Hypoxia Modulates A431 Cellular Pathways Association to Tumor Radioresistance and Enhanced Migration Revealed by Comprehensive Proteomic and Functional Studies (original) (raw)
Abstract
Tumor hypoxia induces cancer cell angiogenesis, invasiveness, treatment resistance, and contributes to poor clinical outcome. However, the molecular mechanism by which tumor hypoxia exerts a coordinated effect on different molecular pathways to enhance tumor growth and survival and lead to poor clinical outcome is not fully understood. In this study, we attempt to elucidate the global protein expression and functional changes in A431 epithelial carcinoma cells induced by hypoxia and reoxygenation using iTRAQ quantitative proteomics and biochemical functional assays. Quantitative proteomics results showed that 4316 proteins were quantified with FDR<1%, in which over 1200 proteins were modulated >1.2 fold, and DNA repair, glycolysis, integrin, glycoprotein turnover, and STAT1 pathways were perturbed by hypoxia and reoxygenation-induced oxidative stress. For the first time, hypoxia was shown to up-regulate the nonhomologous end-joining pathway, which plays a central role in DNA repair of irradiated cells, thereby potentially contributing to the radioresistance of hypoxic A431 cells. The up-regulation of Ku70/Ku80 dimer, a key molecular complex in the nonhomologous end-joining pathway, was confirmed by Western blot and liquid chromatography/tandem mass spectrometry-MRM methods. Functional studies confirmed that up-regulation of glycolysis, integrin, glycoprotein synthesis, and down-regulation of STAT1 pathways during hypoxia enhanced metastastic activity of A431 cells. Migration of A431 cells was dramatically repressed by glycolysis inhibitor (2-Deoxy-d-glucose), glycoprotein synthesis inhibitor (1-Deoxynojirimycin Hydrochloride), and STAT1α overexpression that enhanced the integrin-mediated cell adhesion. These results revealed that hypoxia induced several biological processes involved in tumor migration and radioresistance and provided potential new targets for tumor therapy.
Tumor hypoxia arises when solid tumor expands too rapidly without a concomitant expansion of the supporting vasculature (1). Tumor hypoxia induces resistance to anticancer therapeutics, angiogenesis, genomic instability, invasion/metastasis, and overall poor clinical outcome (2, 3). Given its importance in tumor development, the molecular pathways perturbed by tumor hypoxia are considered attractive targets to be exploited in oncology (4).
Extensive laboratory studies and clinical data have shown that hypoxic conditions contribute to therapeutic resistance in radiotherapy and limit the response of tumor cells to radiation therapy (5–7). DNA damage by irradiation is kept from repair with the presence of oxygen, but it can be repaired by hydrogen without oxygen under hypoxia and thus leads to increased radioresistance (7). Other biological molecules have also been reported to be involved in the radioresistance in tumor cells under hypoxic conditions (8). A major cellular response to hypoxia is the activation of hypoxia induced factor 1 (HIF-1)1, a transcription factor, which has been implicated in the regulation of tumor radiosensitivity (9). HIF-1α expression in uterine cervical cancer tissues positively correlates with the adverse effects of radiotherapy, and its expression in cervical cancer after radiotherapy is associated with increased risk for tumor-related death (10, 11). DNA repair pathways are vital for cellular protection against radiation. Recent studies showed that the activation of DNA repair pathways in some tumors contributed to intrinsic resistance to radiotherapy (12, 13). However, several reports have demonstrated that hypoxia induces the repression of DNA repairing genes at the mRNA level, for example, RAD51, BRCA1/2, XRCC3/4, Ku70, and Ligase IV, but the changes in mRNA level do not always correlate with a subsequent decrease in protein level (14–17). These data thus highlight the importance of determining whether some of the functional consequences of hypoxia were mediated by DNA repair proteins.
Laboratory and clinical studies have also indicated that tumor hypoxia has been implicated in enhancing tumor cell metastatic potential (3, 18–22). Metastasis is the major characteristics of malignant tumors and the main cause for cancer-related mortalities. As such, there have been major efforts to elucidate the molecular mechanisms underlying the distinct steps of cancer metastasis (23–26). Some key molecules, such as adhesion molecules and MMPs, play important role in cancer cell metastasis (27). It has been reported that HIF-1 modulates the expression level of proteins, such as MMPs, plasminogen activator inhibitor (PAI-1), tissue factors, CapG, S100A4, filamentation 1, cadherin, and integrin alpha 5 (2, 28–30), which play important roles in regulating the invasion or metastatic potential of tumor cells.
Hypoxia is now a well recognized cause of radioresistance and metastasis but its precise role in these processes is still poorly defined (24, 31). A better understanding of the mechanisms behind this pathophysiology will lead to a more specific and efficient therapeutic outcome. The knowledge of hypoxia-regulated proteins from proteomic investigation will provide a better and global understanding of the molecular pathways perturbed by hypoxic tumor and give rise to novel biological insights and concepts for exploiting this factor (7).
Proteomic studies to elucidate the molecular events elicited by hypoxic stress (3, 32–34) are less extensive and have, to date, been limited to the use of 2-DE based methods (35). To further our proteomic knowledge of hypoxia-induced tumor evolution, shotgun based isobaric tag for relative and absolute quantification (iTRAQ) quantitative proteomics, which provides a global assessment of the proteins modulated, was used to analyze the cellular proteome changes of A431 epithelial carcinoma cells induced by hypoxia and reoxygenation. More than 4300 proteins with unused Protscore ≥1.68 and FDR<1% were identified, in which over 1200 proteins were significantly modulated by hypoxia (Supplemental Table S1). Further analysis of the quantitative proteomics data set indicated that DNA repair, glycolysis, integrin, glycoprotein turnover, and STAT1 pathways were perturbed by hypoxia. Most of them as listed in Supplemental Table S2 were confirmed and validated using Western blot (WB) or LC-MS/MS-multiple reaction monitoring (MRM) methods. Functional studies revealed the roles of the glycolysis, integrin, glycoprotein synthesis and STAT1 pathways in hypoxia-induced cancer metastasis.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
All reagents were purchased from Sigma Chemical Co. (St. Louis, MO) unless otherwise specified. Antibody against integrin alpha 5 was purchased from Millipore (Billerica, MA). Anti-STAT1 and anti-STAT1 (pS727) were purchased from BD Biosciences Pharmingen (San Jose, CA) and Cell Signaling (Danvers, MA), respectively. Anti-Ku70, Ku80, ITGB1, and 1-Deoxynojirimycin Hydrochloride (1-DJ) were purchased from Santa Cruz (Santa Cruz, CA).
Cell Culture and Hypoxic Condition
The A431 epithelial carcinoma cells were purchased from ATCC and maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum. For all experiments, 2 × 106 cells were seeded on 10 cm dishes in DMEM and incubated under 5% CO2 at 37 °C. On reaching 30–40% confluency, the cells were washed with 1 × PBS for three times and serum-free media for three times before incubating in serum-free media. They were subjected to normoxia (Nx: 21% O2/5% CO2) for 72 h or hypoxia (Hx: <0.1% O2/5% CO2) in a hypoxia chamber for 36 h or 72 h. For Reoxygenation (ReOx), the cells were exposed to Hx for 48 h, changed to a fresh medium, and then exposed to Nx condition for an additional 24 h. At least five independent biological replicates were pooled for the analyzed sample in each condition.
Sample Preparation, iTRAQ Labeling, and LC-MS
The cellular proteins were dissolved in 8 m urea in 50 mm triethylammonium bicarbonate (TEAB) (pH 8.5) solution with protease inhibitor mixture (1:50). Protein concentrations were measured using BCA assay. Two hundred micrograms of proteins from each condition were reduced with 5 mm TCEP at 30 °C for 1 h, alkylated with 10 mm methyl methanethiosulfonate (MMTS) at room temperature, in the dark for 45 min. The samples were diluted to 1 m urea using 50 mm TEAB and digested by trypsin in a 1:50 mass ratio at 37 °C overnight. The solution containing tryptic peptides was adjusted to pH 2–3 with TFA and desalted using Sep-Pak C18 cartridge (Waters, Milford, MA). The peptides were dried using speedvac concentrator and, dissolved in 1 m TEAB (pH 8.5) and labeled with the isobaric tags (Applied Biosystems, Foster City, CA) according to manufacturer's protocol as followed: the Nx-treated sample was labeled with 114; the samples under Hx for 36 h or 72 h were labeled with 115 or 116 respectively; and the ReOx-treated sample was labeled with 117.
The labeled sample was fractionated by a PolySULFOETHYL A column (PolyLC, Columbia, MD; 4.6 × 200 mm, 5 μm particle size, 200-Å pore size) using Shimadzu Prominence UFLC system (Kyoto, Japan). Fractionation was performed with a 60-min linear gradient of 0–500 mm KCl (10 mm KH2PO4, pH 3.0, 25% acetonitrile) at a flow rate of 1 ml/min. A total of 25 fractions were collected and desalted using Sep-Pak C18 cartridges (Waters). Each fraction was dried and reconstituted in 30 μl 3% ACN and 0.1% formic acid for LC-MS/MS analysis. All fractions were analyzed using a QSTAR Elite mass spectrophotometer coupled with an on-line Tempo nano-MDLC system. The sample was injected twice. For each injection, 15 μl iTRAQ labeled peptide mixture was injected and separated on a home-packed nanobored C18 column with a picofrit nanospray tip (75 μm inner diameter×15 cm, 5 μm particles) (New Objectives, Woburn, MA). The detailed parameters were set as previously described (3).
Database Searching and Criteria
Protein identification and quantification were performed using ProteinPilotTM software 2.0.1 with Revision number 67476 (Applied Biosystems) by searching the combined raw data from the two runs of labeled sample against the concatenated “target” (Uniprot human database, downloaded on 12 March 2010, including 95624 sequences and 36307192 residues) and “decoy” (the reverse amino acid sequence for false discovery rate (FDR) estimation) databases. The Paragon and Pro Group algorithms in the ProteinPilot software were used for peptide identification and isoform-specific quantification. User-defined parameters were as follows: (i) sample type, iTRAQ 4plex (peptide-labeled); (ii) cysteine alkylation, MMTS; (iii) digestion, trypsin; (iv) instrument, QStar Elite ESI; (v) special factors, none; (vi) species, none; (vii) specify processing, quantitative; (viii) ID focus, biological modifications; and (ix) search effort, thorough ID. Two missed cleavage sites are permitted and nonspecific cleavage sites are prohibited. The Mass tolerance of precursor ions and fragment ions is both 0.2 Dalton.
To minimize false positive results, a strict cutoff for protein identification was applied with FDR<1% as listed in Supplemental Table S1. The resulting data set was autobias-corrected to get rid of any variations imparted because of unequal mixing during combining different labeled samples. For iTRAQ quantitation, the peptide for quantification was automatically selected by the Pro Group algorithm (at least one peptide with 99% confidence) to calculate the reporter peak area, error factor (EF), and _p_ value. The proteins for further analysis were filtered with _p_ value <0.05, and 1.2-fold changes (>1.20 or <0.83) relative to the normoxic control unless the proteins were known to be of interest from our functional assay. The list of proteins used is shown in Supplemental Table S2.
Semi-quantitative RT-PCR and Western Blotting
Total cellular mRNAs were isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. cDNA was reversely transcribed from 2 μg total RNA in a final volume of 20 μl using RTase (Promega, Madison, WI, USA). Primer sequences were listed in Supplemental Table S3. PCR was performed with KAPA Fast Taq polymerase in a DNA thermal cycler (PTC-200, Bio-Rad, Hercules, CA) according to a standard protocol as follows: 1 cycle of 95 °C for 1 min; 28 cycles of 95 °C for 10 s, annealing at 57 °C for 10 s, 72 °C for 1 s, a final extension at 72 °C for 30 s and holding at 4 °C. The amount of cDNA used for each PCR reaction was 20 ng in a 25 μl reaction volume. The PCR products (5 μl) were analyzed by electrophoresis through 2% agarose gels and visualized with ethidium bromide staining.
Before Western blot analysis, protein concentration of cell lysates dissolved in 1% SDS in 40 mm Tris-HCl (pH 8.0) was quantified using BCA assay, equal protein amount from each sample was separated on 12% SDS-PAGE. For native gel, the solution of 1% Triton X-100 in 20 mm Tris-HCl (pH 7.4), 150 mm NaCl with complete protein inhibitor mixture (1:50) was used in the extraction of protein complexes. Proteins were then transferred to nitrocellulose membrane and immunoblotted using antibodies against STAT1, p-STAT1, Ku70, Ku80, ITGA5 and ITGB1 respectively. The probe proteins were detected with Invitrogen ECL system according to the manufacturer's instructions.
Dynamic MRM Quantification
The same amount of proteins from both normoxic and hypoxic A431 cells were loaded and separated on 12% SDS-PAGE. Based on the proteins molecular weight, each lane was manually excised into six fractions and in-gel digested with trypsin. The peptides were loaded onto 6490 Triple Quadrupole LC-MS/MS system coupled with Infinity 1260 liquid phase separation system (Agilent Technologies Inc, Singapore). The information of all peptides and their transitions of target proteins were obtained from Peptide Atlas (www.peptideatlas.org) database and showed in Supplemental Table S4. All samples were subjected to conventional MRM analysis for confirmation of retention times of peptides. Then dynamic MRM was used to quantify the protein changes.
The mobile phase consisted of solvent A, 0.1% aqueous formic acid and solvent B, acetonitrile with 0.1% formic acid. Peptides were separated by Agilent chip (Large capacity chip II) packed with Zorbax 300SB C18 resin (5 μm, separation: 150 mm × 75 μm; enrichment: 9 mm, 160 nL). Injections were 2 μl of a sample solution and were followed by a 10 min wash period with 100% A, then by elution with a gradient of 0–10% solvent B in 3 min, 10–35% solvent B in 47 min, 35–80% in 2 min, 80–82% solvent B in 3 min and followed by 3% solvent B in 5 min.
LC-MS/MS-MRM analyses of the peptide were done with an electrospray voltage of 1700 V, capillary temperature 185 °C and skimmer offset −5 V. Both Q1 and Q3 were set at unit resolution (FWHM 0.7 Da) and collision gas (N2) pressure in Q2 was held at 1.5 mTorr. Scan width was 0.004 m/z and scan time was 20 ms for the peptide. Collision energy for each peptide was calculated based on the equation: CE = (((m/z)/100.0)*3.6 - 4.0), in which the m/z is the mass to charge ratio of the precursor ion. Peak areas for each peptide were extracted and integrated using Agilent Qualitative Analysis B.04.00 software.
Construction and Transfection of STAT1α Expression Vectors
The full length of STAT1α was obtained from A431 cDNA library prepared as described above using PCR. The primer information is listed in Supplemental Table S3. The gene with NotI and BamH I enzyme sites were inserted into the expression vector of p3xFLAG-CMV-10 and positive clones were confirmed by plasmid digestion with NotI and BamH I and DNA sequencing. For stable transfection, A431 cells were transfected using 20 μl of lipofectamin 2000 (Invitrogen) and 4 μg of p3xFLAG-STAT1α vector or control p3xFLAG vector according to manufacturer's instructions. Cells were then selected with standard medium with 1 mg/ml G418. 2–3 weeks later, the monoclones were picked up and further cultured in medium with 0.5 mg/ml G418. The positive clones were confirmed by WB using anti-Flag, STAT1 and phospho-STAT1 antibodies.
Cell Proliferation and Cell Adhesion Assay
Twenty-four-well plates were coated with 5 μg/ml fibronectin in PBS. Uncoated wells were incubated with 0.5% BSA in DMEM to serve as negative control. Cells were trypsinized and seeded into coated 24-well dish at a density of 2.5 × 105 live cells. After incubation for 20 mins to an hour at 37 °C, the detached cells were removed by serum-free medium and attached cells were fixed with 95% ethanol for 10 mins and stained with 0.5% crystal violet for 25 mins. The plate was reimmersed in deionized water to remove excess dye and then allowed to air-dry for 5 to 10 mins. The adherent cells were photographed by optical microscopy under 100 × magnification and a total of 200 μl of 0.5% Triton X-100 was added to each well to solubilize the cells overnight at room temperature. The absorbance was measured at OD595 using a microplate reader (Tecan MagellanTM, Männedorf, Switzerland). Cell proliferation was assessed by the ability to exclude 5 mg/ml MTT in PBS for 2 h. MTT uptake was quantified at A540 nm, reference A630 nm after solubulization in DMSO.
Cell Staining
To visualize ITGA5 distribution in cells, immunofluorescent staining with anti-integrin a5 antibody was performed. A431 cells were seeded into 20 × 20-mm coverslips, treated for the indicated conditions, fixed by 4% paraformaldehyde, and then permeabilized with 0.2% Triton X-100 in PBS for 10 mins. The cells were incubated with 4% BSA in 0.1% Triton X-100 for 20 mins to block nonspecific binding sites before incubation with anti-ITGA5 in 1% Triton X-100 (Millipore; 1:100) for 2 h at 37 °C. Subsequently, the cells were washed, and then incubated with Alexa Fluor® 488-conjugated secondary antibody for 1 h at room temperature. After that, cells were washed, mounted, and examined by fluorescence microscope.
Scratch-wound Assay
A total of 2 × 106 cells were seeded in a 6-well plate. A wound was made by scratching the cells in a line with a sterile pipette tip in the middle of the well next day. Then cells were washed twice with 1 X PBS and three times with serum free media before incubating in serum-free media. Photographs were captured by a Nikon Eclipse TE2000-U microscope. After incubation in indicated conditions under hypoxia for specific time points shown in Fig. 2, 4, and 6, photographs were taken again. Hypoxic treatment was the same as above mentioned. For 1-DJ wound-scratch assay, the cells were treated by the drug for 48 h and then undergone scratch assay under hypoxia. After hypoxia, the cells were lysed with 1% SDS solution and used for WB to detect their glycoproteins by HRP-labeled lectin. Because 2-DG and 1-DJ are water soluble, no control solvents are used in the experiments. The migration of cells into the wound was recorded and the wound closure was determined.
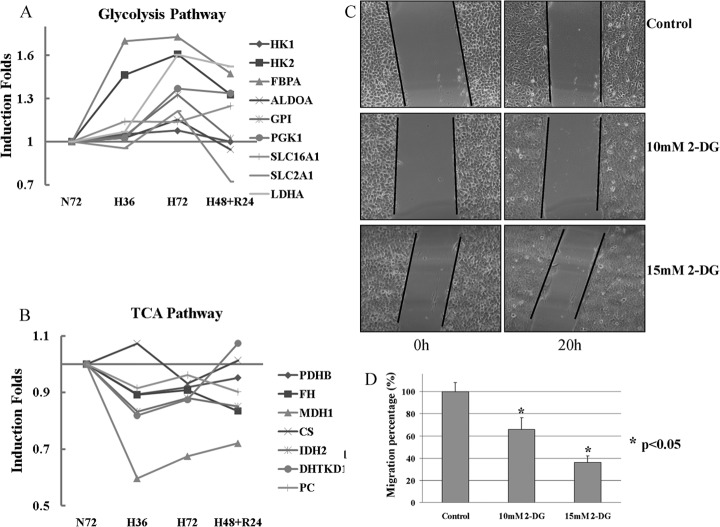
Fig. 2.
Hypoxia activates glycolysis and inhibits Kreb cycles by modulating the enzymes involved in the pathways. A and B, Change pattern of enzymes involved in glycolysis and pyruvate metabolism induced by hypoxia or reoxygenation; C, Images of scratch assay of A431 treated with glycolysis inhibitor, 2-DG, for 20 h, Magnification, ×100; D, The effect of 2-DG on inhibition of cell migration at 10 mm and 15 mm concentration.
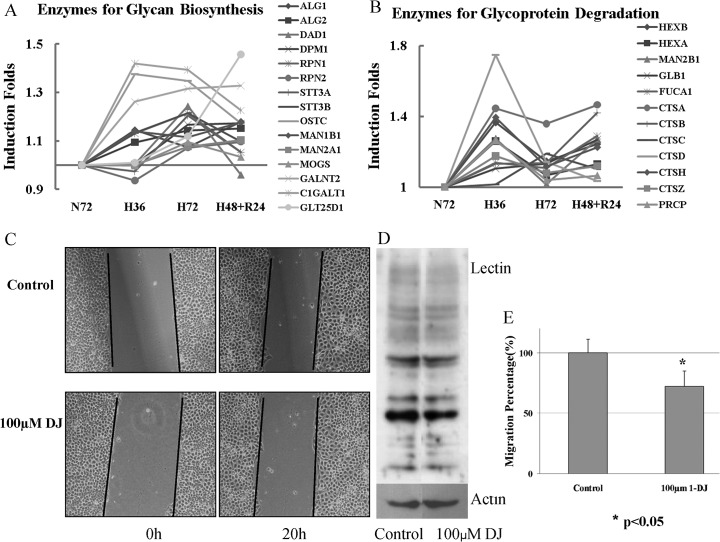
Fig. 4.
Hypoxia speeds up the turnover of glycoproteins in A431. A and B, Change patterns of enzymes involved in glycan biosynthesis and glycoprotein degradation during hypoxia or reoxygenation; Glycoprotein degradation has two steps: proteolysis and glycan degradation. The two sets of enzymes have similar change pattern responsible to hypoxia or reoxygenation. C, Images of scratch assay of A431 treated with 100 μm 1-DJ for 48 h first to suppress glycoprotein synthesis, Magnification, ×100; D, The inhibition of glycoprotein synthesis after 1-DJ treatment was verified in lectin binding assay by WB; E, The effect of 1-DJ in inhibition of cell migration at 100 μm concentration.
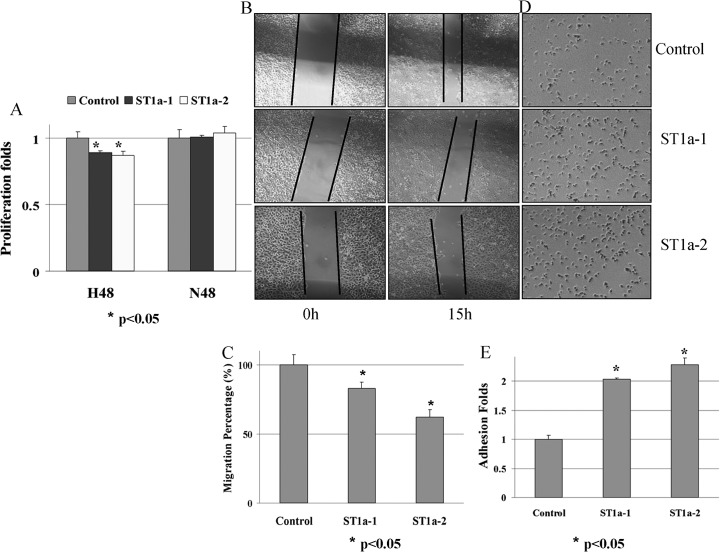
Fig. 6.
Overexpression of STAT1α suppresses the migration and enhances the adhesion of A431. A, MTT assay under Nx and Hx; B and C, Images and quantification of scratch assay of control and STAT1α overexpression A431 cells, Magnification, ×100; D and E, Images and quantification of adhesion assay of control and STAT1 overexpression A431 cells
Metabolite Analysis
For A431 cell metabolite extraction, MeOH/H2O (80:20) solution was used as described in published paper because the solution had been proved to be a more suitable extraction solvent (36). Briefly, after the same amount of A431 cells were treated with or without 2-DG or hypoxia, the growth medium was discarded and cells were rapidly washed three times with cold PBS followed by adding cold MeOH/H2O (80:20), which caused protein precipitation and hence cell disruption. Wells were then lightly scraped with a rubber tipped cell scraper. The dishes were washed two times with MeOH/H2O solution and the wash and the extract solution were combined for centrifuging at 4 °C and 5725 × g for 5 min. The supernatant was collected for dry. All steps were performed on ice. The metabolites were solved in 0.1% formic acid for ESI-Q-TOF analysis. Metabolite levels were normalized to protein content in the pellet after metabolite extraction, which was determined by the 2D-Quant Kit (GE Healthcare, Munich, Germany) (36, 37).
The same amount of metabolites normalized with protein level was directly injected to mass spectrometer with 10 μl/min flow. Negative-ion mode electrospray mass spectra were obtained using an Accurate-Mass Q-TOF LC/MS 6530 (Agilent Technologies). Spray voltage was –1.0 kV and the capillary temperature was set to 185 °C. Spectra were collected in full scan mode in a scan range of 50–400 m/z. The precursors with very close m/z with metabolites in glycolysis pathway were chosen for MS/MS identification. MS/MS spectra were obtained using isolation width 4 m/z, and scan rate 1 spectra/s, with collision energy –9. The m/z and intensity of MS/MS spectra was input to Massbank database for metabolite identification (http://www.massbank.jp/MetaboIdentif.html).
Statistical Analysis
The measurements were expressed as mean ± S.D. The paired t test was used for statistical analysis between two groups. Significant level was set at p < 0.05.
RESULTS AND DISCUSSION
Proteomic Analysis of Normoxic, Hypoxic, and Reoxygenated A431 Cell Lysate
The iTRAQ reagent labeled peptides were quantitatively profiled by LC-MS/MS using QStar Elite MS. The expression level changes of proteins were indicated by the ratios of labeling tag (N72:114; H36:115; H72:116; H48+R24:117). Raw data were analyzed using ProteinPilot 2.0.1 software with autobias correction against the UniProt human database. A total of 4316 proteins were identified with unused score ≥ 1.68 and FDR <1% (Supplemental Table S1). iTRAQ reporter ratios of 1.2 and 0.83 were set as the cutoff of protein changes. Totally 1253 proteins were found responsive to hypoxia. These proteins have at least one ratio with p value<0.05, including 522 up-regulated and 743 down-regulated (Table I and Supplemental table S1). All of the data including wiff raw data from QStar, searched group file from ProteinPilot 2.0.1 and the search results of PeptideSummary and ProteinSummary can be downloaded from ProteomeCommons.org Tranche Network using the following hash (Q85ii60OE5SelJDghF4vp2Gjv/dJndWgdMF8yZlmbqXeYgOCLVMBsRz840rGKEoyovesJyYL3rmak5baMgCRO5UBWKoAAAAAAAAZ9Q== for wiff raw data set A; 1G135KGPNp+aETVXL7j5VSBfJDYGNshh6mDkHTpC7eXYzFWn5gq1xfx69LP/qHPaOpnyDd37dlB+auqnmWCSwFSeczQAAAAAAAAZ9Q== for wiff raw data set B; LjkJyHVkOJU59YzP26G6Q5ozCw6Iy1XQQfNNGoDZaL8wrbE0eav3TNatlUL2z3dhsqz17RXpx7v2pgxYiIHBe9MV0I8AAAAAAAAB0Q== for searched group file; MAFBxhUOXAz2nHHgcMZszGUCRewTyg0hoHJ/a5CWtGTr9/ZxYOeYc5yJgrRW9W8jhJawUjdA9bNfdN0HUiwsXBnzYC8AAAAAAAACGg== for PeptideSummary; StQPOjfbpRPzCc3AP4SCSL6hzckua/anydKvaG0TSvim5DdFDFpXucq+n6MMpvdn7CFycO1R6xhLp7lOOx5OTpHSrNcAAAAAAAACDQ== for ProteinSummary).
Table I. Data analysis of identified proteins modulated by hypoxia.
| 115:114 | 116:114 | 117:114 | Unique protein No. | Total | |
|---|---|---|---|---|---|
| p < 0.05 | 1281 | 1260 | 1035 | 1904a | 1253d |
| Ratio>1.2 | 334 | 255 | 203 | 522b | |
| Ratio<0.83 | 560 | 398 | 288 | 743c |
The high quality iTRAQ quantitative datasets provided invaluable biological insight on hypoxia induced tumor progression. To gain the global insights of these proteins, Ingenuity Pathway Analysis (IPA, Ingenuity® Systems, www.ingenuity.com) platform was used to analyze the data to reveal the molecular and cellular functions and canonical pathways induced by hypoxia stress. All the proteins listed in Supplemental Table S1 were used for IPA. The up-regulated and down-regulated proteins were analyzed separately, and thus these proteins were mapped to different function, pathway and toxic lists. The summary of IPA was listed in Supplemental Fig. S1 including top five function, pathway and tox lists that up-regulated and down-regulated proteins involved. The top changes in function and pathway were those regulating basic cell survival, for example, protein synthesis and degradation, post-transcriptional or translational modification, protein folding and trafficking and energy and material metabolism. The top toxic lists were mainly involved in mitochondrial dysfunction, oxidative stress and HIF signaling, which indicated the impact of hypoxia on cellular functions. The proteins involved in HIF signaling pathways were shown down-regulated after hypoxia because the top one pathway down-regulated under hypoxia is ubiquitination degradation pathway and the increased expression of HIF-1α was primarily because of increased protein stabilization via decreased ubiquitination degradation. The pathways with dominant difference in comparison analysis of down-regulated and up-regulated proteins were chosen for further studies (Supplemental Fig. S2 and Supplemental Table S2). The proteins mapped to STAT1 (down-regulated pathway) and N-glycosylation degradation (up-regulated pathway) were illuminated in Supplemental Fig. S3.
Hypoxia Activated A431 NHEJ Pathway in DNA Repair
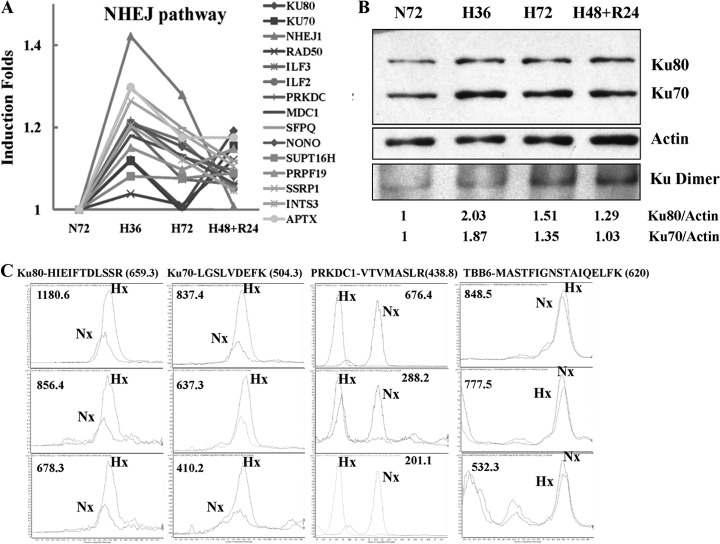
Ionizing radiation causes cell death primarily by DNA double strand breaks (DSBs). The two main pathways involved in the repair of DSBs are homologous recombination (HR) and nonhomologous end-joining (NHEJ) pathways. Of the two, NHEJ pathway generally considered to be an error prone pathway is the dominant repair pathway in radiation-induced DNA damage (38). Our iTRAQ data showed that hypoxia substantially increased A431 proteins involved in DNA repair, especially those responsible for DNA double strand break (DSB) repair in the NHEJ pathway (Fig. 1A). When DNA DSBs occurred, Ku70 and Ku80 bind rapidly with the DNA ends, promoting the inward sliding of Ku70 and Ku80 along the length of DNA, at the same time recruiting PRKDC to assemble a DNA-PK heterotrimer (39). Formation of this complex may also be promoted through interactions with heterodimeric ILF2 and ILF3 (40). Our proteomic data showed that all of these proteins (Ku70, Ku80, PRKDC, ILF2 and ILF3) were up-regulated under hypoxic conditions (Fig. 1A). Subsequent phosphorylation of DNA-PKcs destabilizes and causes the complex to dissociate, allowing the recruitment of several end-processing enzymes including ligase IV, NHEJ1 and Mre11/Rad50/Nbs1 proteins of the MRN complex to DNA. Two of them, NHEJ1 and Rad50 were shown to be up-regulated in our data (39). In addition, the proteins that favor DSBs repair were also up-regulated under hypoxia. These include MDC1, SFPQ, NONO, SSRP1, SUPT16H, INTS3, APTX, and PRPF19. MDC1 and SFPQ-NONO heterodimer have been shown to facilitate the DNA repair in NHEJ pathway (41, 42). SSRP1-SUPT16H dimers, INTS3, APTX, and PRPF19 also have been proven to stabilize DSB repair (43–46). The expression level and heterodimer changes of Ku70 and Ku80 were validated by LC-MS/MS-MRM and Western blot (Figs. 1B and 1C). In dynamic MRM quantification results, TBB6 and ACTN1, housekeeping proteins, and ATP5B were used as internal controls as their expression levels assessed by iTRAQ were not modulated by hypoxia. The peptides of TBB6, ACTN1, and ATP5B with several strong transitions as shown in Supplemental Table S4 were quantified in dynamic MRM with ratios of 0.92 ± 0.07, 0.90 ± 0.07, and 0.96 ± 0.09 respectively. The copy numbers of the two peptides from Ku70 with seven transitions showed significant increase to 2.86 ± 0.78 folds under hypoxia stress. The intensity of four peptides of Ku80 under hypoxic condition was also higher (2.05 ± 0.69) than that of peptides under normoxic condition (Fig. 1C and Supplemental Table S4). Their change ratios are much higher than those of Ku70 and Ku80 in iTRAQ quantification results (with ratios 1.1–1.2). In addition, the increase of another key component of DNA-PK heterotrimer, PRKDC1, and other components of the NHEJ pathway (ILF2, NONO, SP16H, PRP19 SFPQ, SSRP1, and APTX) during hypoxia was also verified by MRM method (Fig. 1C and Supplemental Table S4). The slight changes of Ku70 and Ku80 observed from iTRAQ data were further validated by SDS-PAGE WB that confirmed increased expression (with ratios less than 2.03) of both proteins as shown in Fig. 1B (the upper two panels). As formation of Ku70/Ku80 heterodimer are prerequisites for functional activity, protein extracts from the tumor cells were separated on native gels and analyzed for the presence of Ku70/Ku80 heterodimer using antibodies against either Ku70 or Ku80. Both antibodies detected the same single band on the native gel which indicated the Ku70/Ku80 dimer position (the lowest panel in Fig. 1B). The WB results clearly showed obvious increase of heterodimer under hypoxia. To further confirm the result and avoid the interference from cellular proteins, we isolated chromatin of A431 under normoxia and hypoxia using sucrose gradient, and quantified the chromatin binding proteins using iTRAQ analysis. Result showed that the key proteins in NHEJ pathway, such as Ku70, Ku80, PRKDC1, associated with chromatin increased under hypoxic condition (unpublished data). In 2008, for the first time, a decrease in Ku70 and Ku80 was found to be present in more hypoxic cervical carcinoma tissue by immunohistochemistry; however, the results were not further validated (38). Our quantitative proteomic data for the first time showed that hypoxia increased most components of the NHEJ pathways in A431. As these components have been implicated in mediating ionizing radiation resistance, our data also provided a possible molecular mechanism for hypoxia-induced radiation resistance.
Fig. 1.
Hypoxia activates NHEJ pathway proteins mainly responsible for DNA repair under irradiation. A, Change patterns of NHEJ pathway proteins because of hypoxia or reoxygenation from iTRAQ data; B, The increase of Ku70, Ku80 and their heterodimer induced by hypoxia was confirmed by Western blot; In these WB experiments, the cell proteins were extracted by the solution of 1% Triton X-100 in 20 mm Tris-HCl (pH 7.4), 150 mm NaCl. Ku70 and Ku80 monomers were separated by SDS-PAGE and detected by individual antibody (the upper three panels). For the dimer quantification, the same proteins with those loaded on SDS-PAGE gel were separated in a native gel and the dimer was detected by both antibodies (the lower two panels). C, The increase of three components of DNA-PK heterotrimer, Ku70, Ku80, and PRKDC1 was further proved by MRM. The retention time for PRKDC1 had some shift between Nx and Hx samples for some unknown reasons. The other transitions also occurred with the same shift between the two samples. Hx, Hypoxic sample; Nx, Normoxic sample.
Hypoxia Induced Cell Migration Through Metabolic Switch from TCA Cycle to Glycolytic Pathway
It has been well known that tumor cells exhibit increased rates of glucose uptake and glycolysis (47). Increased glycolysis in tumor cells is regarded as an effect of intratumoral hypoxia. Under hypoxic conditions, cells switch their form of glucose metabolism from the oxygen-dependent tricarboxylic acid (TCA) cycle to glycolysis, an oxygen-independent metabolic pathway. HIF-1 has been shown to activate transcription of genes that encode glucose transporters GLUT1, GLUT3, and all glycolytic enzymes (2, 48). In our iTRAQ data, the enzymes that catalyze the four steps in glycolysis pathway, hexokinase (HK), glucose 6-phosphate isomerase (G6PI), fructose-bisphosphate aldolase (ALDOA and FBPA) and biphosphoglycerate kinase (PGK) were all found to be up-regulated after hypoxia. The enzyme that regulates pyruvic acid efflux, lactate dehygrogenase A (LDHA), was also significantly up-regulated under hypoxia (Fig. 2A). Surprisingly, enzymes that are involved in the TCA cycle such as pyruvate dehydrogenase (PDHB), citrate synthase (CS), isocitrate degydrogenase (IDH2), a-ketoglutarate dehydrogenase (DHTKD1), fumarase (FH), malate dehydrogenase(MDH1) and pyruvate carboxylase (PC) were down-regulated under hypoxia (Fig. 2B). LC-MS/MS-MRM analysis results showed that the enzymes of HK, ALDOA, PGK1, and GPI in glycolysis pathway were also found obviously up-regulated as detected by iTRAQ analysis under hypoxia (Supplemental Table S4). Increased glycolysis has been associated with tumor invasiveness, metastasis and/or resistance to therapies (49). Recently, Cai et al. highlighted this relationship between glycolysis and metastasis in cancer cells by knocking down LDHA or overexpressing PDHB to force pyruvate into TCA cycle rather than the glycolysis process. This inhibited cellular growth and migration of gastric cancer cells (50). Our iTRAQ and MRM quantification data both indicated the increase of LDHA and the decrease of PDHB after hypoxia treatment (Figs. 2A, 2B, and Supplemental Table S4). In support of this link between glycolysis and metastasis, Izumi et al. also reported that knockdown of monocarboxylate transporters (MCT1 and MCT4) led to reduced invasiveness of human lung cancer cells (51). Because MCT1 and MCT4 transport monocarboxylate such as lactate across membranes and are important in maintaining continuous glycolysis, their knockdown would result in reduced glycolysis. In view of this, the up-regulation of monocarboxylate transporter, SLC16A1 (MCT1) under hypoxic conditions as observed in our study (Fig. 2A) would enhance continuous glycolysis. In addition to MCT1, glucose transporter SLC2A1 (GLUT1) was also found up-regulated in iTRAQ and MRM quantification results (Fig. 2A and Supplemental Table S4). Because down-regulation of GLUT1 inhibits cellular glucose uptake, growth, and invasiveness of MG63 cells (52), its up-regulation under hypoxia would enhance cellular glucose uptake, growth, and invasiveness. Based on these data, we hypothesized that cells increased proteins that will enhance glycolysis and promote metastasis during hypoxia.
2-Deoxy-d-glucose (2-DG) is one of the best characterized glycolysis inhibitors in animal model studies and human clinical trials (53, 54). In scratch-wound assay, 2-DG at a higher concentration was used to treat A431 cells and suppress the glycolysis pathway. The cell migration was found to be significantly inhibited when compared with that of the control cells under hypoxia and this reduced migration appeared to show positive correlation to the concentration of 2-DG (Fig. 2C). After exposure of A431 cells to 10 mm or 15 mm 2-DG for 20 h, cell migration into the wounded region of A431 cells was reported to be 65.9 ± 9.4% and 36.3 ± 5.9% as compared with control cells, respectively (Fig. 2D). The inhibition of 2-DG to cell glycolysis pathway was confirmed by metabolite analysis using LC-ESI-Q-TOF. The TIC of control metabolite sample had a slight higher peak than that of 2-DG treated cells, which indicated the loading amount of control and 2-DG treated samples was similar and the normalization with protein content was relatively accurate (Supplemental Fig. S4_A_). However, the EIC peaks of one intermediate of glycolysis pathway, Fructose 1,6-biphosphate (FBP, m/z:339.12), extracted from TICs had obvious difference. The area of EIC peak from control sample is about 12-fold of that of 2-DG treated sample (Supplemental Fig. S4_B_). FBP level in normoxic and hypoxic A431 was also detected using the same method. This glycolysis metabolite in hypoxia sample is up-regulated compared with normoxia sample even with a lower TIC in hypoxia sample during MS analysis (Supplemental Figs. S4_C_ and S4_D_). Supplemental Fig. S4_E_ is referred to the MS/MS fragments of the precursor 339.12. The m/z and intensity of these MS/MS fragments were input to Massbank database for metabolite identification (http://www.massbank.;1jp/MetaboIdentif.html). Only the Fructose 1,6-biphosphate of metabolite was matched. These results clearly showed that 2-DG inhibited, but hypoxia activated glycolysis pathway. Though 2-DG also resulted in the inhibition of glycan biosynthesis of glycoproteins, thus reducing the glycoproteins in cells and cell adhesion, treatment with glycan biosynthesis inhibitor (1-deoxynojirimycin hydrochloride, a glucose analog that inhibits α-glucosidase I and II activity) during the scratch assay for 20 h had no effects in cell migration (data not shown). These results indicate that the activation of glycolysis under hypoxic conditions is associated with enhanced A431 cell migration ability.
Hypoxia Induced Overexpression of Intergrins and Their Ligands: Implication for Increased Metastasis
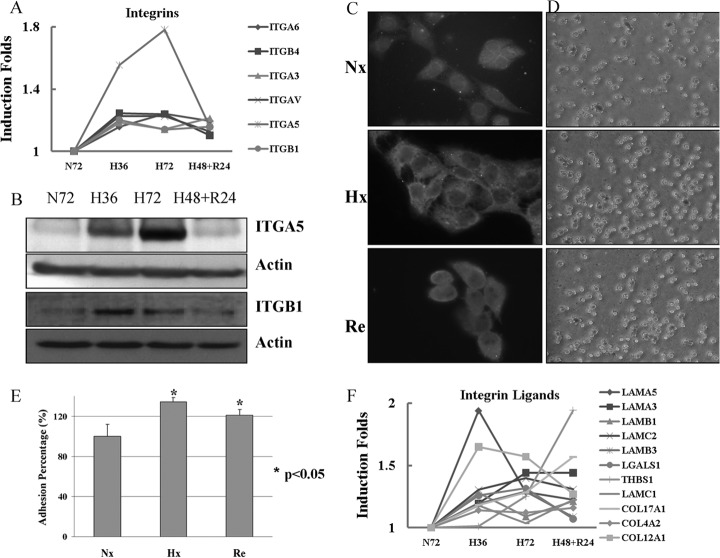
Integrins, a major family of adhesion proteins, are involved in numerous physiological and pathological processes, including inflammation, wound repair, proliferation, differentiation, and apoptosis (55, 56). In particular, they play a crucial role in metastasis. It is well established that fibronectin-integrin complexes played direct and indirect roles in signaling pathways that are involved in adhesion, migration, apoptosis, and growth (57). Expression of fibronectins and integrins is up-regulated in migratory cells in promoting of cell migration (58). Each integrin is composed of two subunits, namely α and β subunits. 18α and 8β integrin subunits assemble into 24 distinct receptors in mammals (59). Our iTRAQ data revealed the up-regulation of several integrin subunits under hypoxic conditions. The changes were partially reversed by reoxygenation (Fig. 3A). ITGα5β1 is one of the important integrin molecules. The increase expression of ITGA5 and ITGB1 subunits under hypoxia was validated by WB and LC-MS/MS-MRM methods (Fig. 3B and Supplemental Table S4). Both results clearly showed that hypoxia induced the expression of the two subunits. Cell staining results not only showed the up-regulation of ITGA5 but also their recruitment to the plasma membrane under hypoxic conditions. ITGA5 appears to be evenly distributed in the normoxic A431 cells, whereas it has been observed to reside in the plasma membrane of both hypoxic and reoxygenated cells (Fig. 3C). LC-MS/MS-MRM experiment also confirmed up-regulation of other integrin molecules (ITGA3, ITGA6, and ITGB4) in A431 cells under hypoxia, indicating hypoxia may increase cell adhesion (Supplemental Table S4). We thus examined the adhesion ability of A431 cells on matrix-coated dishes after pre-exposure to hypoxia. The adhesion assay proved that hypoxia induces increased adhesiveness of A431 cells pre-exposed to hypoxia within 20 mins of seeding onto the fibronectin-coated dishes (Fig. 3D). The adhesion of the hypoxic and reoxygenated cells to fibronectin is 134.5 ± 4.2% and 121.4 ± 5.5% of that of normoxic cells, respectively (Fig. 3E). This increase in cell adhesion was consistent with the increase of several integrin molecules and ITGA5 recruitment to the plasma membrane. It has been reported that up-regulation of ITGA5 and fibronectin mediated by hypoxia, is responsible for the increased invasiveness of cancer cells (60). In hypoxia, the loss of E-cadherin leads to the up-regulation of ITGA5, resulting in the increased migration of extravillous trophoblast cells during early implantation (29). Integrins have a wide binding range to matrix proteins, which include laminin, fibronectin, trombospondin, vitronectin, and the various types of collagen (61). The cell adhesion ligands including several lamimin and collagen subunits were also found to be up-regulated under hypoxia (Fig. 3F). Hypoxia induced factor 1 (HIF-1α) directly regulates the promoter activity of laminin α3 chain of laminin-332 (formerly laminin-5) and contributes to the migration of keratinocyte by increasing the expression of laminin-332 (62).
Fig. 3.
Hypoxia increases the expression level of integrin subunits and their recruitment to plasma membrane and even induces the expression of their partners. A, Change patterns of integrin subunits because of hypoxia or reoxygenation from iTRAQ data; B, The higher expression level of ITGA5 and ITGB1 under hypoxia was confirmed by Western blot. C, Cell immunostaining of ITGA5 clearly indicated that the adhesion molecule was prone to localize at the cell membrane under hypoxia. Magnification, ×630; D and E, Images and quantification of adhesion assay of A431 to FN under Nx, Hx, and Reox condition. Magnification, ×100; F, Change patterns of integrin ligands because of hypoxia or reoxygenation from iTRAQ data.
Hypoxia Induced Higher Turnover Rates of Glycoproteins
Glycoproteins play important roles in a wide range of biological events, including bacterial and viral infections, the metastasis of tumor cells, immune responses, and fertilization and differentiation (63). There are two major types of glycoproteins in mammalian cells, _N_-linked and O-linked glycoproteins. The synthesis and breakdown of _N_-linked glycoproteins have been studied intensively. The biosynthesis of the various types of _N_-linked oligosaccharide structures involves two series of reactions: 1) the formation of the lipid-linked saccharide precursor, Glc3Man9(GlcNAc)2-pyrophosphoryl-dolichol, by the stepwise addition of GlcNAc, mannose and glucose to dolichyl-P, and 2) the removal of glucose and mannose by membrane-bound glycosidases and the addition of GlcNAc, galactose, sialic acid, and fucose by Golgi localized glycosyltransferases to produce different complex oligosaccharide structures (64). Glycosidases are key enzymes involved in the biosynthesis of _N_-linked glycoproteins as they hold critical role in the formation of hybrid and complex types of _N_-linked oligosaccharides (65). Glycoproteins are degraded in lysosomes. Once inside the lysosome, glycoproteins are broken down by a combination of proteases and glycosidases. We found that, for the first time, the turnover of glycoproteins is regulated and promoted by increase of enzymes involved in biosynthesis and breakdown of glycoproteins under hypoxia (Fig. 4A and 4B).
The iTRAQ data showed that the enzymes (ALG1, ALG2 and DPM1), involved in the formation of lipid-linked saccharide precursor by transferring mannose in _N_-linked oligosaccharide formation step 1, were up-regulated under hypoxia. The glycosidase (MA1B1and MAN2A1) responsible for the removal of mannose, MOGS for glucose removal and the glycosyltransferases (Oligosaccharyltransferase complex, OST) in the step 2 of _N_-linked glycan synthesis were also found to be up-regulated. OST exists in different forms, namely RPN1, RPN2, OST48, DAD1, OSTC, KRTCAP2, and either STT3A or STT3B, which catalyzes the transfer of a high mannose oligosaccharide from a lipid-linked oligosaccharide donor to an asparagine residue within an Asn-X-Ser/Thr consensus motif. RPN1, RPN2, OST48, DAD1, OSTC, STT3A, and STT3B were found in higher expression in hypoxia. In addition, the enzymes for O-linked glycan formation (GALNT2, C1GALT1, and GLT25D1) were also up-regulated in hypoxia. The enzymes for glycoprotein catabolism in lysosomes including proteases and glycosidases were induced by hypoxia too. The cathepsins (CTSA/B/C/D/H/Z) for polypeptide degradation and glycosidases (HEXA, HEXB, MAN2B1, GLB1, and FUCA1) for cleavage of N-acetyl hexosamines, α-mannose, β-galactose, and α-fucose, respectively, were found to undergo similar change pattern during hypoxia and reoxgenation (Fig. 4B).
We speculated that the quantitative changes in glycosylation enzymes during hypoxia would affect glycoproteome and that this change would be associated with cell metastasis because of the important roles of glycoproteins in tumor cell metastasis. 1-Deoxynojirimycin (1-DJ), an analog of glucose that contains an NH group substituting for the oxygen atom of the pyranose ring, inhibits Golgi alpha-glucosidase and thus inhibits glycoprotein synthesis. This drug was used to treat A431 and perturb the glycoprotein-turnover under hypoxia. Short term drug treatment (20 h) exhibited no prominent effect on cell migration in scratch assay and this result also was supported by the reported data (66). However, once A431 cells were treated with 100 μm 1-DJ for 48 h, the cells displayed slower migration than nontreated cells under hypoxia (Fig. 4C). The migration into wound of A431 cells treated by 100 μm 1-DJ is only 72.3% ± 12.6% of that of control cells (Fig. 4E). The lectin binding result showed that the synthesis of glycoproteins in A431 was inhibited after 1-DJ and hypoxia treatment (Fig. 4D). These data indicate that hypoxia contributes to A431 enhanced migration by increasing glycoprotein synthesis and modulating glycoprotein turnover.
Hypoxia Promoted A431 Migration by Suppression of STAT1 Pathway
Thus far, studies determining the influence of hypoxia on protein expressions have been restricted to proteins with enhanced expressions. Only a few studies have been done on proteins whose expression is down-regulated by hypoxia (35). STAT1, the first described member of the STAT transcription factor family, is the master transcription factor for IFN-related intracellular signaling (67). STAT1 has two isoforms, STAT1α and STAT1β. The β isoform lacks the 38 residues encompassing the transcription activation domain at C-terminal and is transcriptionally inactive (68). STAT1α is phosphorylated by JAK1/2 kinases at the Tyr701 position and then translocates to the nucleus where it binds to GAS (IFNc-activated sequence) promoter elements, thereby activating several hundred genes (69). Phosphorylation of STAT1α at S727 is essential for activating transcription of target genes and interaction with proteins, such as BRCA1 and MCM-5, potentiating STAT1-mediated transcription (70–73).
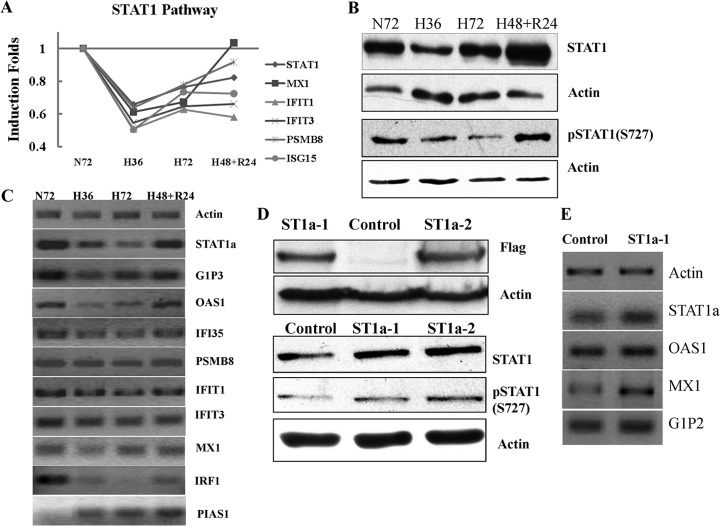
STAT1 and its downstream proteins (MX1, IFIT1, IFIT3, ISG15, and PSMB8) were significantly down-regulated under hypoxia as reflected in our proteomic data (Fig. 5A), the suppression of STAT1, PSMB8, and IFIT1 was confirmed by MRM quantification (Supplemental Table S4). The significant decline of total STAT1 was further confirmed by WB experiment. The active isoform of STAT1 was also shown to be highly suppressed by hypoxia because the anti-pSTAT1(S727) only recognizes STAT1α isoform with transcriptional activity (Fig. 5B). The mRNA level changes of STAT1α detected by its specific primers and its downstream genes were validated by RT-PCR. Fig. 5C clearly indicated their suppressed transcription as a result of STAT1α down-regulation after hypoxia. STAT1 dramatically increases the transcription of IRF-1 through the interaction with NF-κB or in response to IFN (74, 75). IRF-1 transcription was suppressed in response to decreased STAT1 in RT-PCR; however, as a specific inhibitor of STAT1, PIAS1 showed a reverse finding with STAT1 (76). The alteration of STAT1 expression level because of hypoxia at present remains controversial. Several studies have reported that hypoxia led to activation of STAT1, especially in cardiomyocytes (77–80). Paradoxically, Ivanov et al. reported that STAT1 and its downstream genes could be repressed by a pVHL/HIF-1 target DEC1/STRA13 in carcinoma cells under hypoxia (81).
Fig. 5.
Confirmation of STAT1 pathway suppressed by hypoxia and validation of stable cell lines with STAT1α overexpression in A431; A, Change patterns of STAT1 and its downstream proteins because of hypoxia or reoxygenation; B, The down-regulation of STAT1 and pSTAT1α (S727) was confirmed by WB. C, The decrease of functional STAT1 represses the transcription of its downstream genes. D, The two positive stable cell lines with STAT1α overexpression were confirmed by WB using anti-FLAG, STAT1 and pSTAT1α (S727) antibodies. E, The activation of downstream genes of STAT1 in the stable cell lines indicated that the cell lines indeed have more active STAT1α.
To get more evidence of STAT1 functions under hypoxia, two A431 stable cell lines with STAT1α overexpression were screened for further studies. The human cDNA of STAT1α was inserted into p3xFLAG-CMV-10 expression vector and transfected into A431. G418 was used to screen the cell line stably overexpressing STAT1α. The positive clones were confirmed by WB using anti-FLAG and STAT1 antibodies. When compared with the control cell line transfected with empty p3xFLAG-CMV-10, FLAG-STAT1 was only detected in the STAT1α stable cell lines (A431-p3XFLAG-ST1a-1 and 2). The stable cell lines also showed higher expression level of STAT1 and phosphorylated STAT1α, in which A431-p3XFLAG-ST1a-2 has a higher expression of STAT1α than A431-p3XFLAG-ST1a-1 (Fig. 5D). RT-PCR results showed up-regulation of STAT1 downstream genes (Fig. 5E). In summary, the results highly suggested that the stable cell lines with more active STAT1α expressed could be used for functional studies.
MTT was used to evaluate the proliferation/survival of the STAT1α stable cell lines under nomorxia and hypoxia. Under nomorxia, there was no significant difference in the proliferation between control and STAT1α overexpression cell lines. But under hypoxia, STAT1α cell survival suffered a greater decline as compared with control cells and A431-p3XFLAG-ST1a-2 with the highest STAT1α level had a maximum decline (Fig. 6A). These results indicated that A431 cells increased their survivability through the down-regulation of STAT1 under hypoxia. STAT1 has been implicated in the inhibition of cell proliferation in several cell systems by inducing the expression of cyclin kinase inhibitors and caspases in response to extracellular stimuli (82–86). It is also reported that STAT1 can repress HIF-1-mediated transcription after hypoxia (87).
Because hypoxia induces cancer cell migration and causes functional suppression of STAT1, we went on further to examine the effects of STAT1 overexpression on the migration ability of cancer cells under hypoxia. In the scratch-wound assay, statistical results showed that STAT1α overexpression resulted in the repression of A431 cell migration. The migration of A431-p3XFLAG-ST1a-1 and 2 cells is only 82.9 ± 4.3% and 62.0 ± 5.4% of that of A431-p3XFLAG control cells (Figs. 6B and 6C). The migration of A431-p3XFLAG-ST1a-2 is significantly lower than that of A431-p3XFLAG-ST1a-1 because of its higher expression of STAT1α, which shows negative regulation of STAT1 in tumor metastasis. STAT1 has been reported to suppress the metastasis of STAT1-reconstituted RAD-105 cells through the reduction of MMP-2 and MMP-9 in vivo and arrest monocyte migration by modulating RAC/CDC42 pathways (88, 89). Our studies further revealed that STAT1α might suppress A431 cell migration by promoting cell adhesion. In adhesion assay, the adhesion ability of A431-p3XFLAG-ST1a-1 and 2 to fibronectin was much higher than that in control cell. After 30 mins incubation, the adhesion cell amount of A431-p3XFLAG-ST1a-1 and 2 is about 2.03 ± 0.03 and 2.28 ± 0.12 folds of that of A431-p3XFLAG (Figs. 6D and 6E). Cell adhesion receptors play important roles in promoting migration; however, the establishment of strong adhesion inhibits migration, which is fastest at optimum adhesion strength strong enough to support traction yet weak enough to allow rapid detachment of the rear of the cell (58). The above results provide evidences that hypoxia induces cell migration by down-regulation of STAT1 because overexpression STAT1 can inhibit migration by enhancing cell adhesion.
CONCLUSION
In this study, based on iTRAQ quantification data, the pathways associated with tumor radioresistance and migration induced by hypoxia were identified and validated. For the first time, most key molecules in NHEJ pathway, which occurs more often than homologous repair in radiated cells, were found up-regulated under hypoxia and confirmed by LC-MS/MS-MRM or WB. Thus, the proteins in this pathway may provide new targets for molecular therapy in support to radiotherapy (90). In addition, we reported hypoxia perturbed the expression of enzymes in glycoprotein synthesis and degradation to modulate glycoprotein turnover. Inhibiting glycoprotein synthesis by small molecule inhibitors under hypoxia led to decrease in cell migration. Glycosylation undergoes a dramatic transformation during cancer malignancy. But only a few structural changes have been frequently correlated with tumor progression by altering specific enzymes for further modification of glycans in Golgi (91). It is an intriguing question to find whether improvement of glycoprotein turnover contributes to the glycosylation transformation during tumor malignancy. STAT1 and its downstream genes were significantly suppressed by hypoxia. The repression is believed to promote cell migration because overexpression of STAT1α can inhibit migration by enhancing cell adhesion. STAT1 has been reported to be inhibited by HIF-1 (81). How about the NHEJ and glycoprotein turnover pathway proteins? It is our future work to knockdown HIF-1 and to investigate its impact on the two pathways under hypoxia. Our findings add in new dimension to our present understanding to tumor hypoxia and provide potential new therapy targets for tumor therapy.
Supplementary Material
Supplemental Fig. S1
Footnotes
* This work is supported by grants from Nanyang Technological University (RG 157/06, RG 61/06 and RG 51/10).
1 The abbreviations used are:
HIF-1
hypoxia-inducible factor-1
iTRAQ
isobaric tag for relative and absolute quantification
MRM
multiple reaction monitoring
TEAB
triethylammonium bicarbonate
MMTS
methyl methanethiosulfonate
FDR
false discovery rate
EF
error factor
WB
Western blot
NHEJ
Nonhomologous end-joining
NHEJ1
Nonhomologous end-joining factor 1
2-DG
2-Deoxy-D-glucose
2-DE
Two-dimensional electrophoresis
DMEM
Dulbecco's modified Eagle's medium
IPA
Ingenuity Pathway Analysis
DSB
DNA double strand break
TCA
tricarboxylic acid
Ku70
X-ray repair cross-complementing protein 6
Ku80
X-ray repair cross-complementing protein 5
STAT1
Signal transducer and activator of transcription 1
RAD51
DNA repair protein RAD51 homolog 1
BRCA1/2
breast cancer type 1/2 susceptibility protein
XRCC3/4
DNA repair protein XRCC3/4
SCX
Strong Cation Exchange
ITGB1
integrin beta 1
ITGA5
integrin alpha 5
LDHA
L-lactate dehydrogenase A chain
PDHB
Pyruvate dehydrogenase E1 component subunit beta
IFN
interferon
IRF-1
Interferon regulatory factor 1
PIAS1
E3 SUMO-protein ligase PIAS1
JNK/1/2
c-Jun N-terminal kinase1/2
The other abbreviations of proteins used in this article were listed in Supplemental Table S2.
REFERENCES
- 1.Brown J. M., Giaccia A. J. (1998) The unique physiology of solid tumors: opportunities (and problems) for cancer therapy. Cancer Res. 58, 1408–1416 [PubMed] [Google Scholar]
- 2.Harris A. L. (2002) Hypoxia–a key regulatory factor in tumour growth. Nat. Rev. Cancer 2, 38–47 [DOI] [PubMed] [Google Scholar]
- 3.Park J. E., Tan H. S., Datta A., Lai R. C., Zhang H., Meng W., Lim S. K., Sze S. K. (2010) Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol. Cell. Proteomics 9, 1085–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson W. R., Hay M. P. (2011) Targeting hypoxia in cancer therapy. Nat. Rev. Cancer 11, 393–410 [DOI] [PubMed] [Google Scholar]
- 5.Gray L. H., Conger A. D., Ebert M., Hornsey S., Scott O. C. (1953) The concentration of oxygen dissolved in tissues at the time of irradiation as a factor in radiotherapy. Br. J. Radiol. 26, 638–648 [DOI] [PubMed] [Google Scholar]
- 6.Moulder J. E., Rockwell S. (1987) Tumor hypoxia: its impact on cancer therapy. Cancer Metastasis Rev. 5, 313–341 [DOI] [PubMed] [Google Scholar]
- 7.Le Q. T., Giaccia A. J., Brown J. M. (2005) Squamous Cell Head and Neck Cancer: The role of tumor hypoxia in head and neck cancer radiotherapy, pp. 145–163, Cleveland Clinic Foundation [Google Scholar]
- 8.Liu J., Zhang J., Wang X., Li Y., Chen Y., Li K., Zhang J., Yao L., Guo G. (2010) HIF-1 and NDRG2 contribute to hypoxia-induced radioresistance of cervical cancer Hela cells. Exp. Cell Res. 316, 1985–1993 [DOI] [PubMed] [Google Scholar]
- 9.Moeller B. J., Dewhirst M. W. (2006) HIF-1 and tumour radiosensitivity. Br. J. Cancer 95, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markowska J., Grabowski J. P., Tomaszewska K., Kojs Z., Pudelek J., Skrzypczak M., Sobotkowski J., Emerich J., Olejek A., Filas V. (2007) Significance of hypoxia in uterine cervical cancer. Multicentre study. Eur. J. Gynaecol. Oncol. 28, 386–388 [PubMed] [Google Scholar]
- 11.Dellas K., Bache M., Pigorsch S. U., Taubert H., Kappler M., Holzapfel D., Zorn E., Holzhausen H. J., Haensgen G. (2008) Prognostic impact of HIF-1alpha expression in patients with definitive radiotherapy for cervical cancer. Strahlenther Onkol. 184, 169–174 [DOI] [PubMed] [Google Scholar]
- 12.Tribius S., Pidel A., Casper D. (2001) ATM protein expression correlates with radioresistance in primary glioblastoma cells in culture. Int. J. Radiat. Oncol. Biol. Phys. 50, 511–523 [DOI] [PubMed] [Google Scholar]
- 13.Munshi A., Kurland J. F., Nishikawa T., Tanaka T., Hobbs M. L., Tucker S. L., Ismail S., Stevens C., Meyn R. E. (2005) Histone deacetylase inhibitors radiosensitize human melanoma cells by suppressing DNA repair activity. Clin. Cancer Res. 11, 4912–4922 [DOI] [PubMed] [Google Scholar]
- 14.Meng A. X., Jalali F., Cuddihy A., Chan N., Bindra R. S., Glazer P. M., Bristow R. G. (2005) Hypoxia down-regulates DNA double strand break repair gene expression in prostate cancer cells. Radiother. Oncol. 76, 168–176 [DOI] [PubMed] [Google Scholar]
- 15.Bindra R. S., Schaffer P. J., Meng A., Woo J., Maseide K., Roth M. E., Lizardi P., Hedley D. W., Bristow R. G., Glazer P. M. (2004) Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol. Cell. Biol. 24, 8504–8518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bindra R. S., Schaffer P. J., Meng A., Woo J., Maseide K., Roth M. E., Lizardi P., Hedley D. W., Bristow R. G., Glazer P. M. (2005) Alterations in DNA repair gene expression under hypoxia: elucidating the mechanisms of hypoxia-induced genetic instability. Ann. N.Y. Acad. Sci. 1059, 184–195 [DOI] [PubMed] [Google Scholar]
- 17.Bindra R. S., Gibson S. L., Meng A., Westermark U., Jasin M., Pierce A. J., Bristow R. G., Classon M. K., Glazer P. M. (2005) Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res. 65, 11597–11604 [DOI] [PubMed] [Google Scholar]
- 18.Graeber T. G., Osmanian C., Jacks T., Housman D. E., Koch C. J., Lowe S. W., Giaccia A. J. (1996) Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature 379, 88–91 [DOI] [PubMed] [Google Scholar]
- 19.Young S. D., Marshall R. S., Hill R. P. (1988) Hypoxia induces DNA overreplication and enhances metastatic potential of murine tumor cells. Proc. Natl. Acad. Sci. U.S.A. 85, 9533–9537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brizel D. M., Scully S. P., Harrelson J. M., Layfield L. J., Bean J. M., Prosnitz L. R., Dewhirst M. W. (1996) Tumor oxygenation predicts for the likelihood of distant metastases in human soft tissue sarcoma. Cancer Res. 56, 941–943 [PubMed] [Google Scholar]
- 21.Cairns R. A., Hill R. P. (2004) Acute hypoxia enhances spontaneous lymph node metastasis in an orthotopic murine model of human cervical carcinoma. Cancer Res. 64, 2054–2061 [DOI] [PubMed] [Google Scholar]
- 22.Fyles A., Milosevic M., Hedley D., Pintilie M., Levin W., Manchul L., Hill R. P. (2002) Tumor hypoxia has independent predictor impact only in patients with node-negative cervix cancer. J. Clin. Oncol. 20, 680–687 [DOI] [PubMed] [Google Scholar]
- 23.Liotta L. A., Kleinerman J., Catanzaro P., Rynbrandt D. (1977) Degradation of basement membrane by murine tumor cells. J. Natl. Cancer Inst. 58, 1427–1431 [DOI] [PubMed] [Google Scholar]
- 24.Montagner M., Enzo E., Forcato M., Zanconato F., Parenti A., Rampazzo E., Basso G., Leo G., Rosato A., Bicciato S., Cordenonsi M., Piccolo S. (2012) SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature 487, 380–384 [DOI] [PubMed] [Google Scholar]
- 25.Sahai E. (2007) Illuminating the metastatic process. Nat. Rev. Cancer 7, 737–749 [DOI] [PubMed] [Google Scholar]
- 26.Horwitz A. R., Parsons J. T. (1999) Cell migration–movin' on. Science 286, 1102–1103 [DOI] [PubMed] [Google Scholar]
- 27.Yoon S. O., Park S. J., Yun C. H., Chung A. S. (2003) Roles of matrix metalloproteinases in tumor metastasis and angiogenesis. J. Biochem. Mol. Biol. 36, 128–137 [DOI] [PubMed] [Google Scholar]
- 28.Liao S. H., Zhao X. Y., Han Y. H., Zhang J., Wang L. S., Xia L., Zhao K. W., Zheng Y., Guo M., Chen G. Q. (2009) Proteomics-based identification of two novel direct targets of hypoxia-inducible factor-1 and their potential roles in migration/invasion of cancer cells. Proteomics 9, 3901–3912 [DOI] [PubMed] [Google Scholar]
- 29.Arimoto-Ishida E., Sakata M., Sawada K., Nakayama M., Nishimoto F., Mabuchi S., Takeda T., Yamamoto T., Isobe A., Okamoto Y., Lengyel E., Suehara N., Morishige K., Kimura T. (2009) Up-regulation of alpha5-integrin by E-cadherin loss in hypoxia and its key role in the migration of extravillous trophoblast cells during early implantation. Endocrinology 150, 4306–4315 [DOI] [PubMed] [Google Scholar]
- 30.Kim S. H., Xia D., Kim S. W., Holla V., Menter D. G., Dubois R. N. (2010) Human enhancer of filamentation 1 Is a mediator of hypoxia-inducible factor-1alpha-mediated migration in colorectal carcinoma cells. Cancer Res. 70, 4054–4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuonen F., Secondini C., Ruegg C. (2012) Molecular Pathways: Emerging pathways mediating growth, invasion and metastasis of tumors progressing in an irradiated microenvironment. Clin. Cancer Res. [DOI] [PubMed] [Google Scholar]
- 32.Stockwin L. H., Blonder J., Bumke M. A., Lucas D. A., Chan K. C., Conrads T. P., Issaq H. J., Veenstra T. D., Newton D. L., Rybak S. M. (2006) Proteomic analysis of plasma membrane from hypoxia-adapted malignant melanoma. J. Proteome Res. 5, 2996–3007 [DOI] [PubMed] [Google Scholar]
- 33.Sørensen B. S., Horsman M. R., Vorum H., Honoré B., Overgaard J., Alsner J. (2009) Proteins upregulated by mild and severe hypoxia in squamous cell carcinomas in vitro identified by proteomics. Radiother. Oncol. 92, 443–449 [DOI] [PubMed] [Google Scholar]
- 34.Vorum H., Østergaard M., Hensechke P., Enghild J. J., Riazati M., Rice G. E. (2004) Proteomic analysis of hyperoxia-induced responses in the human choriocarcinoma cell line JEG-3. Proteomics 4, 861–867 [DOI] [PubMed] [Google Scholar]
- 35.Kumar G. K., Klein J. B. (2004) Analysis of expression and posttranslational modification of proteins during hypoxia. J. Appl. Physiol. 96, 1178–1186; discussion 1170–1172 [DOI] [PubMed] [Google Scholar]
- 36.Dettmer K., Nürnberger N., Kaspar H., Gruber M. A., Almstetter M. F., Oefner P. J. (2011) Metabolite extraction from adherently growing mammalian cells for metabolomics studies: optimization of harvesting and extraction protocols. Anal. Bioanal. Chem. 399, 1127–1139 [DOI] [PubMed] [Google Scholar]
- 37.Finley L. W., Carracedo A., Lee J., Souza A., Egia A., Zhang J., Teruya-Feldstein J., Moreira P. I., Cardoso S. M., Clish C. B., Pandolfi P. P., Haigis M. C. (2011) SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell 19, 416–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lara P. C., Lloret M., Clavo B., Apolinario R. M., Bordon E., Rey A., Falcon O., Alonso A. R., Belka C. (2008) Hypoxia downregulates Ku70/80 expression in cervical carcinoma tumors. Radiother. Oncol. 89, 222–226 [DOI] [PubMed] [Google Scholar]
- 39.Chen D. J., Nirodi C. S. (2007) The epidermal growth factor receptor: a role in repair of radiation-induced DNA damage. Clin. Cancer Res. 13, 6555–6560 [DOI] [PubMed] [Google Scholar]
- 40.Ting N. S., Chan D. W., Lintott L. G., Allalunis-Turner J., Lees-Miller S. P. (1999) Protein-DNA complexes containing DNA-dependent protein kinase in crude extracts from human and rodent cells. Radiat. Res. 151, 414–422 [PubMed] [Google Scholar]
- 41.Lou Z., Chen B. P., Asaithamby A., Minter-Dykhouse K., Chen D. J., Chen J. (2004) MDC1 regulates DNA-PK autophosphorylation in response to DNA damage. J. Biol. Chem. 279, 46359–46362 [DOI] [PubMed] [Google Scholar]
- 42.Salton M., Lerenthal Y., Wang S. Y., Chen D. J., Shiloh Y. (2010) Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle 9, 1568–1576 [DOI] [PubMed] [Google Scholar]
- 43.Kumari A., Mazina O. M., Shinde U., Mazin A. V., Lu H. (2009) A role for SSRP1 in recombination-mediated DNA damage response. J. Cell. Biochem. 108, 508–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y., Bolderson E., Kumar R., Muniandy P. A., Xue Y., Richard D. J., Seidman M., Pandita T. K., Khanna K. K., Wang W. (2009) HSSB1 and hSSB2 form similar multiprotein complexes that participate in DNA damage response. J. Biol. Chem. 284, 23525–23531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clements P. M., Breslin C., Deeks E. D., Byrd P. J., Ju L., Bieganowski P., Brenner C., Moreira M. C., Taylor A. M., Caldecott K. W. (2004) The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair 3, 1493–1502 [DOI] [PubMed] [Google Scholar]
- 46.Mahajan K. N., Mitchell B. S. (2003) Role of human Pso4 in mammalian DNA repair and association with terminal deoxynucleotidyl transferase. Proc. Natl. Acad. Sci. U.S.A. 100, 10746–10751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boros L. G., Lee W. N., Go V. L. (2002) A metabolic hypothesis of cell growth and death in pancreatic cancer. Pancreas 24, 26–33 [DOI] [PubMed] [Google Scholar]
- 48.Chen C., Pore N., Behrooz A., Ismail-Beigi F., Maity A. (2001) Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. J. Biol. Chem. 276, 9519–9525 [DOI] [PubMed] [Google Scholar]
- 49.Mikuriya K., Kuramitsu Y., Ryozawa S., Fujimoto M., Mori S., Oka M., Hamano K., Okita K., Sakaida I., Nakamura K. (2007) Expression of glycolytic enzymes is increased in pancreatic cancerous tissues as evidenced by proteomic profiling by two-dimensional electrophoresis and liquid chromatography-mass spectrometry/mass spectrometry. Int. J. Oncol. 30, 849–855 [PubMed] [Google Scholar]
- 50.Cai Z., Zhao J. S., Li J. J., Peng D. N., Wang X. Y., Chen T. L., Qiu Y. P., Chen P. P., Li W. J., Xu L. Y., Li E. M., Tam J. P., Qi R. Z., Jia W., Xie D. (2010) A combined proteomics and metabolomics profiling of gastric cardia cancer reveals characteristic dysregulations in glucose metabolism. Mol. Cell. Proteomics 9, 2617–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Izumi H., Takahashi M., Uramoto H., Nakayama Y., Oyama T., Wang K. Y., Sasaguri Y., Nishizawa S., Kohno K. (2011) Monocarboxylate transporters 1 and 4 are involved in the invasion activity of human lung cancer cells. Cancer Sci. 102, 1007–1013 [DOI] [PubMed] [Google Scholar]
- 52.Fan J., Zhou J. Q., Yu G. R., Lu D. D. (2010) Glucose transporter protein 1-targeted RNA interference inhibits growth and invasion of the osteosarcoma cell line MG63 in vitro. Cancer Biother. Radiopharm. 25, 521–527 [DOI] [PubMed] [Google Scholar]
- 53.Maschek G., Savaraj N., Priebe W., Braunschweiger P., Hamilton K., Tidmarsh G. F., De Young L. R., Lampidis T. J. (2004) 2-deoxy-D-glucose increases the efficacy of adriamycin and paclitaxel in human osteosarcoma and non-small cell lung cancers in vivo. Cancer Res. 64, 31–34 [DOI] [PubMed] [Google Scholar]
- 54.Zhong D., Xiong L., Liu T., Liu X., Liu X., Chen J., Sun S. Y., Khuri F. R., Zong Y., Zhou Q., Zhou W. (2009) The glycolytic inhibitor 2-deoxyglucose activates multiple prosurvival pathways through IGF1R. J. Biol. Chem. 284, 23225–23233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilcrease M. Z., Truong L., Brown R. W. (1996) Correlation of very late activation integrin and CD44 expression with extrarenal invasion and metastasis of renal cell carcinomas. Hum. Pathol. 27, 1355–1360 [DOI] [PubMed] [Google Scholar]
- 56.Hynes R. O. (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 57.Chammas R., Brentani R. (1991) Integrins and metastases: an overview. Tumour Biol. 12, 309–320 [DOI] [PubMed] [Google Scholar]
- 58.Janik M. E., Lityńska A., Vereecken P. (2010) Cell migration-the role of integrin glycosylation. Biochim. Biophys. Acta 1800, 545–555 [DOI] [PubMed] [Google Scholar]
- 59.Leitinger B., Hohenester E. (2007) Mammalian collagen receptors. Matrix Biol. 26, 146–155 [DOI] [PubMed] [Google Scholar]
- 60.Ryu M. H., Park H. M., Chung J., Lee C. H., Park H. R. (2010) Hypoxia-inducible factor-1alpha mediates oral squamous cell carcinoma invasion via upregulation of alpha5 integrin and fibronectin. Biochem. Biophys. Res. Commun. 393, 11–15 [DOI] [PubMed] [Google Scholar]
- 61.Hogg N., Landis R. C. (1993) Adhesion molecules in cell interactions. Curr. Opin. Immunol. 5, 383–390 [DOI] [PubMed] [Google Scholar]
- 62.Fitsialos G., Bourget I., Augier S., Ginouvès A., Rezzonico R., Odorisio T., Cianfarani F., Virolle T., Pouysségur J., Meneguzzi G., Berra E., Ponzio G., Busca R. (2008) HIF1 transcription factor regulates laminin-332 expression and keratinocyte migration. J. Cell Sci. 121, 2992–3001 [DOI] [PubMed] [Google Scholar]
- 63.Kajimoto T., Node M. (2009) Inhibitors against glycosidases as medicines. Curr. Top. Med. Chem. 9, 13–33 [DOI] [PubMed] [Google Scholar]
- 64.Elbein A. D. (1991) Glycosidase inhibitors: inhibitors of N-linked oligosaccharide processing. FASEB J 5, 3055–3063 [DOI] [PubMed] [Google Scholar]
- 65.Kornfeld R., Kornfeld S. (1985) Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54, 631–664 [DOI] [PubMed] [Google Scholar]
- 66.Zhao Y., Liu W., Zhou Y., Zhang X., Murphy P. V. (2010) N-(8-(3-ethynylphenoxy)octyl-1-deoxynojirimycin suppresses growth and migration of human lung cancer cells. Bioorg. Med. Chem. Lett. 20, 7540–7543 [DOI] [PubMed] [Google Scholar]
- 67.Shuai K., Schindler C., Prezioso V. R., Darnell J. E., Jr. (1992) Activation of transcription by IFN-gamma: tyrosine phosphorylation of a 91-kD DNA binding protein. Science 258, 1808–1812 [DOI] [PubMed] [Google Scholar]
- 68.Schindler C., Levy D. E., Decker T. (2007) JAK-STAT signaling: from interferons to cytokines. J. Biol. Chem. 282, 20059–20063 [DOI] [PubMed] [Google Scholar]
- 69.Khodarev N. N., Roach P., Pitroda S. P., Golden D. W., Bhayani M., Shao M. Y., Darga T. E., Beveridge M. G., Sood R. F., Sutton H. G., Beckett M. A., Mauceri H. J., Posner M. C., Weichselbaum R. R. (2009) STAT1 pathway mediates amplification of metastatic potential and resistance to therapy. PLoS One 4, e5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wen Z., Zhong Z., Darnell J. E., Jr. (1995) Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82, 241–250 [DOI] [PubMed] [Google Scholar]
- 71.Decker T., Kovarik P. (2000) Serine phosphorylation of STATs. Oncogene 19, 2628–2637 [DOI] [PubMed] [Google Scholar]
- 72.Ouchi T., Lee S. W., Ouchi M., Aaronson S. A., Horvath C. M. (2000) Collaboration of signal transducer and activator of transcription 1 (STAT1) and BRCA1 in differential regulation of IFN-gamma target genes. Proc. Natl. Acad. Sci. U.S.A. 97, 5208–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang J. J., Zhao Y., Chait B. T., Lathem W. W., Ritzi M., Knippers R., Darnell J. E., Jr. (1998) Ser727-dependent recruitment of MCM5 by STAT1αlpha in IFN-gamma-induced transcriptional activation. EMBO J. 17, 6963–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pine R. (1997) Convergence of TNFalpha and IFNgamma signalling pathways through synergistic induction of IRF-1/ISGF-2 is mediated by a composite GAS/kappaB promoter element. Nucleic Acids Res. 25, 4346–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mamane Y., Heylbroeck C., Génin P., Algarté M., Servant M. J., LePage C., DeLuca C., Kwon H., Lin R., Hiscott J. (1999) Interferon regulatory factors: the next generation. Gene 237, 1–14 [DOI] [PubMed] [Google Scholar]
- 76.Liu B., Liao J., Rao X., Kushner S. A., Chung C. D., Chang D. D., Shuai K. (1998) Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. U.S.A. 95, 10626–10631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee M. Y., Joung Y. H., Lim E. J., Park J. H., Ye S. K., Park T., Zhang Z., Park D. K., Lee K. J., Yang Y. M. (2006) Phosphorylation and activation of STAT proteins by hypoxia in breast cancer cells. Breast 15, 187–195 [DOI] [PubMed] [Google Scholar]
- 78.Terui K., Haga S., Enosawa S., Ohnuma N., Ozaki M. (2004) Hypoxia/re-oxygenation-induced, redox-dependent activation of STAT1 (signal transducer and activator of transcription 1) confers resistance to apoptotic cell death via hsp70 induction. Biochem. J. 380, 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stephanou A., Brar B. K., Scarabelli T. M., Jonassen A. K., Yellon D. M., Marber M. S., Knight R. A., Latchman D. S. (2000) Ischemia-induced STAT-1 expression and activation play a critical role in cardiomyocyte apoptosis. J. Biol. Chem. 275, 10002–10008 [DOI] [PubMed] [Google Scholar]
- 80.Stephanou A. (2002) Activated STAT-1 pathway in the myocardium as a novel therapeutic target in ischaemia/reperfusion injury. Eur. Cytokine Netw. 13, 401–403 [PubMed] [Google Scholar]
- 81.Ivanov S. V., Salnikow K., Ivanova A. V., Bai L., Lerman M. I. (2007) Hypoxic repression of STAT1 and its downstream genes by a pVHL/HIF-1 target DEC1/STRA13. Oncogene 26, 802–812 [DOI] [PubMed] [Google Scholar]
- 82.Chin Y. E., Kitagawa M., Su W. C., You Z. H., Iwamoto Y., Fu X. Y. (1996) Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science 272, 719–722 [DOI] [PubMed] [Google Scholar]
- 83.Chin Y. E., Kitagawa M., Kuida K., Flavell R. A., Fu X. Y. (1997) Activation of the STAT signaling pathway can cause expression of caspase 1 and apoptosis. Mol. Cell. Biol. 17, 5328–5337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bromberg J. F., Fan Z., Brown C., Mendelsohn J., Darnell J. E., Jr. (1998) Epidermal growth factor-induced growth inhibition requires Stat1 activation. Cell Growth Differ. 9, 505–512 [PubMed] [Google Scholar]
- 85.Taniguchi T. (1997) Transcription factors IRF-1 and IRF-2: linking the immune responses and tumor suppression. J. Cell. Physiol. 173, 128–130 [DOI] [PubMed] [Google Scholar]
- 86.Lee K. Y., Anderson E., Madani K., Rosen G. D. (1999) Loss of STAT1 expression confers resistance to IFN-gamma-induced apoptosis in ME180 cells. FEBS Lett. 459, 323–326 [DOI] [PubMed] [Google Scholar]
- 87.Hiroi M., Mori K., Sakaeda Y., Shimada J., Ohmori Y. (2009) STAT1 represses hypoxia-inducible factor-1-mediated transcription. Biochem. Biophys. Res. Commun. 387, 806–810 [DOI] [PubMed] [Google Scholar]
- 88.Huang S., Bucana C. D., Van Arsdall M., Fidler I. J. (2002) Stat1 negatively regulates angiogenesis, tumorigenicity and metastasis of tumor cells. Oncogene 21, 2504–2512 [DOI] [PubMed] [Google Scholar]
- 89.Hu Y., Hu X., Boumsell L., Ivashkiv L. B. (2008) IFN-gamma and STAT1 arrest monocyte migration and modulate RAC/CDC42 pathways. J. Immunol. 180, 8057–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dutreix M., Cosset J. M., Sun J. S. (2010) Molecular therapy in support to radiotherapy. Mutat. Res. 704, 182–189 [DOI] [PubMed] [Google Scholar]
- 91.Kim Y. J., Varki A. (1997) Perspectives on the significance of altered glycosylation of glycoproteins in cancer. Glycoconj. J. 14, 569–576 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. S1