Antibiotic-Associated Diarrhea Accompanied by Large-Scale Alterations in the Composition of the Fecal Microbiota (original) (raw)
Abstract
Alterations in the diversity of the gut microbiota are believed to underlie the development of antibiotic-associated diarrhea (AAD). A molecular phylogenetic analysis was performed to document temporal changes in the diversity of fecal bacteria of a patient who developed AAD. Antibiotic administration was associated with distinct changes in the diversity of the gut microbiota, including a marked decrease in the prevalence of butyrate-producing bacteria. Following the discontinuation of the antibiotic, resolution of diarrhea was accompanied by a reversal of these changes, providing the first direct evidence linking changes in the community structure of the gastrointestinal bacteria with the development of AAD.
The normal gut biota represents a complex microbial ecosystem (the “enterome”) that plays a crucial role in homeostasis of the gastrointestinal tract (12). As such, disturbances in the enterome can lead to a variety of pathogenic states. Antibiotic-associated diarrhea (AAD), defined as diarrhea associated with the administration of antibiotics and without another obvious cause, is believed to represent such a condition (3, 9). The frequency of AAD varies among different antibiotics, but AAD can affect up to 25% of the patients receiving a particular antibiotic.
A number of mechanisms underlie the development of AAD. Overgrowth by the toxigenic bacterium Clostridium difficile is a mechanism that has received particular attention, in part because it can occur in nosocomial outbreaks and is responsible for virtually all cases of antibiotic-associated pseudomembranous colitis, which can lead to complications such as paralytic ileus and colonic dilatation and perforation (11, 13). However, it is estimated that only 15 to 25% of all cases of AAD are due to the overgrowth of C. difficile (3).
Another possible mechanism is the loss of beneficial metabolic activities of intestinal microbes (6). Global changes in the composition and quantity of the gut microbiota (even in the absence of overgrowth by pathogenic microorganisms) can result in perturbations of global colonic metabolism that lead to AAD (9). The colon is unable to absorb carbohydrates, and as much as 70 g of undigested carbohydrate reaches the colon each day. Colonic bacteria, especially certain anaerobes, metabolize these carbohydrates as an energy source, producing lactic acid and short-chain fatty acids (SCFAs), the latter being readily absorbed by the colon (5). Loss of these bacteria due to antibiotic treatment can lead to increased amounts of carbohydrate in the colonic lumen, leading to an osmotic diarrhea. Additionally, the SCFA _n_-butyrate is an important source of energy for the mucosa of the distal colon (15). Reduction of the production of SCFA due to reduced levels of colonic anaerobes can thus lead directly to functional disorders of the colonic mucosa (19). In spite of these theoretical considerations, there is little direct evidence for this mechanism of non-C. difficile AAD.
In order to monitor changes in the diversity of the gut ecosystem during antibiotic administration, we conducted a molecular phylogenetic survey of the fecal microbiota from a patient who developed AAD during the administration of a broad-spectrum antibiotic.
CASE REPORT
The patient was a 39-year-old male who was prescribed amoxicillin-clavulanic acid (875 and 125 mg, respectively, twice daily for 10 days) for acute sinusitis. The patient had no history of chronic gastrointestinal disease, had not had antibiotics in the previous year, and was taking no other medications. The patient had a history of taking amoxicillin for dental prophylaxis in the past and never had previous episodes of diarrhea with amoxicillin administration.
Within 24 h of starting amoxicillin-clavulanate, the patient noted the onset of bulky, loose stools. The patient denied abdominal pain, fever, tenesmus, or bright red blood per rectum. The patient continued to have two to three loose bowel movements during the 10-day course of antibiotic administration. Bowel habits returned to baseline within 4 days of the discontinuation of the antibiotic.
MATERIALS AND METHODS
Patient sample collection.
Freshly voided stool was collected from the first daily bowel movement of the patient during antibiotic therapy and on a weekly basis after the discontinuation of antibiotics. The first sample collected was the initial bowel movement after starting antibiotics, which was 12 h after the first oral dose. Four ∼25-mg aliquots of each stool sample were stored at −70°C. For the analysis reported here, an aliquot was selected from the initial collected sample (day 0), the sample from the fourth day of antibiotic administration (day 4), and the sample collected 2 weeks after the final dose of antibiotics (day 24).
DNA purification and construction of 16S ribosomal DNA (rDNA) clone libraries.
Total stool DNA was purified from each selected stool sample using a commercial DNA isolation kit specifically designed to remove inhibitors of PCR from stool samples (Stool DNA kit; QIAGEN, Germantown, Md.). As a contamination control, a sample of autoclaved water was carried through the purification scheme.
PCR employing primers targeting bacterial 16S rRNA genes (8F and 1492R [16]) was performed on each DNA sample. PCR was performed using Pharmacia Ready-To-Go PCR beads (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). Reaction mixtures were set up with 1 μl of template DNA (approximately 100 ng), 20 pmol of each primer, and water to a total volume of 25 μl. This yielded a reaction mixture containing 1.5 U of Taq polymerase, 10 mM Tris-HCl (pH 9.0 at room temperature), 50 mM KCl, 1.5 mM MgCl2, a 200 μM concentration of each nucleotide, and stabilizers, including bovine serum albumin. The reaction mixtures were subjected to amplification in a DNA thermal cycler (Eppendorf Mastercycler gradient) with the following cycling conditions: initial denaturation at 94°C for 3 min followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 48°C for 45 s, and extension at 72°C for 1.5 min. A final extension at 72°C for 10 min was performed. Control amplifications with sterile water and the water subjected to the DNA purification procedure were included in each amplification reaction and never gave visible amplicons.
Amplicons were isolated using a commercial purification kit (GFX; Amersham Pharmacia Biotech) according to the recommendations of the manufacturer. The purified PCR products were ligated into a plasmid vector (pCR 2.1; Invitrogen, Carlsbad, Calif.) that takes advantage of the nontemplated 3′ A overhangs generated during PCR to create three 16S rDNA clone libraries.
DNA sequencing and analysis.
Plasmid purification and DNA sequence determination of 96 randomly selected clones from each library were performed by the Genomic Technology Support Facility at Michigan State University. Each clone was sequenced with a single primer (519R) that typically yielded ∼500 bases of readable sequence. Sequences with numerous ambiguous base calls or with fewer than 350 total bases were excluded from further analysis.
Sequences were analyzed for the formation of chimeras using the Chimera Check program from the Ribosomal Database Project (4). Potential chimeric sequences were excluded from additional analysis. Sequences were aligned to one another and to a phylogenetically diverse collection of rRNA gene sequences using the ARB suite of programs (available through http://www.arb-home.de) running on a Macintosh G4 computer (Apple, Cupertino, Calif.). Alignments were adjusted by hand to account for regions of primary sequence similarity and secondary structure. Regions of ambiguous alignment (primarily stem structures of variable length) were excluded from the final comparison of sequences, such that 388 positions were used in the final phylogenetic analyses.
Partial 16S sequences were initially added to the ARB framework tree using parsimony criteria that added the sequences to the established tree without changing its topology. Subsequent trees, including the tree presented in Fig. 1, were calculated de novo using the ARB neighbor-joining algorithm. Bootstrap analysis was performed by exporting the ARB alignment for analysis with the MEGA2 software package (available at http://www.megasoftware.net). Statistical differences in the composition of clone libraries from each time point were determined using LIBSHUFF (version 1.2) (17).
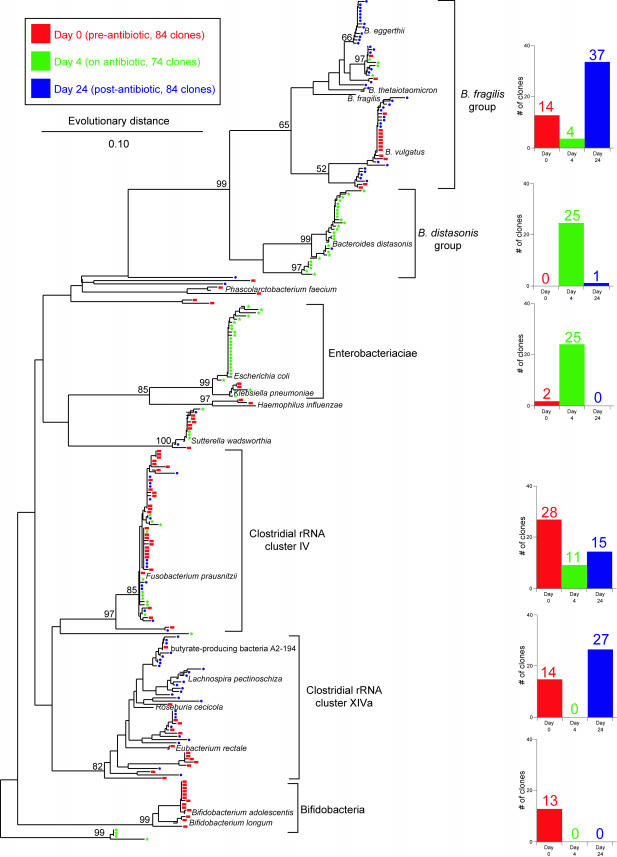
FIG.1.
Phylogenetic tree showing the distribution of 16S rDNA sequences of randomly selected clones from libraries constructed from stool DNA samples obtained from a patient prior to antibiotic therapy (day 0 [red]), during therapy (day 4 [green]), and 2 weeks after discontinuation of therapy (day 24 [blue]). Brackets outline major clusters of organisms, and the adjacent bar graphs document the distribution of clones in each cluster at each time point. Named species are representative type species downloaded from the Ribosomal Database Project and inserted into the tree to provide taxonomic reference points. These reference species do not contribute to the number of clones depicted in the bar graphs. The scale bar represents evolutionary distance (10 substitutions per 100 nucleotides). The tree was constructed using neighbor-joining analysis of a distance matrix obtained from a multiple-sequence alignment performed using the ARB suite of programs. Bootstrap values were calculated using the MEGA2 program.
Nucleotide sequence accession numbers.
The sequences of the cloned inserts were deposited in GenBank under the accession numbers AY457674 to AY457915.
RESULTS
A patient being treated with amoxicillin-clavulanic acid for acute sinusitis was monitored for the development of AAD. The patient developed loose stool within 24 h after the first dose of antibiotics. DNA was purified from stool samples collected (i) from the first voided stool after the initiation of antibiotics (day 0), (ii) 4 days after the initiation of antibiotics (day 4), and (iii) 2 weeks after the last dose of antibiotics (day 24). A 16S rDNA library was constructed from each of three DNA samples following PCR amplification using primers that target bacterial 16S rRNA genes. Ninety-six randomly selected clones from each library were subjected to DNA sequence analysis using a sequencing primer (519R) that yielded an average of 500 bp of readable sequence. Ultimately, 84, 74, and 84 sequences were utilized from each library, respectively, for phylogenetic analysis. Phylogenetic trees were constructed using the ARB suite of programs (Fig. 1).
In the day 0 sample, the majority of the sequences clustered within four bacterial groups (Fig. 1): (i) Bacteroides spp., (ii) Clostridium sp. rRNA cluster IV, (iii) Clostridium sp. rRNA cluster XIVa, and (iv) Bifidobacterium spp. Four days after the initiation of antibiotics, there was a marked shift in the representation of the major bacterial groups. Bacteroides spp. were still a major component of the microbiota, but whereas members of the Bacteroides fragilis cluster were predominant in the day 0 sample, Bacteroides distasonis was the predominant group at day 4. Strikingly, no sequences corresponding to Clostridium rRNA cluster XIVa or to Bifidobacterium spp. were detected at day 4, whereas these two groups represented a third of the sequences detected on day 0. Conversely, 34% of the sequences detected on day 4 were members of the Enterobacteriaceae, which represented only 2% of the day 0 sequences.
Two weeks after the cessation of antibiotics (day 24), there was partial reversal of the changes seen on day 4. B. fragilis was once again the predominant Bacteroides species, and there was reappearance of members of Clostridium rRNA cluster XIVa. No sequences of members of the Enterobacteriaceae were detected on day 24. Interestingly, Bifidobacterium spp., which was one of the major groups seen on day 0 and not present on day 4, did not return by day 24.
LIBSHUFF analysis (17) of each pairwise comparison between the libraries indicated that the libraries were significantly different from one another: P values were <0.001 with the exception of the day 24 library compared to the day 1 library, where the P value was 0.004. Visual inspection of the phylogenetic analysis of cloned sequences (Fig. 1) suggests that the day 24 library is a subset of the day 0 library, with all well-represented phylogenetic groups present at day 0 also present at day 24 (with the exception of the bifidobacteria).
DISCUSSION
AAD is a significant side effect of antimicrobial administration. A critical factor in the etiopathogenesis of AAD is felt to be an alteration in the normal gastrointestinal microbiota (9). This change in the diversity of the microbiota is thought to allow the ingrowth of pathogens such as C. difficile (13). In addition, alteration of the gut microbiota can lead to significant changes in the colonic microenvironment, especially with regards to the concentration and distribution of organic compounds such as carbohydrates, SCFAs, and bile acids (9).
The patient in this study developed AAD that was not associated with evidence of C. difficile, as judged by a negative C. difficile toxin assay (data not shown). It has been proposed that non-_C. difficile_-associated AAD is due to the loss of beneficial metabolic activities of the normal colonic biota. In particular the metabolism of undigested fiber and starch by certain colonic anaerobes to SCFAs (in particular, butyrate) has been hypothesized to prevent osmotic diarrhea as well as provide a supply of the preferred carbon and energy source to the colonic enterocytes (5, 15, 19).
To our knowledge, this study provides the first direct evidence in support of this hypothesis for the etiopathogenesis of non-_C. difficile_-associated AAD. The microbiology of butyrate production in the human colon has been investigated recently (14). The most numerous butyrate-producing bacteria have been found to belong to the clostridial clusters IV and XIVa. Members of these two phylogenetic groups composed almost half of the identified species at day 0 in our patient. This sample was collected approximately 12 h after the first dose of antibiotics. However, since this was the first stool voided after the initiation of antibiotics, it should be a valid representation of the colonic microbiota prior to the administration of antibiotics. This assumption is supported by the fact that the representation and distribution of the major bacterial groups found in this sample are in agreement with previous studies that examined the fecal microbiota of healthy humans (8, 10, 18).
By day 4, after the patient had developed AAD, no members of clostridial cluster XIVa were found, and there was a marked decrease in the number of members belonging to clostridial cluster IV. Recent analysis of the bacteriology of butyrate production in the human gut suggests that the majority of butyrogenic organisms fall within cluster XIVa (2, 14). Thus, our observations provide the first evidence in support of the hypothesis that AAD can result from measurable changes in the composition of the microbiota of the gastrointestinal tract.
The composition of the colonic microbiota was initially determined by culture-based studies (1). More recently, molecular genetic tools have been used to gain a more complete assessment of the diversity of the gut microbiota (7, 8, 10, 18, 20). To our knowledge, the present study is the first to use molecular methods to look at the temporal changes in the diversity of the colonic microbiota brought on by the administration of a broad-spectrum antibiotic. This is a powerful approach that can monitor qualitative and quantitative shifts in the diversity of the enterome under ecologic stresses such as infection or the administration of antibiotics and/or probiotic organisms.
Acknowledgments
Informed consent was obtained from the patient, and human experimentation guidelines of the U.S. Department of Health and Human Services and of MSU were followed in the conduct of the research described herein.
REFERENCES
- 1.Attebery, H. R., V. L. Sutter, and S. M. Finegold. 1972. Effect of a partially chemically defined diet on normal human fecal flora. Am. J. Clin. Nutr. 25**:**1391-1398. [DOI] [PubMed] [Google Scholar]
- 2.Barcenilla, A., S. E. Pryde, J. C. Martin, S. H. Duncan, C. S. Stewart, C. Henderson, and H. J. Flint. 2000. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 66**:**1654-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett, J. G. 2002. Clinical practice. Antibiotic-associated diarrhea. N. Engl. J. Med. 346**:**334-339. [DOI] [PubMed] [Google Scholar]
- 4.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31**:**442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cummings, J. H., E. W. Pomare, W. J. Branch, C. P. Naylor, and G. T. Macfarlane. 1987. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28**:**1221-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunne, C. 2001. Adaptation of bacteria to the intestinal niche: probiotics and gut disorder. Inflamm. Bowel Dis. 7**:**136-145. [DOI] [PubMed] [Google Scholar]
- 7.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64**:**3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi, H., M. Sakamoto, and Y. Benno. 2002. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol. Immunol. 46**:**535-548. [DOI] [PubMed] [Google Scholar]
- 9.Hogenauer, C., H. F. Hammer, G. J. Krejs, and E. C. Reisinger. 1998. Mechanisms and management of antibiotic-associated diarrhea. Clin. Infect. Dis. 27**:**702-710. [DOI] [PubMed] [Google Scholar]
- 10.Hold, G. L., S. E. Pryde, V. J. Russell, E. Furrie, and H. J. Flint. 2002. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 39**:**33-39. [DOI] [PubMed] [Google Scholar]
- 11.Kelly, C. P., and J. T. LaMont. 1998. Clostridium difficile infection. Annu. Rev. Med. 49**:**375-390. [DOI] [PubMed] [Google Scholar]
- 12.McCracken, V. J., and R. G. Lorenz. 2001. The gastrointestinal ecosystem: a precarious alliance among epithelium, immunity and microbiota. Cell. Microbiol. 3**:**1-11. [DOI] [PubMed] [Google Scholar]
- 13.Mylonakis, E., E. T. Ryan, and S. B. Calderwood. 2001. _Clostridium difficile_-associated diarrhea: a review. Arch. Intern. Med. 161**:**525-533. [DOI] [PubMed] [Google Scholar]
- 14.Pryde, S. E., S. H. Duncan, G. L. Hold, C. S. Stewart, and H. J. Flint. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217**:**133-139. [DOI] [PubMed] [Google Scholar]
- 15.Scheppach, W., J. G. Muller, F. Boxberger, G. Dusel, F. Richter, H. P. Bartram, S. U. Christl, C. E. Dempfle, and H. Kasper. 1997. Histological changes in the colonic mucosa following irrigation with short-chain fatty acids. Eur. J. Gastroenterol. Hepatol. 9**:**163-168. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt, T. M., and D. A. Relman. 1994. Phylogenetic identification of uncultured pathogens using ribosomal RNA sequences. Methods Enzymol. 235**:**205-222. [DOI] [PubMed] [Google Scholar]
- 17.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67**:**4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65**:**4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Topping, D. L., and P. M. Clifton. 2001. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81**:**1031-1064. [DOI] [PubMed] [Google Scholar]
- 20.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62**:**2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]