reductants present at concentrations far exceeding NO concentrations. Furthermore, cellular _S_-nitrosation occurs and is sometimes enhanced under anaerobic conditions [5] suggesting that O2 is not required for _S_-nitrosation. Therefore, the biological relevance of the N2O3 pathway might be limited to a small subset of cellular compartments with properties that favor reaction with N2O3.
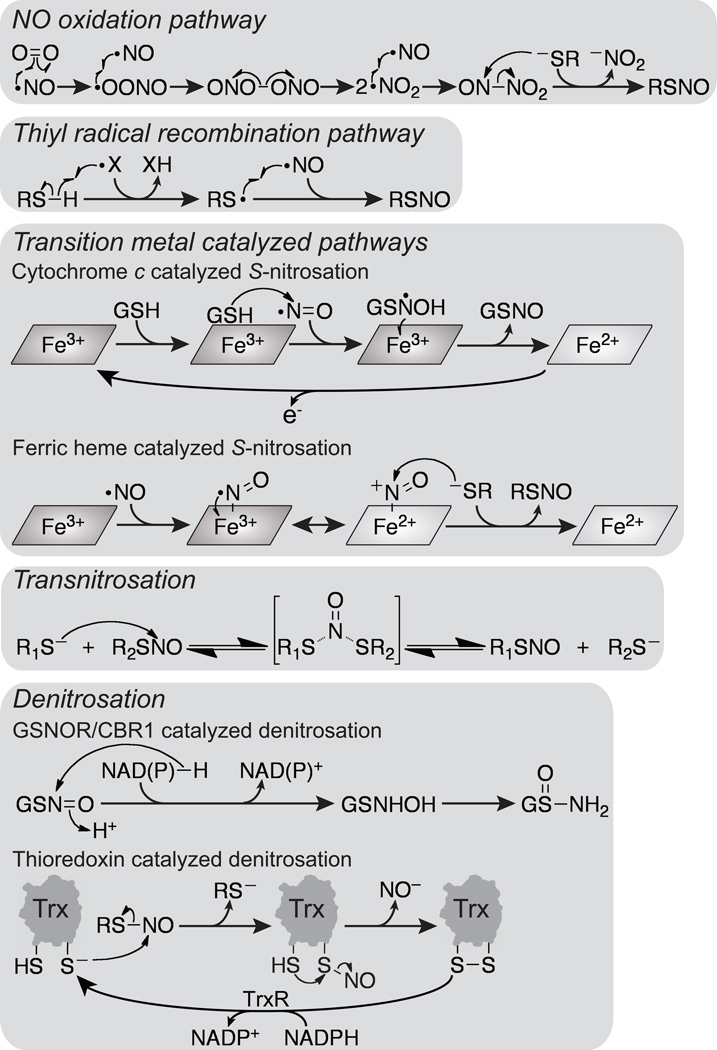
Figure 2.
Mechanisms of reactions discussed in this review. In the NO oxidation pathway NO first reacts with O2 to form a peroxynitrite radical. The peroxynitrite radical then undergoes radical recombination with NO to form ONOONO (or other isomers) followed by homolytic cleavage to form two molecules of NO2 radical. Radical recombination of NO and NO2 radicals results in N2O3 formation, which then reacts with a thiolate (RS−) to form a nitrosothiol (RSNO) and nitrite (NO2−). In the thiyl radical recombination pathway a thiyl radical is formed by hydrogen abstraction by another radical (X•). Radical recombination of the thiyl radical with NO forms a nitrosothiol. Several transition metal catalyzed pathways are possible. In one pathway GSH weakly associates with cytochrome c, and the bound GSH attacks NO to form a GSNOH radical. The ferric heme of cytochrome c then accepts an electron from the GSNOH radical to form ferrous heme and GSNO. Reoxidation of cytochrome c completes the catalytic cycle. In another transition metal catalyzed pathway NO can bind to ferric heme, and the resulting Fe3+–NO complex (which possesses significant Fe2+–NO+ character) then reacts with a thiolate to form a nitrosothiol and ferrous heme. Once formed a nitrosothiol can be transferred through transnitrosation reactions in which a thiolate attacks the nitrogen atom of a nitrosothiol resulting in a nitroxyl disulfide intermediate or transition state that decays to a thiolate and a nitrosothiol. Denitrosation of nitrosothiols can proceed through several mechanisms. GSNOR and CBR1 catalyze hydride transfer from NAD(P)H to GSNO to form a GSNHOH intermediate that spontaneously rearranges to GSONH2. Thioredoxin catalyzed denitrosation proceeds through transnitrosation of a thioredoxin active-site cysteine (C32). Subsequently, the other active-site cysteine (C35) eliminates the nitrosothiol intermediate at C32 to form active-site oxidized thioredoxin and nitroxyl (NO−). The catalytic cycle is completed via thioredoxin reductase catalyzed NADPH-dependent reduction of the thioredoxin active-site disulfide.
The increased hydrophobicity of protein interiors or membranes was proposed to enhance rates of _S_-nitrosation 30-fold [6] due to favorable hydrophobic partitioning of NO and O2. While nitrosothiol formation near membranes or within proteins could take place, _S_-nitrosation would have to proceed faster than diffusion. N2O3 reacts with glutathione (GSH) to form _S_-nitrosoglutathione (GSNO) at a rate of 6.6 × 107 M−1s−1, which is orders of magnitude slower than diffusion [4]. Conversely, thiol deprotonation is disfavored in hydrophobic environments and a thiolate is required for reaction with N2O3 [7]. These two opposing effects must be balanced for hydrophobic phase-catalyzed _S_-nitrosation. Due to the second-order dependence on NO concentrations, N2O3 formation could also be favored near NOS isoforms (see “Direct interaction or colocalization with nitric oxide synthases”).
Thiyl radical recombination with NO
A second mechanism of nitrosothiol formation involves radical recombination of NO with thiyl radicals, yielding nitrosothiols [8]. Among the proposed mechanisms of nitrosothiol formation, only thiyl radical recombination is fast enough (1–3 × 109 M−1s−1 [8]) to compete with NO binding to heme (>1.4 × 108 M−1s−1 for sGC [9]). In isolated systems containing only NO, O2, and a thiol, thiyl radical formation proceeds through radical abstraction by a NO2 radical [3] (Figure 1). However, thiyl radicals can be formed by a variety of mechanisms in vivo and formation of thiyl radicals by NO-independent mechanisms would shift nitrosothiol formation from second-order to first-order in NO concentration and be favored at low O2 concentrations (Figure 2). Kinetic modeling suggested that nitrosothiol formation occurs exclusively through radical recombination as opposed to reaction with N2O3 [10]. However, GSH was the only thiol included in the kinetic model and protein thiols may favor other mechanisms of _S_-nitrosation.
Transition metal catalyzed nitrosothiol formation
A third mechanism is the direct addition of NO to a thiol to form a thionitroxyl radical (RSNOH•) followed by a one-electron oxidation to form a nitrosothiol (Figure 1). While it has been proposed that electron acceptors (e.g. O2 or a transition metal) could oxidize the intermediate thionitroxyl radical, kinetic studies argue against this reaction contributing to nitrosothiol formation in solution [11]. However, the direct addition mechanism could be operative in an enzyme active site. Indeed, cytochrome c (Cyt c) was recently proposed to synthesize GSNO. In the proposed mechanism, initial weak binding of GSH to ferric Cyt c is followed by reaction with NO to form GSNO• (Figure 2), which then reduces the heme to the ferrous state to form GSNO. This reaction is energetically favorable [12] and was found most efficient at the nanomolar NO concentrations expected during NO signaling [13]. However, Cyt c is localized to the mitochondrial intermembrane space and GSNO does not cross the cell membrane [14] and might not cross the outer mitochondrial membrane to the cytosol (Figure 3). Therefore, _S_-nitrosation by Cyt c might be limited to the mitochondrial intermembrane space. Nonetheless, depletion of Cyt c in cells or a whole cell lysates resulted in a decrease in protein _S_-nitrosation [15] consistent with an in vivo role for Cyt c.
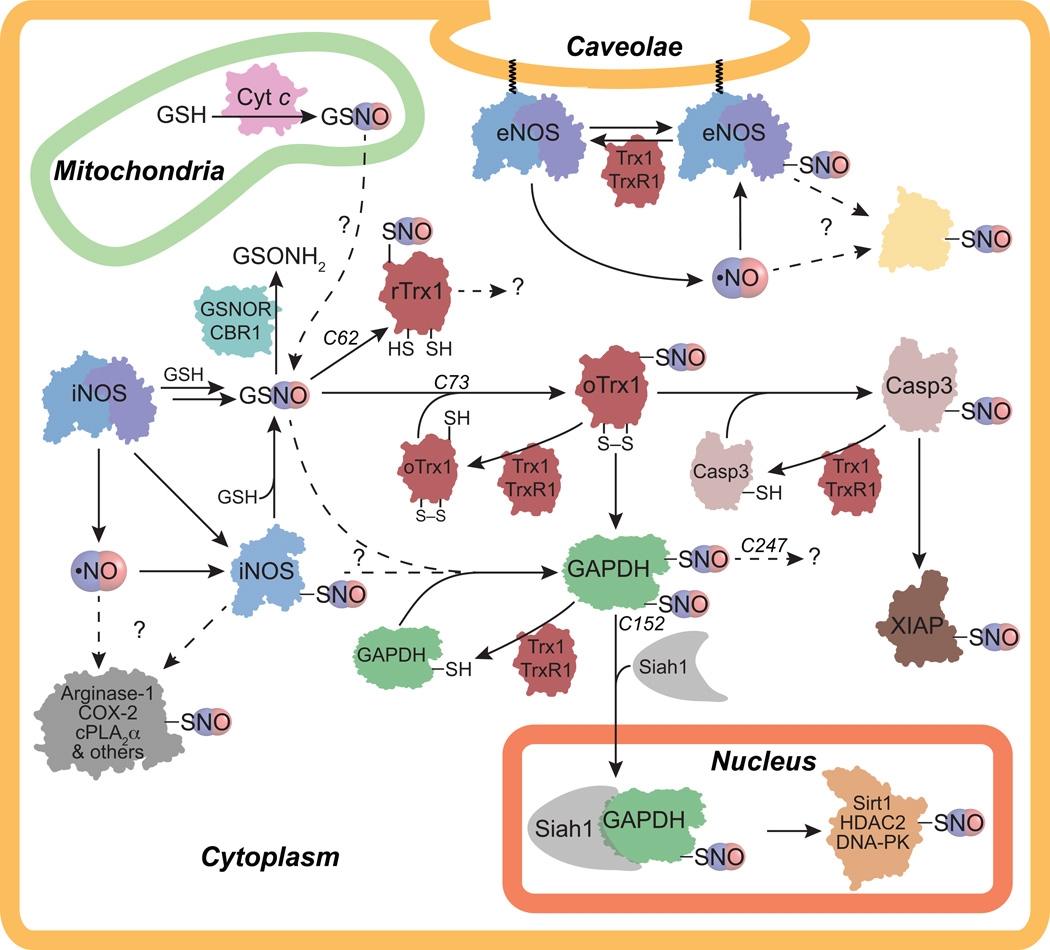
Figure 3.
Potential transnitrosation signaling pathways. Nitric oxide synthase (NOS) activity results in NO formation and NOS auto-_S_-nitrosation. iNOS is cytoplasmic whereas eNOS can be palmitoylated/myristoylated and targeted to the caveolae. Direct interaction with iNOS or eNOS results in _S_-nitrosation of arginase-1, COX-2, cPLA2α, and GAPDH among other proteins although it is unclear if NO, GSNO, or NOS _S_-nitrosation is the source of _S_-nitrosation. GSNO can be generated through a variety of pathways (see Figure 1), but it is unclear if cytochrome c (Cyt c) can contribute to the cytoplasmic GSNO pool as Cyt c is localized to the mitochondrial intermembrane space. Once formed, GSNO transnitrosates active-site reduced thioredoxin 1 (rTrx1) at C62 and active-site oxidized Trx1 (oTrx1) at C73. Although the downstream targets of rTrx1-C62-SNO are unknown, oTrx1-C73-SNO transnitrosates caspase-3 (Casp3) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) among other proteins. _S_-nitrosated Casp3 can then transnitrosate X-linked inhibitor of apoptosis (XIAP). GAPDH is nitrosated at two different cysteines: C152 and C247. GAPDH _S_-nitrosation at C152 recruits Siah1 binding resulting in translocation of the Siah1/GAPDH complex to the nucleus where GAPDH-C152-SNO transnitrosates Sirt1, HDAC2, and DNA-PK. The downstream transnitrosation targets of GAPDH-C247-SNO are unknown. Dashed arrows and question marks indicate areas where further research is particularly needed.
Cu2+ and Fe3+ can catalyze nitrosothiol formation through transient oxidation of NO to NO+ followed by reaction with a thiol. This reaction mechanism was proposed for GSNO formation by ceruloplasmin [3]. However, this reaction must be concerted, as NO+ released into solution would immediately react with water to form NO2−. A more recent study showed that NO2− is the major product of the reaction of NO with ceruloplasmin, and depletion or supplementation of ceruloplasmin is plasma failed to alter plasma nitrosothiol levels [16]. A similar mechanism of nitrosothiol formation involves the binding of NO to ferric heme. The Fe3+–NO will possess significant Fe2+–NO+ character and can react with a thiol resulting in nitrosothiol formation and heme reduction (Figure 2). This has been proposed for ferric hemoglobin, but this process is unlikely to occur in vivo as the concentration of ferric hemoglobin is low and NO much more rapidly reacts with both oxy- and deoxyhemoglobin [12].
Zn2+ ligation has also been proposed to activate thiols towards _S_-nitrosation. Selective _S_-nitrosation of Zn2+ coordinated cysteines has been observed in several proteins including iNOS/eNOS [17], Sirt1 [18], and XIAP [19]. _S_-nitrosation of these proteins generally results in Zn2+ loss and profound effects on protein function. O2 was required for _in vitro S_-nitrosation of Zn2+-thiolate model complexes by NO consistent with N2O3 as the relevant nitrosating agent [20,21].
Transnitrosation
Transnitrosation is a reversible second-order reaction between a nitrosothiol and a thiol (Figure 1). Transnitrosation proceeds through nucleophilic attack of a thiolate anion at the nitrosothiol nitrogen resulting in a nitroxyl disulfide intermediate [22,23] or transition state [24] (Figure 2). The overall mechanism of transnitrosation resembles an SN2 reaction. Factors that control transnitrosation selectivity are discussed under “Protein-protein interaction driven transnitrosation reactions”.
Mechanisms of selectivity in _S_-nitrosation
In order for _S_-nitrosation to be a viable signaling mechanism the formation and decay of nitrosothiols must be tightly controlled. Several mechanisms might impart selectivity to nitrosothiol formation and degradation including cysteine reactivity, localization with NOS isoforms, transnitrosation reactions, control of GSNO concentrations, and denitrosation reactions.
Analysis of known sites of _S_-nitrosation
Some proteins may have a cysteine in a microenvironment that favors _S_-nitrosation. However, the majority of factors that activate a particular cysteine towards _S_-nitrosation will also activate this cysteine towards other cysteine post-translational modifications. Analysis of _S_-nitrosation sites has not yielded a dominant sequence motif [25–27], and factors including the p_K_a value and nearby hydrophobicity were not predictive. Instead these analyses found that most _S_-nitrosation sites contained charged residues within 6–8 Å of the nitrosated cysteine [28,29]. Importantly, the majority of these charged residues were pointed away from the nitrosated cysteine indicating that they were not directly involved in the chemistry. In addition, _S_-nitrosation was enriched in surface-accessible sites [28], suggesting that these charged residues are important for protein-protein transnitrosation reactions.
The lack of a consensus motif for _S_-nitrosation is likely due to the many mechanisms leading to nitrosothiol formation (Figure 1). Indeed, genetic manipulation of NO and GSNO concentrations suggested that distinct proteins are _S_-nitrosated by NO oxidation pathways versus GSNO transnitrosation pathways [30]. Therefore, it may be possible to cluster _S_-nitrosation sites based on their mechanism of formation. To this end, several groups have developed machine-learning techniques to describe and predict the nitrosoproteome. These efforts have resulted in three web servers (SNOsite, GPS-SNO, and CPR-SNO) in which a user can input a protein sequence and generate predicted _S_-nitrosation sites [31–33]. In addition, a centralized database (dbSNO) of >3000 protein _S_-nitrosation site curated from >219 research articles was recently made available [34].
Direct interaction or colocalization with nitric oxide synthases
There are several examples where interaction with NOS results in selective protein _S_-nitrosation due to increased concentration of nitrosating agents near the site of _S_-nitrosation (Figure 3). For example, the formation of an iNOS/cyclooxygenase-2 (COX-2) or iNOS/arginase-1 complex was necessary for COX-2 or arginase-1 _S_-nitrosation, respectively [17,35]. In cases where direct interaction does not occur, a scaffolding protein can facilitate protein _S_-nitrosation. For instance, COX-2 enables the association of cPLA2α with iNOS resulting in cPLA2α _S_-nitrosation [36].
Protein _S_-nitrosation may also be controlled through compartmentalization with NOS isoforms. eNOS is the only NOS isoform that is myristoylated/palmitoylated and targeted to specific intracellular membranes including the caveolae and Golgi apparatus (Figure 3). Therefore, colocalization with eNOS may represent a mechanism for local protein _S_-nitrosation even when direct interaction with eNOS does not occur [37]. Indeed, a general increase in protein _S_-nitrosation surrounding eNOS at the Golgi apparatus has been observed [38].
Both iNOS and eNOS are selectively _S_-nitrosated at the Zn2+-tetrathiolate site [17]. In all cases involving NOS in protein _S_-nitrosation, it is unclear if _S_-nitrosation is due to transnitrosation with _S_-nitrosated NOS or due to an increase in the local concentration of nitrosating agents (Figure 3). Transnitrosation with _S_-nitrosated NOS would represent an elegant mechanism for controlled and compartmentalized nitrosothiol formation and reconcile the apparent paradox between the high diffusability of NO with the localized protein _S_-nitrosation observed in close proximity to NOS [38]. Furthermore, transnitrosation from NOS _S_-nitrosated at the Zn2+-tetrathiolate (or potentially _N_-nitrosated at the tetrahydrobiopterin cofactor [39]) to GSH would allow for controlled GSNO formation [40]. There is evidence that NOS auto-_S_-nitrosation is a controlled mechanism. All three mammalian NOS isoforms contain a hydrophobic tunnel between the heme cofactor and the Zn2+-tetrathiolate. This tunnel was proposed to provide a hydrophobic cavity for generation of nitrosating agents such as N2O3 and shuttle these nitrosating agents to the Zn2+-tetrathiolate. Detailed kinetic analyses were consistent with the ability of NO generated at the iNOS heme cofactor to _S_-nitrosate the Zn2+-tetrathiolate without leaving iNOS [40].
Protein-protein interaction driven transnitrosation reactions
Structural analysis of protein _S_-nitrosation suggested that protein-protein interaction driven transnitrosation is a major mechanism imparting selectivity to protein nitrosothiol formation [28,29]. Several factors control the rate of transnitrosation including steric hindrance of the nitrosothiol or attacking thiolate, the p_K_a of the attacking thiol, and the electrophilicity of the nitrosothiol. However, the p_K_a of the attacking thiol is unlikely to determine which thiols are stably _S_-nitrosated, since a thiol with a low p_K_a is both a better nucleophile and a better leaving group during transnitrosation. Instead low p_K_a thiols might catalyze transnitrosation reactions through two subsequent transnitrosation reactions with a nitrosothiol intermediate analogous to a ping-pong mechanism with a covalent enzyme intermediate. This will increase the rate of transnitrosation and also impart specificity through specific protein-protein interactions. Both glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and thioredoxin 1 (Trx1) have been shown to participate in protein-protein interaction driven transnitrosation reactions [18,25,41,42].
Although transnitrosation is reversible, the reaction can be driven in one direction if _S_-nitrosation results in a conformational change rendering the nitrosothiol inaccessible or if _S_-nitrosation results in translocation of the protein to a different cellular compartment. GAPDH _S_-nitrosation at C152 results in GAPDH binding to Siah1, which contains a nuclear localization sequence and translocation of the GAPDH/Siah1 complex to the nucleus occurs [43] (Figure 3). Once in the nucleus, GAPDH-C152-SNO transnitrosates Sirt1, histone deacetylase 2 (HDAC2), and DNA-dependent protein kinase (DNA-PK) [18]. GAPDH _S_-nitrosation at C247 has also been observed [25,28,44,45], but the transnitrosation targets of GAPDH-C247-SNO have not been identified. In addition, the mechanism of GAPDH _S_-nitrosation at either C152 or C247 is unknown. GSNO is often used to selectively _S_-nitrosate GAPDH at C152 in vitro and docking analysis suggested good affinity for GSNO near C152 [29]. Alternatively, GAPDH was shown to form a complex with NOS isoforms [46] and might be directly transnitrosated by _S_-nitrosated NOS. Interestingly, treatment of cells with interferon-γ and oxidized low-density lipoprotein resulted in selective _S_-nitrosation of C247, whereas treatment with interferon-γ and lipopolysaccharide resulted in selective _S_-nitrosation of C152 [44].
Thioredoxin is another important nexus of _S_-nitrosation signaling. Thioredoxin 1 (Trx1) contains five conserved cysteines: two redox-active catalytic cysteines (C32 and C35) and three non active-site cysteines (C62, C69, and C73). _S_-nitrosation of all three non active-site cysteines of Trx1 has been observed by GSNO, but more recent analyses showed that the redox state of the active-site cysteines is a critical factor in controlling GSNO transnitrosation [25,47]. Specifically, Trx1 is selectively transnitrosated by GSNO at C73 when the active-site cysteines (C32 and C35) are oxidized to a disulfide (oTrx1) [25,47] (Figure 3). Active-site reduced (rTrx1) was found to be selectively transnitrosated at C62 [47], although others could not detect rTrx1 _S_-nitrosation [25].
The redox dependent transnitrosation of Trx1 suggests the intriguing possibility that Trx1 may transnitrosate distinct proteins targets depending on the redox state of the cell. While the possibility that rTrx1-C62-SNO selectively transnitrosates distinct protein targets has not been explored, a recent proteomics study identified 47 putative transnitrosation targets of oTrx1-C73-SNO including GAPDH [25], suggesting crosstalk between the Trx1 and GAPDH transnitrosation pathways (Figure 3). oTrx1-C73-SNO is also known to selectively transnitrosate the active-site cysteine of caspase-3 (C163) [41,42]. Importantly, the direct interaction of Trx1 and caspase-3 (Casp3) is essential for transnitrosation as a Trx1 mutant that lost the ability to bind Casp3 was unable to transnitrosate Casp3 [41,42]. Downstream, Casp3-C163-SNO was found to transnitrosate the X-linked inhibitor of apoptosis (XIAP) [19], and disruption of the Casp3/XIAP interaction abolished XIAP _S_-nitrosation.
Regulation of GSNO concentrations
Since GSNO participates in transnitrosation reactions with proteins, modulation of GSNO levels should directly affect levels of protein _S_-nitrosation. Several enzymes have been reported to degrade GSNO in vitro including protein disulfide isomerase, xanthine dehydrogenase/oxidase, superoxide dismutase, and glutathione peroxidase [48]. However, only two enzymes have been shown to degrade GSNO and affect protein _S_-nitrosation in vivo: GSNO reductase (GSNOR) and carbonyl reductase 1 (CBR1) (Figure 1 and 3).
GSNOR (a.k.a. alcohol dehydrogenase class III or glutathione-dependent formaldehyde dehydrogenase) catalyzes the NADH-dependent reduction of GSNO to form the intermediate, GSNHOH, which rearranges to form glutathione sulfinamide (GSONH2) [49,50] (Figure 1, 2, and 3). GSNOR does not reduce _S_-nitrosocysteine, _S_-nitrosohomocysteine, _S_-nitroso-_N_-acetylpenicillamine, or protein nitrosothiols [51,52]. GSNOR knockout mice have markedly increased levels of both GSNO and _S_-nitrosated proteins [48], and GSNOR inhibitors are currently being developed to treat lung and cardiovascular disorders, asthma, and other inflammatory diseases [53,54].
CBR1 catalyzes the NADPH-dependent reduction of GSNO. CBR1 is specific for GSNO, as the reduction of _S_-nitrosocysteine was not catalyzed by CBR1. In cell lysates CBR1 was shown to account for 30% of the NADPH-dependent GSNO reductase activity [55]. Although CBR1 and CBR3 possess 72% sequence identity, only CBR1 is capable of GSNO reduction [56]. CBR1 and GSNOR possess similar _K_M values for GSNO (~30 µM), but GSNOR exhibits a 5-fold greater _k_cat value [49,55]. The NADH-dependence of GSNOR and NADPH-dependence of CBR1 suggests that the NADH/NAD+ and NADPH/NADP+ ratios may influence the relative GSNO reduction rates of GSNOR versus CBR1.
Denitrosation of protein nitrosothiols
If _S_-nitrosation is a specific cellular post-translational modification, then regulated mechanisms to remove protein nitrosothiols (denitrosation) must exist. Implicit in the transnitrosation mechanism is denitrosation of the original nitrosothiol (Figure 1). Transnitrosation reactions involving GSH will result in GSNO formation, and denitrosation of GSH sensitive protein nitrosothiols can be driven to completion by GSNOR or CBR1. However, only a subset of cellular nitrosothiols are susceptible to transnitrosation by GSH [45,57] suggesting other mechanisms exist to denitrosate protein nitrosothiols. Although many proteins catalyze denitrosation in vitro [48], only thioredoxin has been shown to affect levels of cellular protein nitrosothiols.
Thioredoxin catalyzed denitrosation generates a free thiol at the original _S_-nitrosation site, active-site oxidized thioredoxin, and putatively NO− [58] (Figure 2). Active-site reduced Trx1 is then regenerated through the action of thioredoxin reductase 1 (TrxR1). There is evidence that Trx1/TrxR1 catalyzes denitrosation of proteins such as eNOS [17], Casp3 [59,60], Casp8 [61], Casp9 [59], protein Tyr phosphatase 1B [59], GAPDH [59,62], and _N_-ethylmaleimide-sensitive factor [62] (Figure 2). In addition, recent proteomics studies identified >100 putative substrates of Trx1 denitrosation [28,63,64]. Trx1 denitrosation specificity might be determined by direct protein-protein interactions between Trx1 and _S_-nitrosated proteins. Compartmentalization might also regulate specificity in Trx catalyzed denitrosation. For example, both the cytosolic Trx1 and mitochondrial Trx2 can catalyze protein denitrosation [59] suggesting these Trx isoforms may denitrosate distinct cytoplasmic or mitochondrial targets.
Trx1 possesses two seemingly opposed functions catalyzing both transnitrosation and denitrosation of proteins. This is best exemplified with Casp3 where different cysteine residues in Trx1 catalyze the transnitrosation (C73) or denitrosation (C32) of Casp3. Inhibition of TrxR results in an increase in protein _S_-nitrosation and Trx1 active-site oxidation (oTrx1) [25,57,59]. However, conversion of rTrx1 to oTrx1 will both inactivate the denitrosation by C32 and activate transnitrosation by C73. In either case an increase in protein _S_-nitrosation is expected. Further complicating manners is the observation that oTrx1-C73-SNO is denitrosated by rTrx1 [25,65] (Figure 3). Therefore, it is difficult to distinguish the transnitrosating and denitrosating activities of Trx1.
Outlook
It is becoming increasingly clear that NO signaling involves protein targets other than sGC. _S_-nitrosation is an attractive candidate for these cGMP-independent pathways, but several fundamental questions have yet to be resolved to establish _S_-nitrosation as a viable signaling mechanism and not simply a byproduct of nitrosative stress. How are nitrosothiols initially formed in the cell? What determines the specificity of _S_-nitrosation to individual cysteine residues in proteins? How are these processes regulated? This review has summarized how these critical questions are beginning to be addressed, but many details have yet to be uncovered.
Highlights.
- Nitric oxide can signal through _S_-nitrosation of cysteine residues.
- _S_-nitrosation can occur through NO oxidation to N2O3 or radical recombination.
- Cytochrome c was recently discovered to catalyze the synthesis of GSNO.
- Colocalization with NOS isoforms can confer selectivity to _S_-nitrosation.
- Another significant mechanism of selectivity is protein-protein transnitrosation.
Acknowledgements
We thank Dr. Sarah Wynia Smith for her critical reading of this manuscript. Financial support was provided by a NIH National Institute of General Medical Sciences postdoctoral fellowship 5F32GM095023 (B.C.S).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Derbyshire ER, Marletta MA. Structure and regulation of soluble guanylate cyclase. Annu Rev Biochem. 2012;81:533–559. doi: 10.1146/annurev-biochem-050410-100030. [DOI] [PubMed] [Google Scholar]
- 2.Anand P, Stamler JS. Enzymatic mechanisms regulating protein S-nitrosylation: implications in health and disease. J Mol Med. 2012;90:233–244. doi: 10.1007/s00109-012-0878-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broniowska KA, Hogg N. The Chemical Biology of S-Nitrosothiols. Antioxid Redox Signal. 2012 doi: 10.1089/ars.2012.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim CH, Dedon PC, Deen WM. Kinetic analysis of intracellular concentrations of reactive nitrogen species. Chemical research in toxicology. 2008;21:2134–2147. doi: 10.1021/tx800213b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosworth CA, Toledo JC, Zmijewski JW, Li Q, Lancaster JR. Dinitrosyliron complexes and the mechanism(s) of cellular protein nitrosothiol formation from nitric oxide. Proc Natl Acad Sci USA. 2009;106:4671–4676. doi: 10.1073/pnas.0710416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Möller MN, Li Q, Vitturi DA, Robinson JM, Lancaster JR, Denicola A. Membrane “Lens” Effect: Focusing the Formation of Reactive Nitrogen Oxides from the •NO/O 2Reaction. Chemical research in toxicology. 2007;20:709–714. doi: 10.1021/tx700010h. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Andrekopoulos C, Xu Y, Joseph J, Hogg N, Feix J, Kalyanaraman B. Decreased S-nitrosation of peptide thiols in the membrane interior. Journal of Biological Chemistry. 2009;47:962–968. doi: 10.1016/j.freeradbiomed.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madej E, Folkes LK, Wardman P, Czapski G, Goldstein S. Thiyl radicals react with nitric oxide to form S-nitrosothiols with rate constants near the diffusion-controlled limit. Free Radic Biol Med. 2008;44:2013–2018. doi: 10.1016/j.freeradbiomed.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Brandish PE, Ballou DP, Marletta MA. A molecular basis for nitric oxide sensing by soluble guanylate cyclase. Proc Natl Acad Sci USA. 1999;96:14753–14758. doi: 10.1073/pnas.96.26.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lancaster JR. Nitroxidative, Nitrosative, and Nitrative Stress: Kinetic Predictions of Reactive Nitrogen Species Chemistry Under Biological Conditions. Chemical research in toxicology. 2006;19:1160–1174. doi: 10.1021/tx060061w. [DOI] [PubMed] [Google Scholar]
- 11.Keszler A, Zhang Y, Hogg N. Reaction between nitric oxide, glutathione, and oxygen in the presence and absence of protein: How are S-nitrosothiols formed? Free Radic Biol Med. 2010;48:55–64. doi: 10.1016/j.freeradbiomed.2009.10.026. The authors combine detailed kinetic analysis and simulation to study the reaction between glutathione and nitric oxide under aerobic conditions and conclude that both the N2O3 and thiyl radical recombination pathways occur simultaneously. This study also provides evidence against two previously proposed phenomena: direct addition of NO to a thiol followed by oxidation and catalysis of _S_-nitrosation by protein hydrophobic pockets.
- 12.Koppenol WH. Nitrosation, Thiols, and Hemoglobin: Energetics and Kinetics. Inorg. Chem. 2012;51:5637–5641. doi: 10.1021/ic202561f. This paper concisely describes the various thermodynamic and kinetic factors that come into play in _S_-nitrosation reactions.
- 13.Basu S, Keszler A, Azarova NA, Nwanze N, Perlegas A, Shiva S, Broniowska KA, Hogg N, Kim-Shapiro DB. A novel role for cytochrome c: Efficient catalysis of S-nitrosothiol formation. Free Radic Biol Med. 2010;48:255–263. doi: 10.1016/j.freeradbiomed.2009.10.049. This reference along with reference 15 examines the reaction between glutathione, nitric oxide, and cytochrome c and provides evidence for efficient GSNO production. While there are details still to be addressed, this reaction represents the first potentially catalytic mechanism of GSNO synthesis.
- 14.Zhang Y, Hogg N. S-Nitrosothiols: cellular formation and transport. Free Radic Biol Med. 2005;38:831–838. doi: 10.1016/j.freeradbiomed.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 15.Broniowska KA, Keszler A, Basu S, Shapiro DBK, Hogg N. Cytochrome c-mediated formation of S-nitrosothiol in cells. Biochem J. 2012;442:191–197. doi: 10.1042/BJ20111294. This reference along with reference 13 examines the reaction between glutathione, nitric oxide, and cytochrome c and provides evidence that depletion of cytochrome c from cells and cell lysates decreases GSNO production.
- 16.Shiva SS, Wang XX, Ringwood LAL, Xu XX, Yuditskaya SS, Annavajjhala VV, Miyajima HH, Hogg NN, Harris ZLZ, Gladwin MTM. Ceruloplasmin is a NO oxidase and nitrite synthase that determines endocrine NO homeostasis. Nat Chem Biol. 2006;2:486–493. doi: 10.1038/nchembio813. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell DA, Michel T, Marletta MA. Effects of S-nitrosation of nitric oxide synthase. Advances in Experimental Biology. 2007;1:151–179. 456. [Google Scholar]
- 18.Kornberg MD, Sen N, Hara MR, Juluri KR, Nguyen JVK, Snowman AM, Law L, Hester LD, Snyder SH. GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol. 2010;12:1094–1100. doi: 10.1038/ncb2114. This paper provides the first evidence for transnitrosation of nuclear proteins by _S_-nitrosated GAPDH. Prior to this study the mechanisms by which nuclear proteins are _S_-nitrosated was largely unknown.
- 19.Nakamura T, Wang L, Wong CCL, Scott FL, Eckelman BP, Han X, Tzitzilonis C, Meng F, Gu Z, Holland EA, et al. Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol Cell. 2010;39:184–195. doi: 10.1016/j.molcel.2010.07.002. This study shows that caspase 3 _S_-nitrosates X-linked inhibitor of apoptosis (XIAP) through a protein-protein interaction driven transnitrosation reaction. This is the first example of XIAP participating in transnitrosation.
- 20.Kozhukh J, Lippard SJ. Zinc Thiolate Reactivity toward Nitrogen Oxides: Insights into the Interaction of Zn(2+) with S-Nitrosothiols and Implications for Nitric Oxide Synthase. Inorg. Chem. 2012 doi: 10.1021/ic3007684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varonka MS, Warren TH. S-Nitrosothiol and Nitric Oxide Reactivity at Zinc Thiolates. Inorg. Chem. 2009;48:5605–5607. doi: 10.1021/ic900664r. [DOI] [PubMed] [Google Scholar]
- 22.Houk KN, Hietbrink BN, Bartberger MD, McCarren PR, Choi BY, Voyksner RD, Stamler JS, Toone EJ. Nitroxyl Disulfides, Novel Intermediates in Transnitrosation Reactions. J Am Chem Soc. 2003;125:6972–6976. doi: 10.1021/ja029655l. [DOI] [PubMed] [Google Scholar]
- 23.Perissinotti LL, Turjanski AG, Estrin DA, Doctorovich F. Transnitrosation of Nitrosothiols: Characterization of an Elusive Intermediate. J Am Chem Soc. 2005;127:486–487. doi: 10.1021/ja044056v. [DOI] [PubMed] [Google Scholar]
- 24.Lai C-H, Chou P-T. The theoretical comparison between two model NO carriers, MeSNO and MeSeNO. J Mol Model. 2008;14:1–9. doi: 10.1007/s00894-007-0246-z. [DOI] [PubMed] [Google Scholar]
- 25.Wu C, Liu T, Chen W, Oka S-I, Fu C, Jain MR, Parrott AM, Baykal AT, Sadoshima J, Li H. Redox regulatory mechanism of transnitrosylation by thioredoxin. Mol. Cell Proteomics. 2010;9:2262–2275. doi: 10.1074/mcp.M110.000034. This paper along with reference 45 indicates that the cysteine selectivity of thioredoxin 1 (Trx1) transnitrosation by GSNO is controlled by the redox state of the Trx1 active site. The authors then performed the first proteomics study to identify selective protein targets of Trx1 transnitrosation.
- 26.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci USA. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greco TM, Hodara R, Parastatidis I, Heijnen HFG, Dennehy MK, Liebler DC, Ischiropoulos H. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc Natl Acad Sci USA. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doulias P-T, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H. Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc Natl Acad Sci USA. 2010;107:16958–16963. doi: 10.1073/pnas.1008036107. The authors use a novel organomercury resin proteomic approach to identify endogenously _S_-nitrosated proteins. Analysis of the _S_-nitrosation sites suggested that protein-protein interaction driven transnitrosation reactions largely dictate the specificity of endogenous _S_-nitrosation.
- 29.Marino SM, Gladyshev VN. Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of trans-nitrosylation. J Mol Biol. 2010;395:844–859. doi: 10.1016/j.jmb.2009.10.042. In this study the authors analyze a manually curated data set of known _S_-nitrosation sites. They propose that charged residues within 8 Å of the _S_-nitrosation site are engaged in protein-protein interaction driven transnitrosation reactions, which explains the observation that cysteine reactivity does not correlate with sites of _S_-nitrosation.
- 30.Foster MW, Liu L, Zeng M, Hess DT, Stamler JS. A Genetic Analysis of Nitrosative Stress. Biochemistry. 2009;48:792–799. doi: 10.1021/bi801813n. [DOI] [PubMed] [Google Scholar]
- 31.Li Y-X, Shao Y-H, Jing L, Deng N-Y. An efficient support vector machine approach for identifying protein S-nitrosylation sites. Protein Pept. Lett. 2011;18:573–587. doi: 10.2174/092986611795222731. [DOI] [PubMed] [Google Scholar]
- 32.Xue Y, Liu Z, Gao X, Jin C, Wen L, Yao X, Ren J. GPS-SNO: Computational Prediction of Protein S-Nitrosylation Sites with a Modified GPS Algorithm. PLoS ONE. 2010;5:e11290. doi: 10.1371/journal.pone.0011290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee T-Y, Chen Y-J, Lu T-C, Huang H-D, Chen Y-J. SNOSite: Exploiting Maximal Dependence Decomposition to Identify Cysteine S-Nitrosylation with Substrate Site Specificity. PLoS ONE. 2011;6:e21849. doi: 10.1371/journal.pone.0021849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee T-Y, Chen Y-J, Lu C-T, Ching W-C, Teng Y-C, Huang H-D, Chen Y-J. dbSNO: a database of cysteine S-Nitrosylation. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts436. [DOI] [PubMed] [Google Scholar]
- 35.Dunn J, Gutbrod S, Webb A, Pak A, Jandu SK, Bhunia A, Berkowitz DE, Santhanam L. S-nitrosation of arginase 1 requires direct interaction with inducible nitric oxide synthase. Mol. Cell. Biochem. 2011;355:83–89. doi: 10.1007/s11010-011-0841-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu L, Han C, Lim K, Wu T. Activation of cytosolic phospholipase A2alpha through nitric oxide-induced S-nitrosylation. Involvement of inducible nitric-oxide synthase and cyclooxygenase-2. J Biol Chem. 2008;283:3077–3087. doi: 10.1074/jbc.M705709200. [DOI] [PubMed] [Google Scholar]
- 37.Straub AC, Billaud M, Johnstone SR, Best AK, Yemen S, Dwyer ST, Looft-Wilson R, Lysiak JJ, Gaston B, Palmer L, et al. Compartmentalized Connexin 43 S-Nitrosylation/Denitrosylation Regulates Heterocellular Communication in the Vessel Wall. Arterioscler Thromb Vasc Biol. 2011;31:399–U353. doi: 10.1161/ATVBAHA.110.215939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwakiri Y. S-nitrosylation of proteins: A new insight into endothelial cell function regulated by eNOS-derived NO. Nitric Oxide. 2011;25:95–101. doi: 10.1016/j.niox.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenfeld RJ, Bonaventura J, Szymczyna BR, Maccoss MJ, Arvai AS, Yates JR, Tainer JA, Getzoff ED. Nitric-oxide Synthase Forms N-NO-pterin and S-NO-Cys: Implications for activity, allostery, and regulation. J Biol Chem. 2010;285:31581–31589. doi: 10.1074/jbc.M109.072496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith BC, Fernhoff NB, Marletta MA. Mechanism and Kinetics of Inducible Nitric Oxide Synthase Auto- S-nitrosation and Inactivation. Biochemistry. 2012;51:1028–1040. doi: 10.1021/bi201818c. In this study a detailed kinetic model of iNOS auto-_S_-nitrosation was developed. The results suggested that a portion of the NO synthesized at the heme cofactor reacts with the iNOS Zn2+-tetrathiolate without being released into solution, potentially utilizing a protein tunnel. The results were also consistent with a role for NOS _S_-nitrosation in the physiological generation of nitrosothiols.
- 41.Mitchell DA, Marletta MA. Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nat Chem Biol. 2005;1:154–158. doi: 10.1038/nchembio720. [DOI] [PubMed] [Google Scholar]
- 42.Mitchell DA, Morton SU, Fernhoff NB, Marletta MA. Thioredoxin is required for S-nitrosation of procaspase-3 and the inhibition of apoptosis in Jurkat cells. Proc Natl Acad Sci USA. 2007;104:11609–11614. doi: 10.1073/pnas.0704898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of GAPDH: Views from different subcellular compartments. Cellular Signalling. 2011;23:317–323. doi: 10.1016/j.cellsig.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jia J, Arif A, Willard B, Smith JD, Stuehr DJ, Hazen SL, Fox PL. Protection of Extraribosomal RPL13a by GAPDH and Dysregulation by S-Nitrosylation. Mol Cell. 2012;47:656–663. doi: 10.1016/j.molcel.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paige JS, Xu G, Stancevic B, Jaffrey SR. Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chemistry & Biology. 2008;15:1307–1316. doi: 10.1016/j.chembiol.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakravarti R, Aulak KS, Fox PL, Stuehr DJ. GAPDH regulates cellular heme insertion into inducible nitric oxide synthase. Proc Natl Acad Sci USA. 2010;107:18004–18009. doi: 10.1073/pnas.1008133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barglow KT, Knutson CG, Wishnok JS, Tannenbaum SR, Marletta MA. Site-specific and redox-controlled S-nitrosation of thioredoxin. Proc Natl Acad Sci USA. 2011;108:E600–E606. doi: 10.1073/pnas.1110736108. This study along with reference 25 indicates that the cysteine selectivity of thioredoxin 1 (Trx1) transnitrosation by GSNO is controlled by the redox state of the Trx1 active site. The development and utility of a novel mass spectrometry technique to directly measure the kinetics of protein _S_-nitrosation is also described.
- 48.Benhar M, Forrester MT, Stamler JS. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat Rev Mol Cell Biol. 2009;10:721–732. doi: 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- 49.Hedberg J, Griffiths W, Nilsson S, Hoog J. Reduction of S-nitrosoglutathione by human alcohol dehydrogenase 3 is an irreversible reaction as analysed by electrospray mass spectrometry. Eur J Biochem. 2003;270:1249–1256. doi: 10.1046/j.1432-1033.2003.03486.x. [DOI] [PubMed] [Google Scholar]
- 50.Staab CA, Alander J, Brandt M, Lengqvist J, Morgenstern R, RCG m, J-OHÃ g. Reduction of S-nitrosoglutathione by alcohol dehydrogenase 3 is facilitated by substrate alcohols via direct cofactor recycling and leads to GSH-controlled formation of glutathione transferase inhibitors. Biochem J. 2008;413:493–504. doi: 10.1042/BJ20071666. [DOI] [PubMed] [Google Scholar]
- 51.Jensen DE, Belka GK, Bois Du G. S-Nitrosoglutathione is a substrate for rat alcohol dehydrogenase class III isoenzyme. Biochemical Journal. 1998;331:659. doi: 10.1042/bj3310659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 53.Sanghani PC, Davis WI, Fears SL, Green S-L, Zhai L, Tang Y, Martin E, Bryan NS, Sanghani SP. Kinetic and cellular characterization of novel inhibitors of s-nitrosoglutathione reductase. J Biol Chem. 2009;284:24354–24362. doi: 10.1074/jbc.M109.019919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun X, Wasley JWF, Qiu J, Blonder JP, Stout AM, Green LS, Strong SA, Colagiovanni DB, Richards JP, Mutka SC, et al. Discovery of S-Nitrosoglutathione Reductase Inhibitors: Potential Agents for the Treatment of Asthma and Other Inflammatory Diseases. ACS Med. Chem. Lett. 2011;2:402–406. doi: 10.1021/ml200045s. This study describes the initial disclosure of potent GSNOR inhibitors, which show promise in the treatment of asthma and other inflammatory diseases. The lead compound, N6022, is currently in phase II clinical trials for the treatment of asthma.
- 55.Bateman RL, Rauh D, Tavshanjian B, Shokat KM. Human Carbonyl Reductase 1 Is an S-Nitrosoglutathione Reductase. Journal of Biological Chemistry. 2008;283:35756–35762. doi: 10.1074/jbc.M807125200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Staab CA, Hartmanova T, El-Hawari Y, Ebert B, Kisiela M, Wsol V, Martin H-J, Maser E. Studies on reduction of S-nitrosoglutathione by human carbonyl reductases 1 and 3. Chem-Biol Interact. 2011;191:95–103. doi: 10.1016/j.cbi.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 57.Lopez-Sanchez LM, Corrales FJ, Lopez-Pedrera C, Aranda E, Rodriguez-Ariza A. Pharmacological Impairment of S-Nitrosoglutathione or Thioredoxin Reductases Augments Protein S-Nitrosation in Human Hepatocarcinoma Cells. Anticancer Res. 2010;30:415–421. [PubMed] [Google Scholar]
- 58.Stoyanovsky DA, Tyurina YY, Tyurin VA, Anand D, Mandavia DN, Gius D, Ivanova J, Pitt B, Billiar TR, Kagan VE. Thioredoxin and Lipoic Acid Catalyze the Denitrosation of Low Molecular Weight and Protein S-Nitrosothiols. J Am Chem Soc. 2005;127:15815–15823. doi: 10.1021/ja0529135. [DOI] [PubMed] [Google Scholar]
- 59.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sengupta R, Billiar TR, Atkins JL, Kagan VE, Stoyanovsky DA. Nitric oxide and dihydrolipoic acid modulate the activity of caspase 3 in HepG2 cells. FEBS Lett. 2009;583:3525–3530. doi: 10.1016/j.febslet.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sengupta R, Billiar TR, Kagan VE, Stoyanovsky DA. Nitric oxide and thioredoxin type 1 modulate the activity of caspase 8 in HepG2 cells. Biochem Biophys Res Commun. 2010;391:1127–1130. doi: 10.1016/j.bbrc.2009.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ito T, Yamakuchi M, Lowenstein CJ. Thioredoxin Increases Exocytosis by Denitrosylating N-Ethylmaleimide-sensitive Factor. Journal of Biological Chemistry. 2011;286:11179–11184. doi: 10.1074/jbc.M110.201780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benhar M, Thompson JW, Moseley MA, Stamler JS. Identification of S-nitrosylated targets of thioredoxin using a quantitative proteomic approach. Biochemistry. 2010;49:6963–6969. doi: 10.1021/bi100619k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tello D, Tarín C, Ahicart P, Bretón-Romero R, Lamas S, Martínez-Ruiz A. A “fluorescence switch” technique increases the sensitivity of proteomic detection and identification of S-nitrosylated proteins. Proteomics. 2009;9:5359–5370. doi: 10.1002/pmic.200900070. [DOI] [PubMed] [Google Scholar]
- 65.Sengupta RR, Ryter SWS, Zuckerbraun BSB, Tzeng EE, Billiar TRT, Stoyanovsky DAD. Thioredoxin catalyzes the denitrosation of low-molecular mass and protein S-nitrosothiols. Biochemistry. 2007;46:8472–8483. doi: 10.1021/bi700449x. [DOI] [PubMed] [Google Scholar]