Single-Neuron Sequencing Analysis of L1 Retrotransposition and Somatic Mutation in the Human Brain (original) (raw)
. Author manuscript; available in PMC: 2013 Apr 26.
Summary
A major unanswered question in neuroscience is whether there exists genomic variability between individual neurons of the brain, contributing to functional diversity or to an unexplained burden of neurological disease. To address this question, we developed a method to amplify genomes of single neurons from human brains. Since recent reports suggest frequent LINE-1 (L1) retrotransposition in human brains, we performed genome-wide L1 insertion profiling of 300 single neurons from cerebral cortex and caudate nucleus of 3 normal individuals, recovering >80% of germline insertions from single neurons. While we find somatic L1 insertions, we estimate <0.6 unique somatic insertions per neuron and most neurons lack detectable somatic insertions, suggesting that L1 is not a major generator of neuronal diversity in cortex and caudate. We then genotyped single cortical cells to characterize the mosaicism of a somatic AKT3 mutation identified in a child with hemimegalencephaly. Single-neuron sequencing allows systematic assessment of genomic diversity in the human brain.
Introduction
It is unlikely that the genomes of any two cells in the body are identical, due to somatic mutations during replication and other mutagenic forces (Frumkin et al., 2005). The complexity and diversity of neuronal cell types in the brain has also led to suggestions that a somatic mutational mechanism may have been harnessed evolutionarily to diversify neuronal function (Muotri and Gage, 2006; Rehen et al., 2005). Endogenous retrotransposition of LINE-1 elements has been proposed as one potential mechanism generating neuronal genome diversity (Singer et al., 2010). Human-specific LINE-1 (L1Hs) retrotransposons comprise the only known active autonomous transposon family in humans, with ~80–100 active L1Hs elements per individual (Hancks and Kazazian, 2012), and somatic L1Hs insertions have been found both in cancerous and normal cells (Iskow et al., 2010; Lee et al., 2012a; Miki et al., 1992; van den Hurk et al., 2007). Recent studies observed rare retrotransposition of an L1Hs reporter in rodent brain in vivo (Muotri et al., 2005; Muotri et al., 2010) and human neural progenitors in vitro (Coufal et al., 2009), while other studies found evidence for more widespread somatic L1Hs insertions in the human brain by qPCR (Coufal et al., 2009) and bulk DNA sequencing (Baillie et al., 2011). qPCR estimates of these events in human brain approach 80 somatic insertions per cell (Coufal et al., 2009).
Although L1 retrotransposition and other somatic mutations could contribute to functional genomic diversity, they can also cause disease (Erickson, 2010; Hancks and Kazazian, 2012). Therefore, any potential somatic mutational mechanism must be balanced by the need for genome stability. Somatic mutations cause not only cancers but also several malformations of the brain (Gleeson et al., 2000; Riviere et al., 2012), emphasized by the recent identification of somatic mutations affecting genes of the PI3K-AKT3-mTOR pathway in hemimegalencephaly (HMG) (Lee et al., 2012b; Poduri et al., 2012), a severe epileptic brain malformation. However, the rates and types of somatic mutations occurring during normal brain development, and how much of the unexplained burden of neurogenetic disease may be caused by somatic mutations, are completely unknown (Erickson, 2010).
Systematically studying somatic mutations requires sequencing genomes of single cells (Kalisky et al., 2011), since the signals of somatic mutations present in a minority of cells can be missed due to sequencing error or insufficient sequencing depth. Single-cell sequencing overcomes this limitation, as shown by studies of single human cancer cells and single sperm that have yielded important new insight into tumor evolution and genetic heterogeneity (Hou et al., 2012; Navin et al., 2011; Wang et al., 2012; Xu et al., 2012). However, similar technologies have yet to be applied to the study of somatic mutation in normal human tissues such as brain, or to diseases other than cancer.
Here we describe a method to amplify genomes of single neurons from post-mortem and surgically resected human brain, enabling interrogation of a wide-range of somatic mutations by high-throughput sequencing. We performed genome-wide L1Hs insertion profiling of 300 single neurons from cerebral cortex and caudate nucleus of three neurologically normal individuals, and confirmed that somatic L1Hs retrotransposon insertions are present in the normal human brain. Our quantitative analysis of >200,000 L1Hs insertion sites in these 300 single neurons suggests a rate not higher than 0.6 unique somatic insertions per neuron, and possibly as low as 0.04 (1 insertion in 25 neurons), consistent with observed in vitro rates for human neural progenitors but substantially less than previous qPCR-based estimates for human brain (Coufal et al., 2009). We then sequenced single cells from HMG brain tissue harboring a known somatic AKT3 point mutation (c.49G→A; p.E17K) (Poduri et al., 2012), showing that our method can characterize the mosaicism of pathogenic somatic brain mutations. These single-cell studies provide a foundation for studying genomic variability among cells in the human brain, both in normal development and neurologic disease.
Results
High-throughput isolation and amplification of single neuronal genomes from human brains
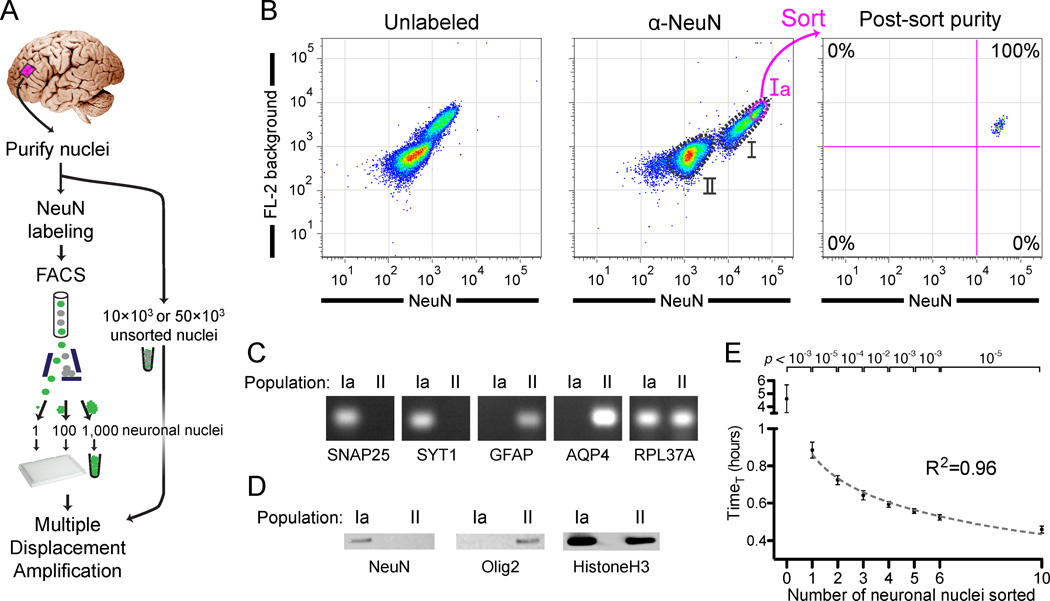
We purified nuclei from post-mortem human frontal cortex and caudate and labeled them with a neuron-specific antibody (NeuN) for sorting using fluorescence-activated cell sorting (FACS) (Figure 1A) (Matevossian and Akbarian, 2008; Spalding et al., 2005). Large nuclei with neuronal nuclear morphology (Parent and Carpenter, 1996) were readily apparent by microscopy (Figure S1A). NeuN immunoreactivity (Figure S1B) (Mullen et al., 1992) labels essentially all neuronal nuclei in cortex and caudate (Wolf et al., 1996), corresponding to 25–35% of all nuclei (population I) (Figures 1B and S1C). Consistent with their increased size on microscopy (Figure S1B), NeuN+ nuclei also had larger forward (FSC) and side (SSC) scatter (correlates of size) by flow cytometry compared to NeuN− nuclei (Figure S1D). Whereas for nuclei isolated from the caudate we performed a simple sort of the NeuN+ population (population I, Figure S1C), we further enriched nuclei from the cortex for pyramidal neuronal nuclei. Since neighboring cortical pyramidal neurons tend to have shared clonal origins due to their primarily radial migration (Magavi et al., 2012), enriching for pyramidal neuronal nuclei increases the chance of identifying clonal somatic mutations shared by multiple neurons. The largest neuronal nuclei in cortex correspond primarily to pyramidal projection neurons (Gittins and Harrison, 2004; Mills, 2007), and indeed their nuclei often show a pyramidal shape (Figure S1A). We therefore sorted cortical nuclei within the top 25% NeuN/FL-2 fluorescence of population I (population Ia), which were the largest nuclei in population I (Figure S1D). We confirmed the neuronal and non-neuronal identities of the sorted populations by RT-PCR and western blot analysis of additional neuronal (SNAP25 and SYT1) and non-neuronal (GFAP, AQP4, and Olig2) markers (Figures 1C and 1D). For every sort, a portion of the sorted nuclei was reanalyzed by FACS, confirming that nuclei remained intact during sorting and that sort purity was >98% (Figures 1B and S1C).
Figure 1. Isolation and genome amplification of single human neuronal nuclei.
(A) Schematic of the method.
(B) Fluorescence-activated cell sorting of cortical nuclei stained with NeuN shows two separable populations: NeuN+ (population I) and NeuN− (population II). A subset of population I (Ia) consisting of large neuronal nuclei was sorted and reanalyzed, confirming sort purity. Two populations of nuclei are sometimes apparent without NeuN staining, due to the increased background staining of the larger population I nuclei. Fluorescence decrease of the sorted population on reanalysis is always observed due to photobleaching and washing of non-specific staining in the first sort.
(C) RT-PCR confirming the neuronal and non-neuronal identities of populations Ia and II, respectively, by assaying for expression of nuclear RNA for two neuronal (SNAP25 and SYT1), two astroglial (GFAP and AQP4), and input control (RPL37A) genes. RT-PCR and western blot experiments (Figures 1C and 1D) were performed with NeuN/Mef2c double labeling in which all NeuN+ nuclei were Mef2c+ (data not shown).
(D) Western blot analysis of NeuN and Olig2 (an oligodendrocyte marker), confirming neuronal and non-neuronal identity, respectively, of populations Ia and II.
(E) Quantitative MDA reactions monitored in real-time confirm accurate sorting of the desired number of nuclei. The time to amplify to a threshold above background (TimeT, analogous to qPCR CT value) is plotted on the y-axis (error bars ±1SD, n=7 or 8 reactions per condition). Points were fit to a semi-log line of slope −4.3, corresponding to 1.7-fold amplification per unit time.
See also Figure S1.
We used multiple displacement amplification (MDA) (Dean et al., 2002) for whole genome amplification of single nuclei because it produces large yields of high molecular weight amplicons, most of which are >30kb (Hou et al., 2012 and data not shown), allowing study of both single-nucleotide mutations and ~6kb full-length L1Hs insertions. We optimized MDA reactions for increased yield (Figure S1E), producing 15–20µg of amplified DNA from single cells. We also measured exogenous (non-human) DNA contamination in the reagents of the MDA reaction (Blainey and Quake, 2011), finding negligible (< 1fg) exogenous DNA (Figures S1F and S1G). Additional controls (see following section) excluded operator human DNA contamination. Quantitative MDA (qMDA) reactions (Zhang et al., 2006) further showed that, as the number of nuclei sorted in a well increased, the time-to-threshold-amplification decreased in a step-wise manner (p <0.01 for each additional nucleus) (Figure 1E), confirming that the desired number of nuclei was correctly sorted in each well. We concluded that our procedure can sort and amplify single neuronal genomes from human brains with high purity and in a high-throughput manner.
Genome-wide coverage and amplification dropout rates of single neuronal genomes
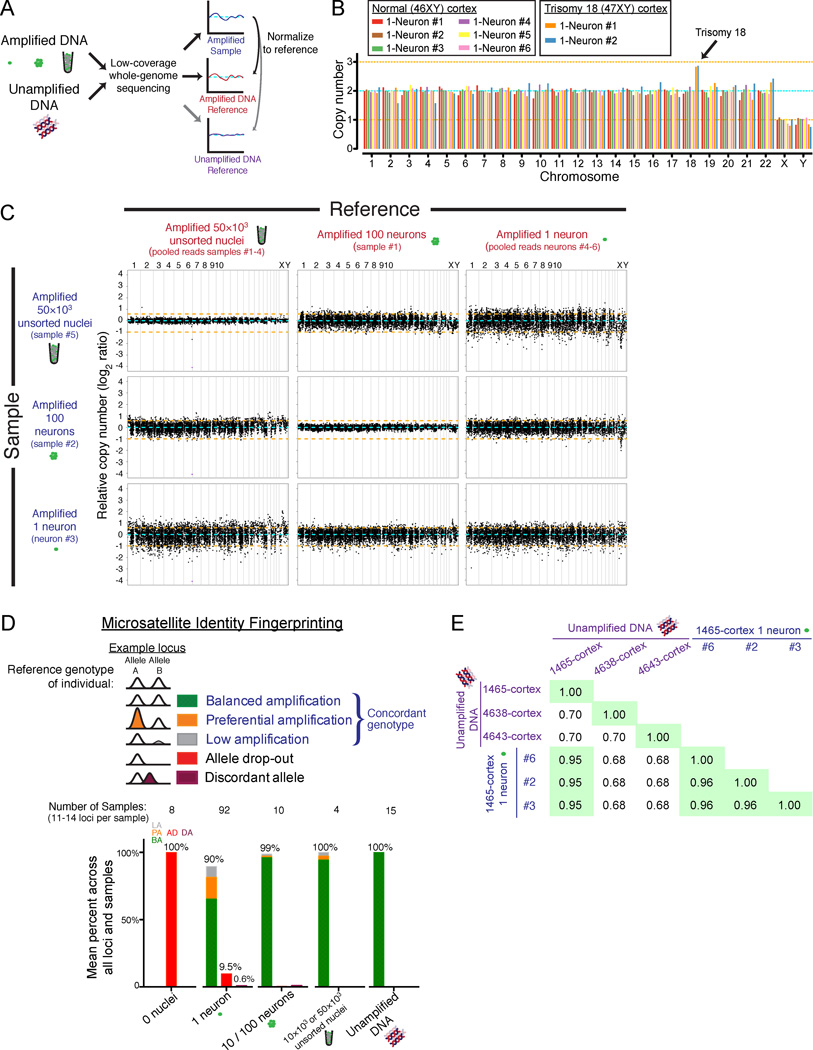
We next evaluated the genome-wide coverage and reproducibility of our single neuronal genome amplification. In an initial 4-locus multiplex PCR quality control, 97% of sorted single neurons amplified at least 3 of the 4 loci, indicating that their genomes were successfully amplified and suitable for further experiments. We then performed low-coverage whole-genome sequencing (Figure 2A) of eight randomly chosen single neurons (0.35× average coverage), six from a normal individual (46XY) and two from a trisomy 18 individual, as well as unamplified and MDA-amplified bulk reference samples. The two neurons from the trisomy 18 individual showed the expected increase in chromosome 18 copy number, and the six single neurons from the normal individual were all euploid, confirming that intact nuclei were sorted and that all chromosomes were amplified (Figure 2B). Counting sequencing reads across the genome in bins ~500kb in size (Navin et al., 2011) revealed a systematic, regional amplification bias for all MDA samples, compared to unamplified bulk DNA, regardless of the number of nuclei amplified (Figure S2A). This regional bias in MDA amplification could be controlled for using any of the MDA samples as a reference (Figure 2C), indicating that most of the regional variability in amplification is inherent to MDA rather than the number of nuclei amplified. Bias in amplification relative to GC content was also similar for all MDA samples types (Figure S2B).
Figure 2. Single-neuron genome-wide coverage, amplification bias, and identity fingerprinting.
(A) Schematic of the low-coverage genome sequencing method.
(B) Chromosome copy numbers of single cortical neurons from normal (UMB1465, 46XY) and trisomy 18 (UMB866, 47XY,+18) individuals. Copy numbers are normalized to the median copy number of each chromosome across the 8 single neurons, with autosomes adjusted to a median copy number of 2. Orange lines denote ±1 copy.
(C) Higher-resolution copy number profiling in 6,000 equal-read bins of ~500kb in size shows that MDA bias can be corrected by normalization to an MDA-amplified reference. Orange lines denote ±1 copy, and purple points indicate off-scale bins.
(D) Identifiler fingerprinting confirms the single neurons derive from the correct individuals, and measures allele preferential amplification (PA), low amplification (LA), allele dropout (AD), and discordant allele (DA) rates.
(E) Fraction of genotypes by SNP microarray that are concordant between 3 single neurons and bulk DNA confirms the single neurons derive from the correct individual.
See also Figure S2 and Table S1.
In order to use single-neuron sequencing for somatic mutation detection, amplified genomes must reflect the diploid genotype (both alleles) of genomic loci. We therefore quantified the fraction of genomic loci that failed to amplify one (allelic dropout, AD) or both alleles (locus dropout, LD). Loss of one allele, AD, was measured with a panel of 16 polymorphic microsatellite markers (Identifiler fingerprinting) and by SNP microarray genotyping. AD measured by Identifiler of 92 single neurons across 1,183 heterozygous loci was 9.5% (Figure 2D), whereas AD measured by SNP microarray (for >60,000 loci that are heterozygous in the bulk DNA and called with high confidence in both the reference and sample) was 8–9% in 3 single neurons (Figure S2C and Table S1A), consistent with previous estimates (Hou et al., 2012). Some dropout tended to recur at specific loci even in MDA-amplified 100- and 1000-neuron samples (Figure S2D), probably reflecting difficulty of MDA to amplify specific loci. Loss of both alleles, LD (locus dropout), was 2.3% in the 92 single neurons assayed by Identifiler. In addition, LD was separately estimated by counting the percentage of low-coverage sequencing bins with less than 1/16 the copy number relative to an unamplified DNA reference, and was 2.0% for 1-neuron samples (Figure S2E). These low rates of AD (~10%) and LD (~2%) demonstrate comprehensive and reproducible amplification of single neuronal genomes, and suggest that genome-wide profiling of L1 insertions in single neurons could capture up to 90% of retrotransposon insertions per cell. These genotyping controls also excluded operator contamination, since all amplified single neuronal genomes tested were concordant with the bulk reference (Figures 2D, 2E and Tables S1B–C).
Genome-wide L1Hs profiling in single neurons
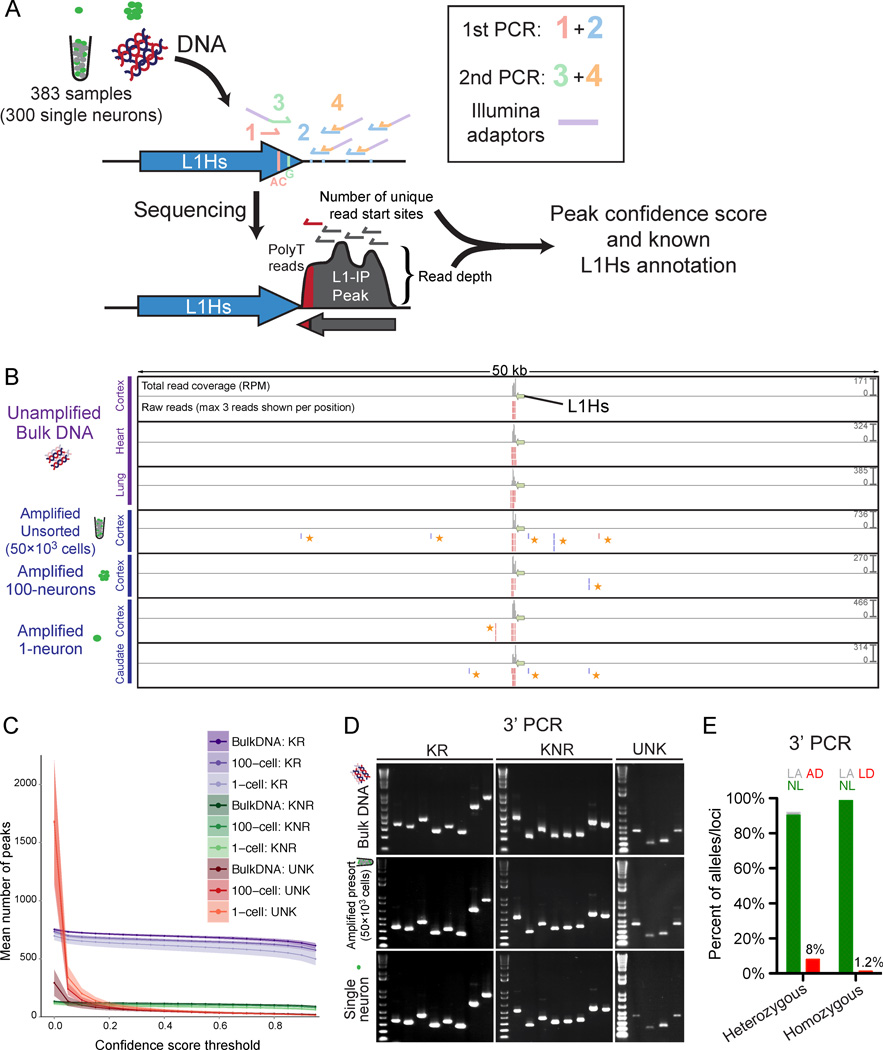
We performed genome-wide L1Hs insertion profiling (L1-IP) of single neurons by adapting the method of Ewing and Kazazian (2010) for high-throughput multiplexed sequencing. All known active and disease-causing L1Hs sub-families possess two sequences diagnostic of L1Hs (Hancks and Kazazian, 2012; Ovchinnikov et al., 2002), and a comprehensive study of somatic insertions in the setting of cancer found that 110/111 somatic insertions (with evidence of a target site duplication and poly-A tail) contained both sequences (Lee et al., 2012a). L1-IP targets these L1Hs-specific sequences and amplifies genomic DNA flanking L1Hs insertions containing these diagnostic sequences (Figures 3A, 3B and S3A).
Figure 3. Genome-wide L1Hs insertion profiling (L1-IP) in single neurons.
(A) Schematic of the L1-IP method. Primers 1 and 3 (L1Hs-AC and ILMN-Adaptor1_L1Hs-G, respectively) are specific to L1Hs diagnostic nucleotides. Primer 2 represents 8 different 5bp arbitrary seed primers, each containing the same barcode. Primer 4 (ILMN-SeqAdaptor2) incorporates an Illumina adaptor. See Table S3 for primer sequences.
(B) L1-IP sequencing reads for one representative known reference insertion (L1Hs-KR-chr11_115209613). For each sample, a total read coverage track and a raw reads track are shown. Each read coverage track is scaled to the maximum peak height of the sample (scale on the right, in reads per million mapped reads, RPM). In the raw reads track, up to 3 reads are shown for each position. The green arrow marks the L1Hs insertion. Plus and minus strand reads are red and blue, respectively. Low-level MDA-chimera reads (yellow asterisks) are seen in the local region of the true insertion only in MDA-amplified samples.
(C) The number of peaks found above different confidence score thresholds corresponding to known reference insertions (KR), known non-reference insertions (KNR), and unknown peaks (UNK). Data shown is the mean for all bulk (n=31), 100-cell (n=15) and 1-cell (n=303) samples from all 3 individuals (includes 15, 5 and 3 technical replicates, respectively). Shading around each line shows ±SD. KR and KNR insertions used for peak annotation are in Table S5.
(D) Representative gel images of 3’ junction PCR (3’PCR) of 20 different germline insertions (8 KR, 8 KNR, and 4 UNK).
(E) 3’PCR quantification of AD and LD in 1-neuron samples (n=83), of 3 heterozygous and 3 homozygous L1Hs insertions. AD and LD are quantified for heterozygous and homozygous insertions, respectively. NL, normal amplification; LA, low amplification; AD, allelic-dropout; LD, locus-dropout.
See also Figures S3, S4, S5, and S6.
We profiled from each of 3 neurologically normal individuals: 50 single neurons from cerebral cortex and 50 from caudate nucleus (i.e. 300 MDA-amplified single neurons total), unamplified bulk DNA from 5–6 tissues (cortex, caudate, cerebellum, heart, liver, lung), MDA-amplified 50,000-cell, 10,000-cell, 1,000-cell, and 100-neuron samples, as well as technical replicates to assess reproducibility (Figures S3B and S3C), for a total of 383 samples (see Table S2 for sample details). A custom data analysis pipeline classified detected peaks as known reference insertions present in the human genome reference (KR), known non-reference insertions identified in previous studies (KNR), or unknown (UNK) candidate insertions, and assigned a confidence score ranging from 0 to 1 (low-quality to high-quality peaks) based on the number of reads and the number of unique read start sites per peak (Figure 3C). The confidence score was derived from a logistic regression model of germline insertions reproducibly found in bulk DNA samples of the individual (Figure S3D, and see Extended Experimental Procedures for details of the analysis pipeline).
MDA is known to produce rare, low-level chimeric sequences due to local, occasional mispriming of single-stranded amplicons to each other during amplification (Lasken and Stockwell, 2007). These chimeras were seen in MDA-amplified samples as an excess of background reads and peaks with low read depth, and one or few unique read start sites, in the local ~20kb flanks of some, though not all L1 insertions (Figures 3B and S4A–D). Since chimeras form at different sites in different MDA reactions, they are not recurrent between samples (Figures S5A and S5B), and cloning of chimeras (representative example in Figures S5A–C) confirmed their MDA-derived mechanism of formation. Their low confidence scores (Figure S4B) allowed most MDA-chimera peaks to be filtered with minimal reduction in sensitivity for bona fide insertions (Figure 3C).
We first assessed the sensitivity of L1-IP to detect L1Hs insertions genome-wide. In 1-neuron samples, the sensitivity of L1-IP for KR insertions (mostly homozygous) present in bulk DNA of the individual was 81±6% (SD), with a confidence score threshold of 0.5 (Figure S6A), and of 300 1-neuron samples in this study, only 4 were low quality outliers (Figure S6B). Sensitivity increased to 87% when relaxing the confidence threshold to 0.1, though at this lower confidence score, more insertions with weaker evidence supporting them were also detected. Since somatic insertions are expected to be present in a single copy, sensitivity for single copy insertions in 1-neuron samples was assessed with chrX KR/KNR insertions in individual 1465 (male) and was only slightly lower at 75±10%, with a confidence score threshold of 0.5. We further confirmed that we detect the expected absolute number of insertions: the mean number of KR, KNR and UNK insertions (with confidence score > 0.5) per bulk DNA sample was 689, 113, and 43, respectively (Figure S6C), compared to 628 KR and 152 KNR/UNK insertions found on average in a previous study (Ewing and Kazazian, 2010). 605, 87 and 47 KR, KNR, and UNK peaks were found on average in 1-neuron samples (Figure S6C). A plot of L1Hs peaks found in bulk DNA, a 100-neuron sample, and two representative single neurons is shown in Figure 4.
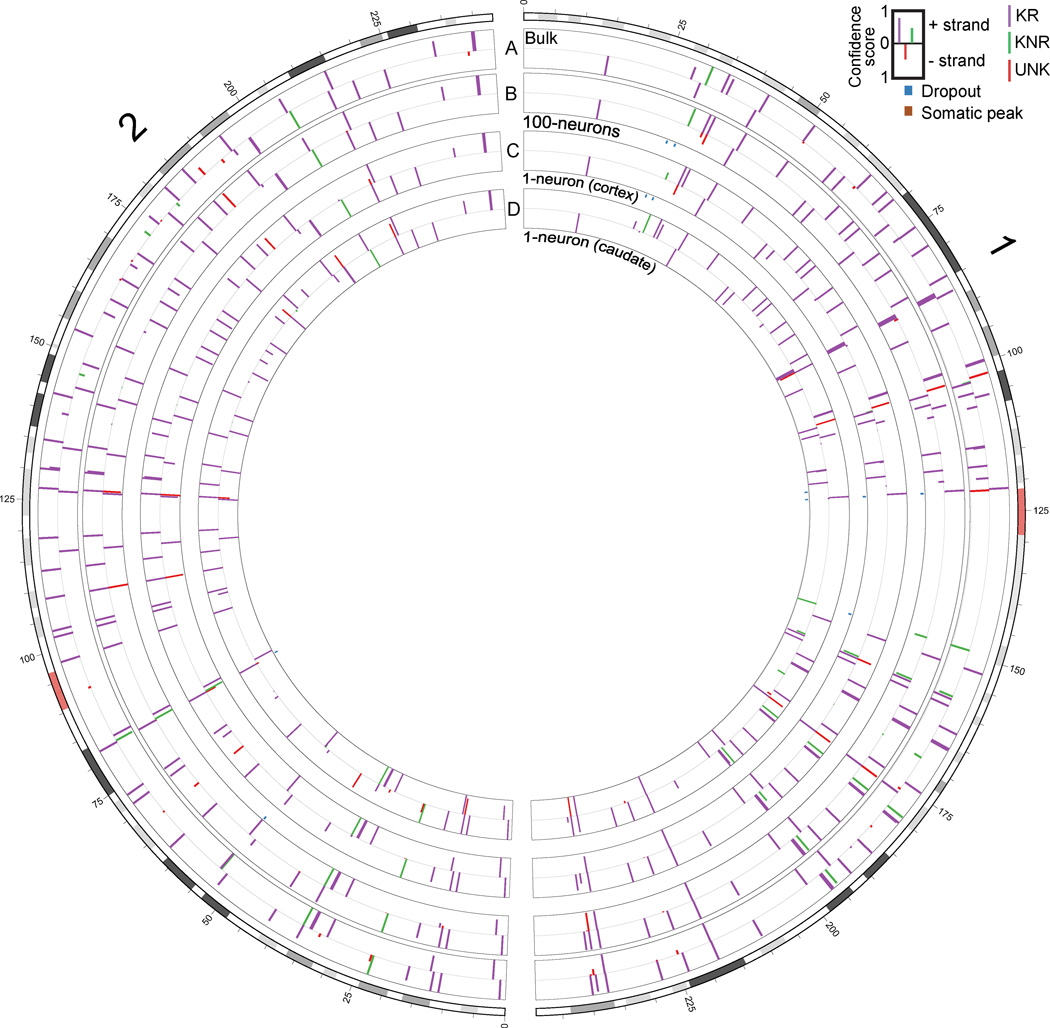
Figure 4. Chromosome L1-IP profile of single neurons.
Circos plot (Krzywinski et al., 2009) of chromosomes 1 and 2, from representative L1-IP samples from individual 1465: (A) bulk DNA, (B) cortex 100-neurons #1, (C) cortex 1-neuron #2, and (D) caudate 1-neuron #1. Peaks are shown for loci where at least one of the samples has a peak confidence score >0.5. Bulk DNA track shows the mean confidence score across all bulk DNA samples of individual 1465. KR, KNR, and UNK peaks are colored as indicated in the key. Below 100-neuron and 1-neuron sample tracks are annotations for peaks present with a score >0.5 in bulk DNA but absent in the sample (‘Dropout’), and peaks absent from bulk DNA but present in the sample with a score >0.5 and at least 20kb away from the nearest KR/KNR insertion in the individual to exclude MDA-chimera peaks (‘Somatic peak’). Figures for all chromosomes can be found in Supplemental Data 1.
In order to validate L1-IP predicted insertions, we optimized a 3’ junction PCR validation method (3’PCR) (Figure S6D), and further used it to directly measure allelic dropout (AD) and locus dropout (LD) of L1Hs insertions in amplified single neurons. The technical sensitivity of the 3’PCR validation method (i.e. 3’PCR detection rate of true germline insertions) was important to determine first, in order to estimate at what rate true insertions found by L1-IP fail to validate by 3’PCR. This was assayed by 3’PCR of 64 known germline insertions (33 KR and 31 KNR) in unamplified bulk DNA, and amplified unsorted-50k and 1-neuron samples. In 1-neuron samples, 3’PCR detected 94% of known germline insertions with the first primer attempted (the remainder were validated successfully with redesigned primers), and this detection rate was not significantly different between amplified and unamplified samples (Figures 3D and S6E). 3’PCR can therefore sensitively detect L1Hs insertions in amplified single neuronal genomes. 3’PCR also successfully validated, in both bulk and 1-neuron samples, 12 out of 12 unknown (UNK) germline candidate insertions that we tested (Figures 3D, S6E and Table S3), confirming that L1-IP can identify unknown germline insertions. AD of L1Hs insertions was then estimated by 3’PCR of 3 heterozygous insertions in a larger number of 83 single neurons (Figures 3E and S6F–G), finding 8.0% AD (20/249 alleles), consistent with previous estimates. LD estimated by 3’PCR of 3 homozygous insertions in the same cells (Figures 3E and S6G) was 1.2% (3/249 alleles). We concluded that L1-IP’s high sensitivity to detect germline insertions in single neurons, our robust 3’PCR validation method, and direct confirmation of <10% L1Hs allelic dropout, allows us to confidently search for somatic L1Hs insertions genome-wide in single neurons.
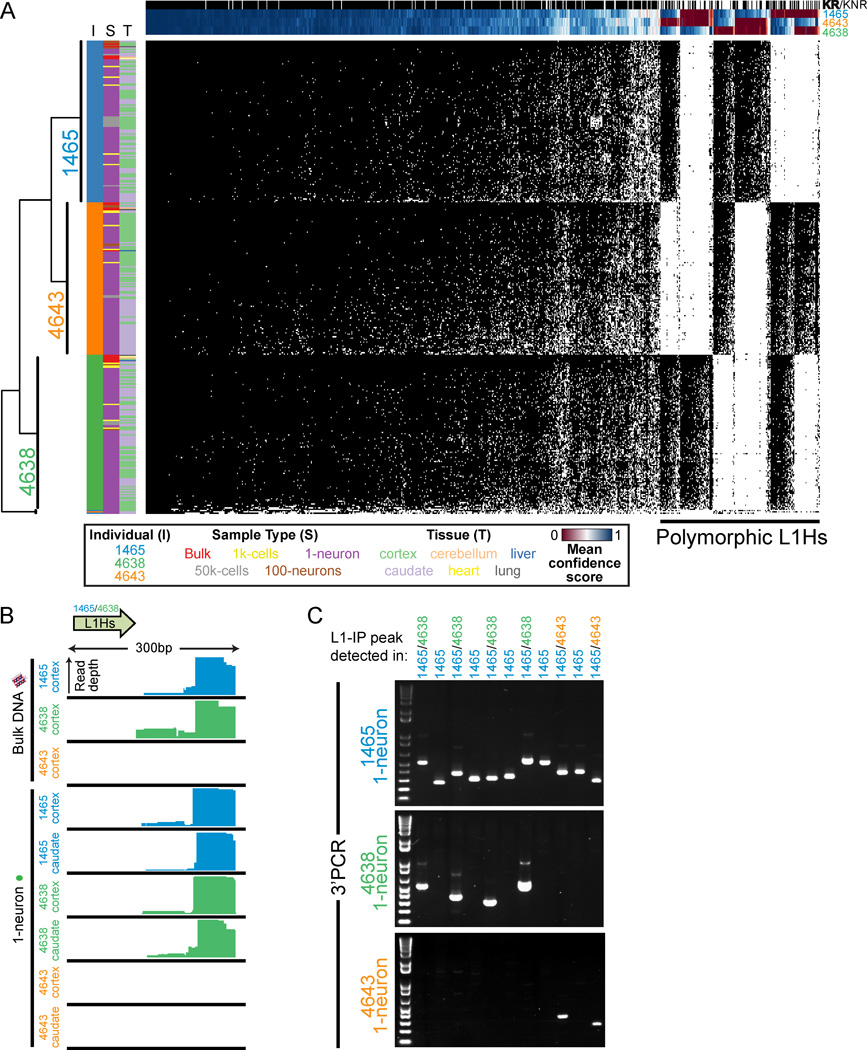
Identity fingerprinting of single neurons by L1Hs profile
L1-IP can reliably detect population-polymorphic L1Hs insertions in single neurons (Figures 5A–C), serving as a fingerprint for each individual. All possible permutations of insertion polymorphisms among the 3 individuals were found (every possible pair of individuals and individual-specific), and as expected, KR and KNR insertions were enriched in fixed and polymorphic insertions, respectively (Figure 5A). Hierarchical clustering of all samples in the study according to L1Hs genotype correctly clustered all samples by individual except for 3 low-quality 1-neuron samples (Figure 5A). Importantly, since both population-polymorphic and somatic insertions belong to the same L1Hs subfamilies and have the same L1Hs diagnostic nucleotides (Beck et al., 2010; Lee et al., 2012a), detection of population-polymorphic L1Hs insertions in single neuronal genomes further illustrates that L1-IP has the potential to capture somatic insertions.
Figure 5. Single-neuron fingerprinting with L1-IP.
(A) Unbiased hierarchical clustering of all samples sequenced in this study (excluding technical replicates) by transposon profile. Each row represents a sample, and each column represents a specific L1Hs insertion. Data is shown for all KR and KNR insertions with an average score of at least 0.5 in at least one individual’s samples. Black and white squares indicate presence or absence, respectively, of the insertion using a confidence score threshold of 0.5. All samples cluster correctly by individual except for 3 low-quality 1-neuron samples that cluster in a separate branch (bottom branch). Additional row annotations are colored for individual (I), sample type (S), and tissue (T), illustrating correct clustering by individual. Column annotations show annotation for KR (black) and KNR (white) insertions, and mean confidence scores across all samples of each individual. Samples also cluster by individual when including all insertions including unknown peaks (data not shown).
(B) L1-IP read coverage for a representative polymorphic known non-reference insertion (L1Hs-KNR-1158).
(C) Representative gel images of 3’PCR of 11 polymorphic germline insertions with 1-neuron DNA. 3’PCR products are only detected in individuals predicted by L1-IP to have the insertion. All polymorphic insertions tested are listed in Table S3.
Somatic L1Hs insertion rate in cortex and caudate neurons
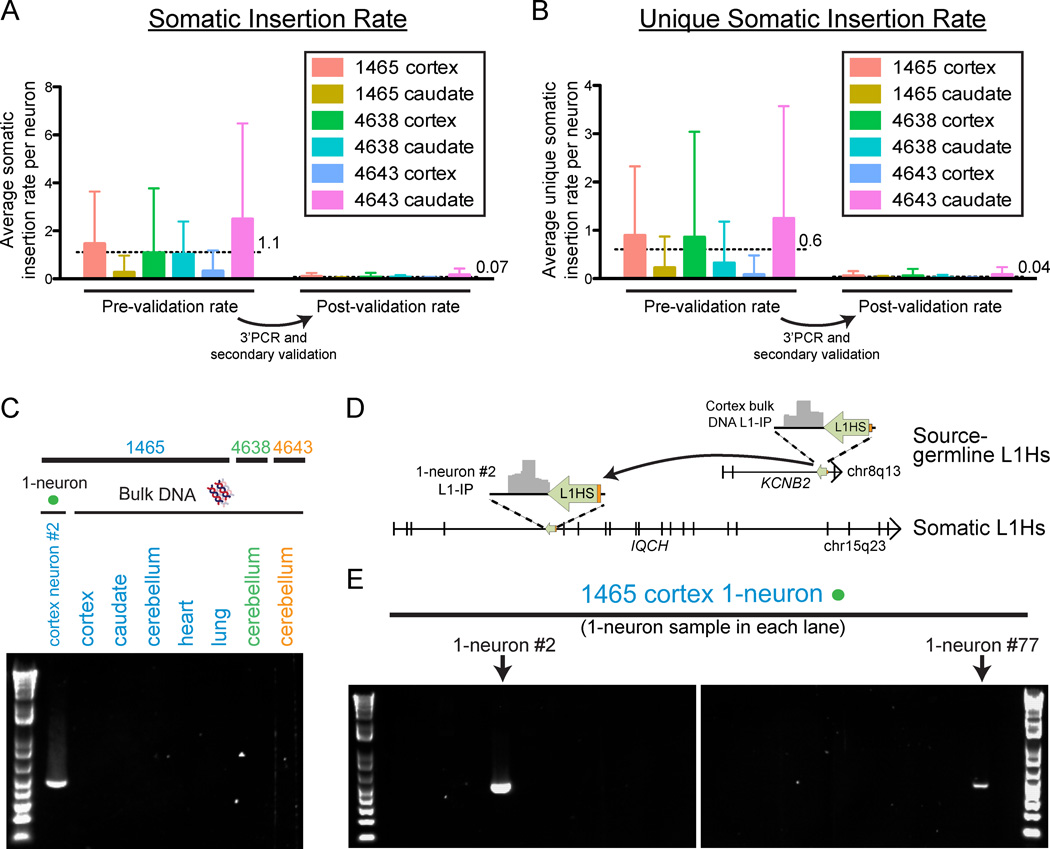
Our single-neuron L1-IP data allowed us to quantify the number of cortex- and caudate-specific somatic insertions in single-neuron samples and estimate an upper bound for the number of somatic L1Hs insertions per neuron (defined as absent from bulk DNA samples of the individual excluding the brain region being analyzed). Rather than using the same confidence score threshold across all samples, we adjusted the confidence score threshold for each single-neuron sample to maintain a constant sensitivity for KNR germline insertions. This controls for variability in single-neuron sample quality and allows for more accurate correction of insertion rates for sensitivity. A KNR reference was specifically chosen as it would be expected to better estimate sensitivity for single-copy somatic events than a mostly homozygous KR reference set. We excluded insertions found within 20kb of known (KR/KNR) insertions, leading to a minimal reduction in sensitivity (by excluding 1.5% of the genome, i.e. 45.5/3137Mb) with a substantial gain in specificity by filtering most, though not all, MDA chimera peaks (Figure S4A). At a sensitivity threshold that detects 50% of KNR insertions, we found an average of 1.1±2.3 (SD) somatic insertion candidates per neuron (corrected for sensitivity) (Figure 6A), and 68% of 1-neuron samples had no detectable somatic insertions. Additionally, we counted the number of unique somatic insertions per neuron (i.e. not present in other single neurons sequenced from the individual) and found 0.6±1.5 candidate unique insertions per neuron (Figure 6B); 82% of 1-neuron samples had no detectable unique somatic insertions.
Figure 6. Quantification of somatic L1Hs insertions, and validation of a somatic insertion, in single neurons.
(A) Mean number (±SD) of somatic insertion candidates per single neuron in each tissue in the study, corrected for sensitivity. The insertion rates per neuron are shown before and after 3’PCR and secondary validation. Horizontal dashed lines and adjacent numbers indicate the mean number of insertions across all single neurons from all tissues. Low-quality samples that did not achieve the necessary KNR detection rate with a confidence score >0.5 were excluded from the analysis in a quality control check (‘QC-fail’ in Table S2). The number of cells included in each analysis were n=50, 45, 45, 50, 50, and 44 for 1465-cortex, 1465-caudate, 4638-cortex, 4638-caudate, 4643-cortex, and 4643-caudate, respectively, after removing low-quality samples failing quality control.
(B) Mean number (±SD) of unique somatic insertion candidates (i.e. present in only one single neuron sample of the individual) per single neuron in each tissue, corrected for sensitivity.
(C) Gel images of 3’PCR validation of a somatic L1Hs insertion found by L1-IP in individual 1465 cortex 1-neuron #2 (L1-IP peak ID chr15_67625710_plus_0_0).
(D) Location of the somatic L1Hs insertion (L1-IP peak ID chr15_67625710_plus_0_0) in antisense orientation in intron 4 of the gene IQCH, and the corresponding L1-IP peak in 1465-cortex 1-neuron #2. The insertion’s target site duplication coordinates are chr15: 67,625,702–67,625,714 (hg19). A 5’ transduction (orange) identified the source L1Hs on chr8: 73,787,792–73,793,823.
(E) Representative gel images from a 3’PCR screen of 83 1-neuron samples from individual 1465 cortex (24 1-neuron samples shown) for the somatic insertion in Figures 6C and 6D. The two cortical 1-neuron samples (#2 and #77) found to have the insertion are shown. 1-neuron #77 was found to have the insertion only in the 3’PCR screen since it was not profiled by L1-IP. 3’PCR product sequencing and full-length cloning confirmed the insertion had identical 5’ and 3’ breakpoints and TSD in both neurons (#2 and #77).
See also Figure S7.
The above upper bound estimate for the somatic insertion rate controls for sensitivity (false negative rate), but is likely an overestimate as it does not take into account specificity (i.e. false positive MDA chimera and other artifactual peaks still remaining after our sensitivity threshold and local 20kb filtering). We therefore screened for false positive candidates by carrying out 3’PCR validation and secondary validations of the 16 highest-scoring candidate somatic insertions from each tissue (96 total). Initial review of L1-IP raw data revealed that at least half of the candidates were likely MDA-chimeras or other recognizable technical artifacts that cannot be systematically filtered. These include peaks caused by read alignment errors, chimeras of older L1Pa insertions, and loci with systematic low-level reads present at sub-threshold levels in many unamplified bulk and MDA-amplified samples of unrelated individuals, but stochastically passing threshold as somatic candidates in one or a few single neuron samples (see Table S3 for annotation of the 96 candidates). Indeed, only 17 of the 81 candidates (21%) for which we could design primers passed 3’PCR validation (Figure S7A), significantly less than the 94% validation rate for known insertions (Figure S6E). Secondary validation sequencing of 3’PCR products and review of L1-IP raw data revealed that 12 of the remaining 17 candidates were chimeras or non-specific PCR products. Therefore, most of the somatic candidates are likely false positives, and the true somatic L1Hs insertion rate may be significantly lower than our upper-bound estimate prior to validation. The post-validation somatic and unique somatic insertion rate estimates are 0.07±0.15 and 0.04±0.10 insertions per neuron, respectively (Figures 6A and 6B).
The remaining 5 somatic candidates were studied further by attempting to clone their full-lengths, and screening for their presence by 3’PCR across all single neurons sorted from the individual in which they were found. We successfully cloned the full-length of one of the five somatic insertion candidates (Figure S7B). This insertion was detected in our L1-IP data in intron 4 of the gene IQCH (IQ motif containing H, chromosome 15), in neuron #2 from the cortex of individual 1465, and is a full-length, intact 6.1kb L1Hs with all the hallmarks of a bona fide L1Hs insertion: a target site duplication (TSD) (13bp), a poly-A tail (~71bp), and a 5’ transduction (101bp) allowing us to trace its source to a full-length, population-polymorphic KR L1Hs on chromosome 8 (Figures S7C and S7D). The full-length sequence of the somatic insertion (Table S3) precisely matched the sequence of the source L1Hs. The insertion was not detected by standard 3’PCR in brain and non-brain bulk tissues from the individual (Figure 6C) and was found in 2/83 (2.4%) cortical and 0/59 caudate single neurons tested (Figures 6D and 6E). The insertion was detected at low-levels in L1-IP data of some 50k-unsorted nuclei samples (Figure S7E), as expected for a low-level mosaic insertion, and with further optimization of our 3’PCR protocol (increased DNA input and higher-cycle PCR) we were able to amplify the insertion from these bulk samples as well (Figure S7F). The remaining four candidates were each found by 3’PCR only in the single neuron in which they were identified by L1-IP. Three of the four had poly-A tails by 3’PCR product sequencing (the fourth had an indeterminate poly-A tail since the breakpoint was within a genomic poly-A) (Table S3). Our results illustrate the ability of single-cell sequencing to identify somatic L1Hs insertions and highlight the potential of single-cell sequencing to identify very low-level mosaic mutations in human tissue.
Single-cell sequencing quantifies mosaicism of a somatic brain mutation causing hemimegalencephaly
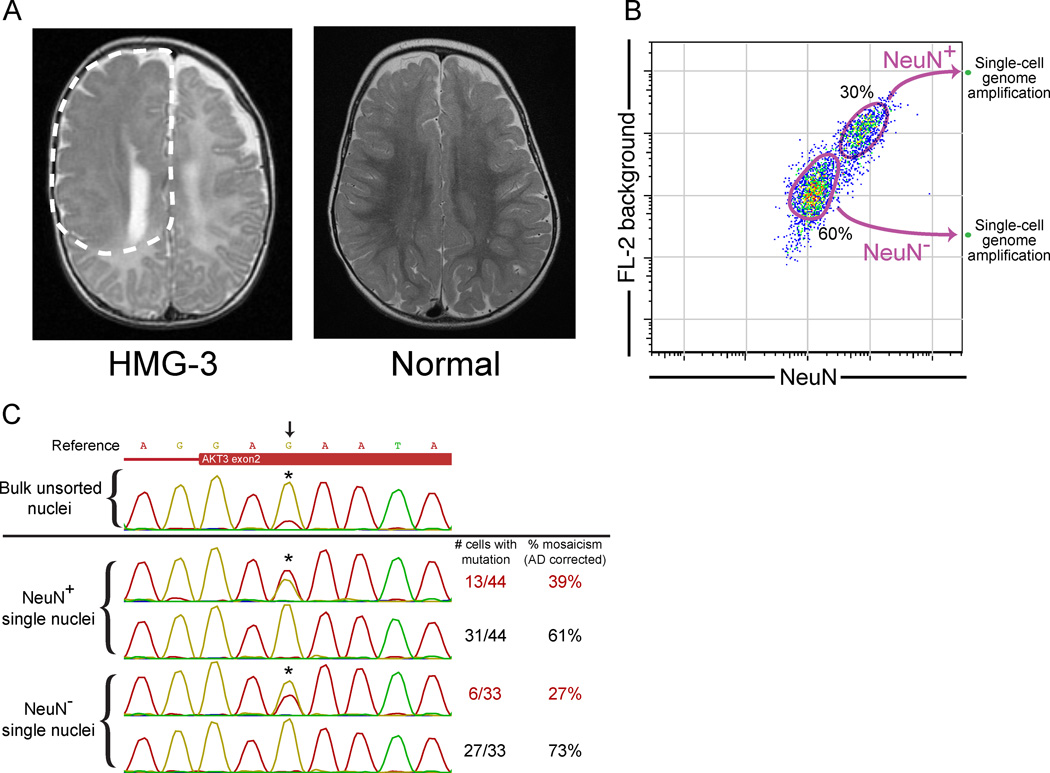
Given the low rate of L1 retrotransposition in neocortical progenitors of normal brains, we next studied the ability of single-neuron sequencing to characterize a pathogenic somatic point mutation in the brain. An open question regarding the pathophysiology of hemimegalencephaly is the lineage (developmental origin) of the pathologic cells (Flores-Sarnat et al., 2003). We recently identified a child with isolated hemimegalencephaly (HMG) caused by a somatic missense (E17K) point mutation in AKT3 present in the brain but not the blood (case HMG-3, Poduri et al., 2012) (Figure 7A). Due to intractable epilepsy, the malformed hemisphere was surgically removed, allowing application of our single-cell method to genotype single sorted cells from this surgical sample and study the origin of the pathologic cells.
Figure 7. Single-cell analysis of a somatic brain AKT3 mutation causing hemimegalencephaly.
(A) An axial T2-weighted image from the MRI of the hemimegalencephaly patient, HMG-3, with a somatic AKT3 E17K mutation shows the enlarged right hemisphere with abnormally thick and malformed cerebral gray matter and abnormal signal of the white matter (white dashed line). On the right is an MRI image of a normal brain.
(B) Single-cell FACS sorting of HMG-3 resected cortex.
(C) Representative Sanger sequencing traces of a bulk unsorted nuclei sample and single-cell samples from NeuN+ and NeuN− populations. The calculated % mosaicism for single-cell samples (corrected for allelic dropout) is shown. Arrow and asterisks mark the site of the AKT3 c.49G→A (E17K) mutation. See Table S4 for percent mosaicism of all samples from HMG-3.
Previous analysis of resected bulk tissue indicated that the mutation was present at ~35% mosaicism based on cloning of PCR products (Poduri et al., 2012). Interestingly, 39±7% (SE; corrected for AD) of single sorted neuronal (NeuN+) nuclei contained the mutation (Figures 7B, 7C, and Table S4), similar to the mosaicism in unsorted bulk tissue containing both neuronal and non-neuronal cells. This suggested that the mutation was also present in non-neuronal cells, consistent with the abnormality of both gray matter and white matter in this patient by MRI (Poduri et al., 2012; Figure 7A). Indeed, we confirmed the presence of the mutation in single non-neuronal (NeuN−) nuclei, at an average percent mosaicism (corrected for AD) of 27±8% (Figure 7C and Table S4). These data indicate that the mutation was present in an early neocortical progenitor capable of giving rise to both neuronal- and non-neuronal cells throughout the majority of the hemisphere. The low mosaicism in neurons also indicates that mutant and non-mutant neurons are extensively intermingled in the abnormal hemisphere, presumably reflecting diverse clonal origins of cortical neurons in this pathological condition.
Discussion
Here we present a single-cell sequencing study of the central nervous system, and perform genome-wide analysis to trace patterns of somatic mutation in human brain. We confirmed that somatic retrotransposon insertions can be detected in normal human brain. However, our analysis of L1 insertions found that somatic insertions are rare in normal human cortical and caudate neurons, suggesting that L1 retrotransposition is not a major source of neuronal diversity in cerebral cortex and caudate nucleus. Finally, we used single-cell analysis to study the mosaicism of a somatic AKT3 mutation, highlighting the potential of single-cell sequencing for cell lineage analysis in human brain.
L1Hs retrotransposition in human cerebral cortex and caudate
Our validation of a somatic L1Hs insertion with all the hallmarks of a bona fide retrotransposition event, including a 5’ transduction identifying its source, confirms that somatic L1Hs insertions are present in the normal human brain. The very low-level mosaicism of this insertion, and its detection only in cortical neurons, further suggests that it may have occurred during cortical development. The source L1Hs on chromosome 8 from which the somatic insertion originated lies in antisense orientation within an intron of the gene KCNB2, and is a full-length insertion with both open reading frames intact. Although it is present in the human genome reference, it is polymorphic in the population and was present only in individual 1465, but not the other individuals in this study (data not shown). In addition to this source L1Hs, only one other L1Hs element has been previously confirmed to be active somatically in humans (van den Hurk et al., 2007). Further single-cell studies will help delineate the spectrum of somatic activity of L1Hs elements in different tissues and developmental stages.
Our quantitative analysis of retrotransposition indicates that somatic L1Hs events are rare in adult human cortical pyramidal neurons and caudate neurons. We find that, although we can detect hundreds of known germline insertions in single neurons, >80% of neurons show no unique somatic insertions (i.e. present in one neuron but not multiple neurons). Somatic L1Hs insertions present in multiple neurons but not all neurons, as seen for the full-length somatic insertion we identified, are also rare. On the other hand, we cannot exclude greater rates of L1Hs activity in other cell types or regions of the human brain, or activity of Alu and SVA retrotransposons in the cortex and caudate. Variability in the number of highly active “hot” L1s per individual (Beck et al., 2010) may also lead to variability in somatic retrotransposition rates among individuals; however, the low number of somatic insertions in 300 neurons from 3 individuals precludes it from being an essential source of neuronal diversity in cortex and caudate that is common in humans.
Our results are generally consistent with the rates of ~1/10,000 to ~1/100 insertion events per human neural progenitor measured in an in vitro L1RP reporter assay (Coufal et al., 2009). This rate is far lower than the rate measured by quantitative PCR (Coufal et al., 2009; Muotri et al., 2010) which estimated a relative copy number increase of L1 of ~5–10% and an absolute estimate of ~80 somatic L1 insertions per cell in human brain. Studies employing targeted capture of L1 sequences from human brain (Baillie et al., 2011) also reported widespread L1 retrotransposition. These methods are less direct, and do not analyze individual neurons, but instead analyze pooled DNA from bulk tissue. Compared to sequencing of bulk tissue (Baillie et al., 2011), our approach of single-cell sequencing has the additional advantage that potential artifacts, such as chimeric reads, are easier to recognize because they are present at lower read depth relative to true insertions. The identification of mammalian species that appear to have lost all L1 activity (Cantrell et al., 2008) further suggests that L1 retrotransposition is not a universal requirement for mammalian neurogenesis. Recent L1 profiling of 26 glial brain tumors did not reveal any somatic insertions (Iskow et al., 2010; Lee et al., 2012a), indicating that somatic L1 insertions may be uncommon in glial progenitors as well. While our study suggests that somatic L1 retrotransposition in the human cortex and caudate is rare, it remains possible that neuronal L1 retrotransposition may occur at higher rates in other brain regions, such as the hippocampus, and/or play a role as a mutagen in the human brain in neurological disease.
Somatic mutations causing cortical malformations can occur in neuroglial progenitors
Our analysis of a somatic retrotransposon insertion and a somatic AKT3 mutation, each found in more than one cortical neuron as well as at low levels in bulk DNA, suggests that both occurred in progenitor cells of the brain, and that other focal brain malformations of unknown etiology may be similarly caused by progenitor mutations during development. The somatic AKT3 mutation in hemimegalencephalic brain was found in both neuronal and non-neuronal cells, further indicating that the mutation occurred in a neuroglial progenitor. Moreover, the normal-appearing basal ganglia of this patient by MRI (data not shown) would be consistent with a mutation occurring in a neuroglial progenitor in the developing neocortex, but not involving the ventral telencephalon, though caudate tissue was not available for testing.
Our study suggests potential future applications of somatic mutations as cell lineage markers in post-mortem human brain. Although retrotransposon insertions appear too rare for systematic study of cell lineages, and the specific AKT3 mutation assayed here clearly changes the behavior of cells carrying the mutation (Poduri et al., 2012), deeper sequencing of single cells might eventually identify diverse, nonfunctional mutations, including mutations at highly mutable sites like microsatellite repeats (Frumkin et al., 2005; Salipante et al., 2008), that may allow more systematic interrogation of lineage relationships even in human post-mortem brain.
Experimental Procedures
Full protocols can be found in Extended Experimental Procedures.
Tissue sources
Fresh-frozen post-mortem tissues of 3 normal individuals and a trisomy 18 fetus (UMB1465, UMB4638, UMB4643, and UMB866) were obtained from the NICHD Brain and Tissue Bank at the University of Maryland. Hemimegalencephalic brain tissue from case HMG-3 (Poduri et al., 2012) was obtained following neurosurgical resection of the affected right hemisphere.
Single neuronal nuclei isolation and genome amplification
Nuclei were purified by sucrose cushion ultra-centrifugation and labeled with NeuN antibody (Millipore, MAB377) for flow cytometry as previously described (Matevossian and Akbarian, 2008; Spalding et al., 2005). Single nuclei were sorted with a FACSAria II cell sorter into 96- or 384-well plates and amplified by MDA (Dean et al., 2002). Low-coverage sequencing libraries were made with the NEXTflex DNA-seq kit (Bioo Scientific).
Genome-wide L1Hs-insertion profiling
L1Hs-insertion profiling libraries (L1-IP) were made by modification of the method of Ewing and Kazazian (2010) for a high-throughput workflow and high-level (up to 32-plex) multiplexing. Libraries were sequenced on HiSeq 2000 sequencers (Illumina). A custom data analysis pipeline was created to call and classify L1-IP peaks.
L1Hs insertion validation
3’ junction PCR (3’PCR) was performed with one primer specific to L1Hs (L1Hs-AC-22) and a 5’ peak flank primer (upstream to the L1-IP peak), to verify the presence of the predicted insertion. Long-range PCR with 5’ and 3’ peak flank primers was performed to clone the entire length of candidate insertions.
Supplementary Material
01
02
03
04
05
06
Acknowledgements
G.D.E. and X.C. performed all experiments, with assistance from L.B.H, P.C.E., H.S.L, J.J.P., and K.D.A. G.D.E. and E.L. analyzed the L1-IP data with input from P.J.P. G.D.E. and X.C. analyzed all other data. G.D.E., X.C., and C.A.W conceived and designed the project with input from E.C.G and A.P. G.D.E., X.C., and C.A.W wrote the manuscript.
We thank Peter V. Kharchenko, Tim W. Yu, Vijay S. Ganesh and Nathan Silberman for helpful discussions; Hal Schneider, Richard Bennett, R. Sean Hill, and Christina Kourkoulis for technical assistance; Robert Johnson from the NICHD Brain and Tissue Bank; the Orchestra research computing support team (Harvard Medical School); and the Hematologic Neoplasia Flow Cytometry Core (Dana-Farber Cancer Institute). Brain image in Figure 1A adapted with permission from http://brainmuseum.org, supported by the US National Science Foundation. C.A.W. is supported by the Manton Center for Orphan Disease Research and grants from the NINDS (RO1 NS079277 and R01 NS35129). C.A.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Sequencing data from this study are deposited in the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under the accession number SRA056303.
References
- Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blainey PC, Quake SR. Digital MDA for enumeration of total nucleic acid contamination. Nucleic Acids Res. 2011;39:e19. doi: 10.1093/nar/gkq1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell MA, Scott L, Brown CJ, Martinez AR, Wichman HA. Loss of LINE-1 activity in the megabats. Genetics. 2008;178:393–404. doi: 10.1534/genetics.107.080275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O'Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson RP. Somatic gene mutation and human disease other than cancer: an update. Mutat Res. 2010;705:96–106. doi: 10.1016/j.mrrev.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Ewing AD, Kazazian HH., Jr High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010;20:1262–1270. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Sarnat L, Sarnat HB, Davila-Gutierrez G, Alvarez A. Hemimegalencephaly: part 2. Neuropathology suggests a disorder of cellular lineage. J Child Neurol. 2003;18:776–785. doi: 10.1177/08830738030180111101. [DOI] [PubMed] [Google Scholar]
- Frumkin D, Wasserstrom A, Kaplan S, Feige U, Shapiro E. Genomic variability within an organism exposes its cell lineage tree. PLoS Comput Biol. 2005;1:e50. doi: 10.1371/journal.pcbi.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittins R, Harrison PJ. Neuronal density, size and shape in the human anterior cingulate cortex: a comparison of Nissl and NeuN staining. Brain Res Bull. 2004;63:155–160. doi: 10.1016/j.brainresbull.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Gleeson JG, Minnerath S, Kuzniecky RI, Dobyns WB, Young ID, Ross ME, Walsh CA. Somatic and germline mosaic mutations in the doublecortin gene are associated with variable phenotypes. Am J Hum Genet. 2000;67:574–581. doi: 10.1086/303043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH., Jr Active human retrotransposons: variation and disease. Curr Opin Genet Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, Li F, Wu K, Liang J, Shao D, et al. Single-Cell Exome Sequencing and Monoclonal Evolution of a JAK2-Negative Myeloproliferative Neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisky T, Blainey P, Quake SR. Genomic analysis at the single-cell level. Annu Rev Genet. 2011;45:431–445. doi: 10.1146/annurev-genet-102209-163607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasken RS, Stockwell TB. Mechanism of chimera formation during the Multiple Displacement Amplification reaction. BMC Biotechnol. 2007;7:19. doi: 10.1186/1472-6750-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, 3rd, Lohr JG, Harris CC, Ding L, Wilson RK, et al. Landscape of Somatic Retrotransposition in Human Cancers. Science. 2012a;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Huynh M, Silhavy JL, Kim S, Dixon-Salazar T, Heiberg A, Scott E, Bafna V, Hill KJ, Collazo A, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet. 2012b;44:941–945. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magavi S, Friedmann D, Banks G, Stolfi A, Lois C. Coincident generation of pyramidal neurons and protoplasmic astrocytes in neocortical columns. J Neurosci. 2012;32:4762–4772. doi: 10.1523/JNEUROSCI.3560-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matevossian A, Akbarian S. Neuronal nuclei isolation from human postmortem brain tissue. J Vis Exp. 2008 doi: 10.3791/914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–645. [PubMed] [Google Scholar]
- Mills SE. Histology for pathologists. 3rd edn. Philadelphia: Lippincott Williams & Wilkins; 2007. [Google Scholar]
- Mullen RJ, Buck CR, Smith AM. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MCN, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Gage FH. Generation of neuronal variability and complexity. Nature. 2006;441:1087–1093. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov I, Rubin A, Swergold GD. Tracing the LINEs of human evolution. Proc Natl Acad Sci USA. 2002;99:10522–10527. doi: 10.1073/pnas.152346799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent A, Carpenter MB. Carpenter's human neuroanatomy. Williams & Wilkins; 1996. [Google Scholar]
- Poduri A, Evrony GD, Cai X, Elhosary PC, Beroukhim R, Lehtinen MK, Hills LB, Heinzen EL, Hill A, Hill RS, et al. Somatic Activation of AKT3 Causes Hemispheric Developmental Brain Malformations. Neuron. 2012;74:41–48. doi: 10.1016/j.neuron.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, Almeida BS, Kingsbury MA, Cabral KM, McConnell MJ, Anliker B, et al. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25:2176–2180. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere JB, Mirzaa GM, O'Roak BJ, Beddaoui M, Alcantara D, Conway RL, St-Onge J, Schwartzentruber JA, Gripp KW, Nikkel SM, et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet. 2012;44:934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salipante SJ, Thompson JM, Horwitz MS. Phylogenetic fate mapping: theoretical and experimental studies applied to the development of mouse fibroblasts. Genetics. 2008;178:967–977. doi: 10.1534/genetics.107.081018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. 2010;33:345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding KL, Bhardwaj RD, Buchholz BA, Druid H, Frisen J. Retrospective birth dating of cells in humans. Cell. 2005;122:133–143. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- van den Hurk JAJM, Meij IC, Seleme MdC, Kano H, Nikopoulos K, Hoefsloot LH, Sistermans EA, de Wijs IJ, Mukhopadhyay A, Plomp AS, et al. L1 retrotransposition can occur early in human embryonic development. Hum Mol Genet. 2007;16:1587–1592. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- Wang J, Fan HC, Behr B, Quake SR. Genome-wide Single-Cell Analysis of Recombination Activity and De Novo Mutation Rates in Human Sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf HK, Buslei R, Schmidt-Kastner R, Schmidt-Kastner PK, Pietsch T, Wiestler OD, Blumcke I. NeuN: a useful neuronal marker for diagnostic histopathology. J Histochem Cytochem. 1996;44:1167–1171. doi: 10.1177/44.10.8813082. [DOI] [PubMed] [Google Scholar]
- Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, Li F, Tsang S, Wu K, Wu H, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Martiny AC, Reppas NB, Barry KW, Malek J, Chisholm SW, Church GM. Sequencing genomes from single cells by polymerase cloning. Nat Biotechnol. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04
05
06