A Mechanism for Controlled Access to GWAS Data: Experience of the GAIN Data Access Committee (original) (raw)
Abstract
The Genetic Association Information Network (GAIN) Data Access Committee was established in June 2007 to provide prompt and fair access to data from six genome-wide association studies through the database of Genotypes and Phenotypes (dbGaP). Of 945 project requests received through 2011, 749 (79%) have been approved; median receipt-to-approval time decreased from 14 days in 2007 to 8 days in 2011. Over half (54%) of the proposed research uses were for GAIN-specific phenotypes; other uses were for method development (26%) and adding controls to other studies (17%). Eight data-management incidents, defined as compromises of any of the data-use conditions, occurred among nine approved users; most were procedural violations, and none violated participant confidentiality. Over 5 years of experience with GAIN data access has demonstrated substantial use of GAIN data by investigators from academic, nonprofit, and for-profit institutions with relatively few and contained policy violations. The availability of GAIN data has allowed for advances in both the understanding of the genetic underpinnings of mental-health disorders, diabetes, and psoriasis and the development and refinement of statistical methods for identifying genetic and environmental factors related to complex common diseases.
Main Text
Genome-wide association studies (GWASs), a proven strategy for identifying common genetic variants associated with health and disease, produce vast amounts of data suitable for addressing a multitude of research questions. The Genetic Association Information Network (GAIN) was established in 2006 to investigate the genetic basis of common disease by the creation of a network of six collaborative GWASs in attention-deficit hyperactivity disorder (MIM 143465]), bipolar disorder (MIM 125480), diabetic nephropathy in type I diabetes (MIM 222100), major depression (MIM 608516), psoriasis (MIM 177900), and schizophrenia (MIM 181500).1 GAIN’s driving principle is that maximum public benefit can be achieved if innovative research is pursued through rich and readily accessible genomic data in a manner that promotes the utmost respect and protection for research-participant interests, as well as for the contributions of the investigators who submit data for broad sharing.2 Therefore, researchers can access GAIN GWAS data through the database of Genotypes and Phenotypes (dbGaP),3 developed by the National Center for Biotechnology Information (NCBI). dbGaP provides two levels of access—open and controlled—to GAIN data. The open-access (public) website provides broad release of nonsensitive data (e.g., study overviews and original study protocols). The controlled-access website provides individual-level phenotypic and genotypic data for GAIN GWASs, but only after a researcher is approved to access the data by the GAIN Data Access Committee (DAC). GAIN has been at the leading edge of implementation of dbGaP’s controlled-access data-request system and has served as a precursor to the National Institutes of Health (NIH)-wide GWAS-data-sharing model.
There are many potential benefits of wide-spread data sharing, and these include opportunities to (1) replicate GWAS findings in large data sets, (2) develop and test statistical methods for GWASs and other genomics research, and (3) maximize the use of GWAS data for the discovery of health-related genetic variants and their translation into effective therapeutic strategies. However, many concerns were raised about this new data-sharing model and its potential risks to study participants and investigators, as summarized in the preamble to the NIH policy for sharing GWAS data (GWAS policy). The concerns included (1) the potential for nonresearch uses of the data (e.g., by law-enforcement agencies, employers, or insurance companies) or for purposes beyond the permitted scope of approved research, (2) the adequacy of data quality control and oversight of data-submission and -access procedures, (3) potential risks to participant privacy, and (4) difficulties in enforcing compliance with publication and privacy policies. Although studies have shown that participants generally support broad data sharing of their individual-level genotype and phenotype data,4 they also expect to be asked whether their data can be included in controlled-access databases such as dbGaP.5 A recent commentary called for greater transparency with regard to the nature and extent of dbGaP data sharing for enhancing the trustworthiness of the resource for data submitters and their institutions.6
GAIN data have been available through dbGaP for over 5 years, and nearly 1,000 project requests (PRs) have been received and reviewed by the GAIN DAC. Here, we hope to provide greater transparency of the GAIN controlled-access data-sharing model by describing (1) the GAIN data sets and their data-use limitations (DULs), (2) the function of the GAIN DAC and key aspects of its governance, (3) the number and type of PRs for GAIN data sets, and (4) the limitations and strengths of this new controlled-access data-sharing model.
GAIN Study Data Sets
The six GAIN studies are presented in Table 1. Most of these studies include more than one data set, or group of participants with distinct DULs on permissible research (see Table 2 for a glossary of terms). In conjunction with their institutional review boards, the principal investigators (PIs) of each of the GAIN studies developed the DULs on the basis of the participants’ informed consent. These DULs, such as the GAIN major-depression study’s limitation to “genetic studies of psychiatric health and related somatic conditions,” might require interpretation by the DAC. Although the GAIN PI defined these terms clearly for the DAC’s use, knowledge of phenotypes composing or related to psychiatric disorders is somewhat specialized and makes essential the inclusion of a DAC member with psychiatric expertise (e.g., Thomas Lehner, National Institute of Mental Health). On numerous occasions, the PIs of the GAIN studies joined the DAC meetings via conference call to provide their perspective on the appropriate interpretation of DUL statements. GAIN studies with more than one data set, such as the GAIN schizophrenia study, often involved a “tiered” consent in which participants could opt to allow their data and samples to be used for any genetic studies (i.e., for “general research use” [GRU]) or limit the use to genetic studies of schizophrenia and related conditions (SARCs). Approved users are able to download only the dbGaP data sets for which they are approved—for example, an investigator approved to access only the GAIN schizophrenia GRU data set cannot download data from the SARC or major-depression data sets.
Table 1.
GAIN Studies and Data Sets
| GAIN Study (dbGaP Accession Number) | Publication Embargo Date | DULs for GAIN Study Data Set(s)a | Sample Size | Number of Approved PRs (through 12/31/11) |
|---|---|---|---|---|
| ADHDb (phs000016) | 3/26/08 | Limited to genetic studies of the pathophysiology or etiology of ADHD or its complications. | 2,758 (924 trios) | 155 |
| Nephropathy in type I diabetes (phs000018) | 7/9/08 | Limited to research on type 1 diabetes and its complications. Complications include nephropathy, cardiovascular disease, retinopathy, neuropathy, and mortality. Phenotypes related to diabetes and its complications, such as body mass index, blood pressure, lipids, and hemoglobin A1C, may also be studied. | 1,825 (904 cases, 881 controls, and 40 others) | 107 |
| Major depressive disorder (phs000020) | 7/9/08 | Limited to genetic studies of psychiatric health and related somatic conditions. Psychiatric health refers to DSM-IV or ICD-10 psychiatric disorders. “Related somatic conditions” refers to general medical disorders whose risks have been elevated in individuals with psychiatric disorders (e.g., cardiovascular disease or migraine). | 3,741 (1,821 cases, 1,822 controls, and 98 others) | 206 |
| Psoriasis (phs000019) | 8/13/08 | GRU | 1,677 (950 cases, 692 controls, and 35 others) | 151 |
| Limited to genetic studies of autoimmune disease. | 1,198 (449 cases, 734 controls, and 15 others) | 73 | ||
| Schizophreniac (phs000021) | 12/3/08 | GRU | 4,591 (1,217 EA cases, 1,442 EA controls, 953 AA cases, and 979 AA controls) | 421 |
| Limited to genetic studies of schizophrenia and related conditions, which include those with evidence of genetic relationships to schizophrenia or schizoaffective disorder, such as acute psychoses, bipolar disorder, major depressive disorder, or “cluster A” personality disorders (schizotypal, schizoid, and paranoid personality disorders). | 475 (187 EA cases and 288 AA cases) | 227 | ||
| Bipolar disorderd (phs000017) | 12/1/08 | GRU | 1,767 (1,081 EA controls and 656 AA controls) | 286 |
| Bipolar and related disorders. | 841 (691 EA cases and 150 AA cases) | 216 | ||
| Bipolar disorder only. | 653 (388 EA cases and 265 AA cases) | 180 |
Table 2.
Glossary of Terms
| Term | Abbreviation | Definition |
|---|---|---|
| Annual report | AR | A report submitted to the DAC on the anniversary of access approval. It summarizes the analysis of NIH genomic data sets obtained through the PRs and any significant findings derived from the work. |
| Approved user | AU | Post-DAC approval will include the PI, home-institution collaborators who are named in the “Senior/Key Person Profile” portion of the PR, the IT director or designee named in the “Senior/Key Person Profile” portion of the PR, and trainees or staff to these investigators. |
| Data-management incident | DMI | Occurs when any of the terms of the NIH DUC agreement have been compromised. |
| Data set | - | Group of participants with distinct DULs on permissible research. One or more data sets can make up a GAIN study. |
| Data Use Certification | DUC | The agreement that outlines the terms of access for NIH genomic data. The DUC is signed by the PI and signing official upon submission of a PR to the NIH. |
| Database of Genotypes and Phenotypes | dbGaP | Currently serves as the primary NIH GWAS data repository. |
| GAIN study | - | One of six studies contributed to the NIH GWAS data repository (dbGaP) as part of the GAIN Consortium activities. Each study focuses on one disease and consists of one or more data sets. |
| Institutional signing official | SO | Someone who has the authority to sign on behalf of the PI’s research institution and who is credentialed through the eRA system as such. |
| Information-technology director | IT director | Someone with the authority to verify the IT data-security capacities at an institution or a higher-level division of an institution (e.g., the school of medicine). |
| Participant Protection and Data Management Steering Committee | PPDM | The principal trans-NIH committee within the GWAS governance structure. It is charged with providing ongoing review and development of specific policies and procedures related to issues of participant protection in GWAS data submission, data management, and data distribution and with promoting communication across the various NIH DACs. |
| Project request | PR | A request for dbGaP data sets. It contains the SF 424 (R&R) cover pages and requested attachments, if any. |
| Senior Oversight Committee | SOC | The principal NIH GWAS governance committee that advises the NIH director on the policies, procedures, and issues regarding the ongoing implementation and monitoring of the GWAS policy and data-sharing practices for other genomic data deposited to dbGaP. |
The GAIN DAC
The GAIN DAC is composed of eight senior scientists who are from several NIH institutes and who have appropriate scientific, bioethics, and human-subjects research expertise. Many of the DAC members have previously served on or chaired institutional review boards and bring this important perspective to the evaluation of PRs. Because dbGaP was developed and is maintained by the NIH, it is considered a federal database and all members of the DAC must be federal employees. DAC members are asked to serve 3 year terms, although some have been members since the inception of the committee in 2007. Before a new member begins voting on PRs, he or she attends several committee meetings as an orientation to the GAIN data sets and their DULs and the review process. For the first few years of its existence, the GAIN DAC met in person on a weekly basis to develop operational procedures and discuss each submitted PR in great detail.
The GAIN DAC reviews requests for the ten GAIN data sets housed in dbGaP. A PR may include multiple data-set requests. The GAIN DAC’s primary charge is to determine whether the proposed research described in each PR is consistent with the DULs for each of the requested data sets. For example, a researcher who is submitting a PR for schizophrenia research and who has requested the GAIN schizophrenia data sets (GRU and SARC) and the diabetic nephropathy in type I diabetes (DN) data set would be denied access to the DN data set because schizophrenia is not considered a complication of type I diabetes and the DN DUL requires that it be. Figure 1 outlines the process for PR submission and review. PRs for GAIN data are submitted online through dbGaP via the Electronic Research Administration (eRA) Commons, an online interface that allows grant applicants, grantees, federal staff at NIH, and other federal grantor agencies access to administrative information relating to research grants. Detailed information describing the process for PR submission and DAC preliminary review can be found in the Supplemental Data, available online.
Figure 1.
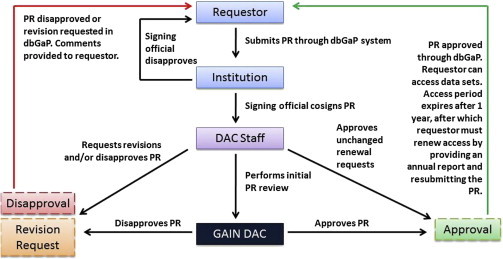
Overview of the GAIN Data-Access Review Process
After a researcher (requestor) submits a PR via dbGaP, the PR is reviewed and signed by the signing official at the requestor’s institution. The PR undergoes a preliminary review by the GAIN DAC staff before being sent to the GAIN DAC for a vote. If the PR is approved, the requestor (approved user) can access data sets from dbGaP. If the PR is disapproved or a revision to the PR is requested, the requestor can revise and resubmit the PR to begin the evaluation process again.
Once the PR is received and a preliminary review by GAIN DAC staff is complete, the PR is made available to the DAC members through a secure site and initial voting takes place electronically. DAC members review each request for consistency with the DULs and vote to approve, disapprove, or discuss the request at the next DAC meeting. PRs that receive unanimous electronic votes for approval or disapproval do not require further discussion. PRs without unanimous approval via electronic voting are discussed at the DAC meeting, and a vote to approve or disapprove the PR is held at the end of discussion. All new PRs must be reviewed and voted on by the DAC, either electronically or in person, to be approved. PRs submitted for an additional year of access after the original access period has expired are reviewed by the DAC chair and are only discussed by the entire DAC if the scope of the research has changed since the last approval.
Key Aspects of GAIN Data-Access Governance
Figure 2 illustrates the governance structure for the GAIN data-access process. The GAIN Steering Committee was responsible for overall guidance to the project and helped establish data-access principles and policies that the GAIN DAC then implemented. The Steering Committee paid particular attention to participant protections, both during data submission from the original studies and during data access and use by outside investigators.1 The NIH established the Advisory Committee to the Director (ACD) Working Group on Participant and Data Protection (ACD PDP) to serve as a source of independent advice about GAIN participant protection and data-management policies in order to support the use of GAIN data in a manner consistent with participant consent and robust standards for protecting participant privacy and confidentiality. The ACD PDP included ethicists, scientists, statisticians, and representatives from the general public (see Supplemental Data for the ACD PDP roster). For the first few years of the GAIN program, the ACD PDP met semiannually and reported back to the NIH ACD. Minutes from these public meetings are available online. In December 2007, the ACD PDP reported that the policies for data access and review were robust. They had also considered the potential scope of secondary use of data for research and determined that some uses—for example, methodological studies—are acceptable. Additional recommendations from the ACD PDP included that the NIH should (1) seek a Freedom of Information Act exemption #3 provision to provide additional privacy protections for those participants whose data are stored in dbGaP and (2) develop a system for addressing public inquiries about GWASs and the repository. The NIH agreed with these recommendations and, for the latter, took steps to enhance the website and the materials available to improve transparency and accessibility of the information within it. In addition to disseminating materials, the NIH has maintained a dedicated email address to receive questions or comments about the GWAS policy. The ACD PDP concluded its work in the summer of 2009 because GAIN oversight was fully integrated into the NIH governance structure developed through the NIH GWAS policy. Communication with the ACD on matters regarding GAIN (and broader GWAS) data sharing has continued as appropriate through that group’s semiannual meetings. The GWAS-policy-oversight structure includes multiple committees, including the NIH Participant Protection and Data Management Steering Committee (PPDM) and the NIH GWAS Senior Oversight Committee (SOC). The PPDM is a trans-NIH committee charged with providing ongoing review of specific policies and procedures related to issues of participant protection in data management and data distribution and with promoting communication across the institutes and centers. The SOC is the principle governance committee that advises the NIH director on the policies, procedures, and issues regarding the ongoing implementation and monitoring of the GWAS policy and data-sharing practices for other genomic data deposited in dbGaP.
Figure 2.
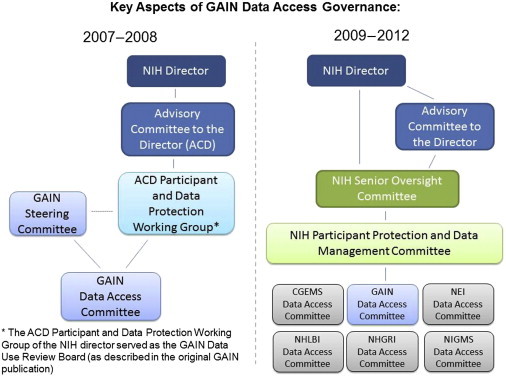
Summary of Key Aspects of Governance for Controlled Access to GAIN Data Sets through dbGaP
Prior to the implementation of the GWAS policy, both the GAIN Steering Committee and the Advisory Committee to the Director (ACD) Working Group on Participant and Data Protection (ACD PDP) provide oversight and input on policy questions related to GAIN data use, privacy concerns, and other topics, as appropriate. Once the governance structure for the GWAS policy is operational, oversight of GAIN is integrated with the other NIH DACs (only 5 of the 13 other DACs are shown here).
Summary of GAIN PRs
PR submission rates and types of proposed research remained roughly constant between 2007 and 2011 (Table 3), signifying a continued interest in GAIN data even though dozens of new study data sets are available via dbGaP. Seventy-nine percent (749/946) of PRs submitted to the DAC were approved. The 749 PRs approved by the DAC reflect 2,022 approved data-set requests. The average number of GAIN data sets requested per PR was three. The schizophrenia GRU data set was the most frequently requested of the ten GAIN data sets. This is not surprising because it is the largest of the GAIN data sets (n = 4,591), includes both white and African American participants, and does not limit research use to a set of particular conditions. Ten percent (78/749) of PRs were partially approved, meaning that some of the requested data sets were determined not to be appropriate for the research proposed. The majority of disapproved or partially approval PRs occurred because the proposed research did not fit with the DULs of the requested data sets or, more frequently, the proposed data uses were not clearly described. The remaining PRs were not approved for reasons such as the request was submitted by a graduate student and not a senior researcher (NIH policy expects submission by a senior researcher) or the request alluded to collaboration with investigators at another institution but provided insufficient detail. Sharing data within such an interinstitution collaborative group is permissible if (1) the collaborators and their institutions are listed in the research-use statement and (2) each collaborator submits a separate PR from his or her own institution and is granted approval to conduct the research. If a requester chooses to revise and resubmit a PR, the PR is reviewed at the next DAC meeting. Thirty-two percent (301/946) of PRs were not sent to the DAC because they either were missing information and were disapproved by the DAC chair or were previously approved and resubmitted without any modifications to the proposed research-use statement for an additional 1 year access period.
Table 3.
GAIN DAC Voting Decisions for Submitted PRs through 12/31/2011
| Year | Number of PRs Submitted | PRs to DACa | Average Data Sets per PR | Approved | Disapproved | Partially Approvedb | Returned for Revisionc | Withdrawnd |
|---|---|---|---|---|---|---|---|---|
| 2007 | 75 | 65 (87%) | 1.75 | 57 (76%) | 16 (21%) | 2 (3%) | 0 (0%) | 0 (0%) |
| 2008 | 226 | 176 (78%) | 2.92 | 190 (84%) | 26 (12%) | 8 (4%) | 1 (0%) | 1 (0%) |
| 2009 | 234 | 134 (57%) | 3.62 | 159 (68%) | 32 (14%) | 23 (10%) | 19 (8%) | 1 (0%) |
| 2010 | 196 | 143 (73%) | 3.05 | 129 (66%) | 32 (16%) | 26 (13%) | 4 (2%) | 5 (3%) |
| 2011 | 215 | 127 (59%) | 2.97 | 136 (63%) | 33 (15%) | 19 (9%) | 7 (3%) | 20 (10%) |
| Total | 946 | 645 (68%) | 3.04 | 671 (71%) | 139 (15%) | 78 (8%) | 31 (3%) | 27 (3%) |
The GAIN DAC aims to review submitted PRs as quickly as possible. For the entire 5 year period ending December 31, 2011, the median number of days from PI submission to institutional approval (via the PIs signing official) was 3 days, and that from institutional approval to the DAC’s decision was 12 days. Since November 2009, the DAC has met weekly instead of two to three times per month to accommodate the steady number of PRs that continue to be submitted, enabling a reduction in the median decision time from 14 days in 2007 and 2008 to 8 days in 2011 (Figure S1).
Requesting investigators came from a variety of backgrounds and included both GAIN PIs and their collaborators. Twenty-five of the 946 submitted PRs were from GAIN investigators and collaborators requesting access to their own data sets. GAIN, unlike most NIH-funded studies under the GWAS policy, requires that the original GAIN study PIs request their genotyping data from dbGaP.1 PRs were also submitted from investigators from a variety of institutions, including public nonprofit (12%), private for-profit (10%), foreign (25%), and domestic (53%) academic research institutions. Four hundred unique PIs from 214 unique institutions requested access to GAIN data sets. The aims of each PR vary. The most common proposed research use was to study GAIN-specific phenotypes (54% of submitted requests), and this was followed by method development (26%) and adding controls to other studies (17%). No proposals were received for nonresearch uses.
Monitoring the Use of GAIN Data
Approved users agree to submit annual reports summarizing research progress, publications and presentations, any problems that arose during use of the data, and future plans for using the data (Box 1). Annual reports are first reviewed by GAIN DAC staff and then by the DAC chair. Although 94% (708/757) of the annual reports due to the GAIN DAC as of December 31, 2011, have been submitted and reviewed, only 39% (274/708) of reports were submitted on time and 40% were submitted more than 30 days past their due date (data not shown). As part of an effort to increase annual reporting compliance, the GAIN DAC worked with dbGaP, other DAC members, and the PPDM to develop an automated series of email reminders to approved users and their signing officials about pending and late annual reports. As part of this new system, if a report is not submitted after these email notifications, access to dbGaP is suspended until the report is submitted or the project is terminated. Furthermore, if an investigator has an outstanding annual report, he or she will not be approved for any additional requests for NIH GWAS data sets until the delinquent reports are submitted. This new effort has improved timely submission of GAIN annual reports in 2012 (compared to 39% in previous years, 52% were submitted on time), and only 20% were submitted more than 30 days past their due date (it was 40% in previous years).
Box 1. Annual Report Elements.
- •
Summary of research progress - •
Proposed plans for further research utilizing currently approved NIH GWAS data sets - •
List of all completed or accepted scientific presentations that include (or will include) findings generated from the individual-level NIH GWAS data accessed through dbGaP - •
List of manuscripts submitted - •
Description of any intellectual property generated as a result of using the NIH GWAS individual-level data - •
Summary information on any inappropriate data-release incidents or other data-security issues - •
General comments on process and suggestions for improving dbGaP, NIH GWASs, study-specific data access, or NIH GWAS policy or procedures in general
Approved users are also asked to comment on the data-access-request process and dbGaP system in their annual reports. Comments from GAIN approved users are collated and shared with the PPDM and dbGaP staff on a regular basis and have led to improvements in dbGaP. For example, dbGaP genotype-data files are now available in a format compatible with PLINK, a commonly used genetic-analysis software program, and PIs can now assign an authorized downloader from their lab to select, package, and download files for the PI and approved project team. Lastly, the GAIN DAC generates semiannual reports—which include data-access and use statistics, summaries of annual reports submitted, trends in access requests, and other information—for the PPDM and SOC. These reports enable the NIH to monitor the data-access system and any issues that arise from investigator or DAC perspectives and that might merit policy or practice updates.
Another key aspect of monitoring the use of GAIN data involves identifying data-management incidents (DMIs), compromises of any of the terms of the NIH Data Use Certification (DUC) agreement, as early as possible and developing systems to prevent similar DMIs from occurring again. As part of the DUC agreement, approved users agree to notify the GAIN DAC of any unauthorized data sharing, breaches of data security, or inadvertent data releases that might compromise data confidentiality within 24 hr of when the incident is identified. Furthermore, the regular review of annual reports enables GAIN DAC staff to identify potential DMIs not previously reported by approved users.
Just over 2% (9/400) of GAIN approved users have been associated with eight DMIs. Once a DMI is identified, the approved user’s signing official is immediately notified and the approved user and his or her research team are informed by the DAC chair to stop using GAIN data until the incident can be investigated and reviewed by appropriate GWAS governance committees. dbGaP also temporarily suspends the approved user’s account to ensure that additional files cannot be downloaded. The governance committees involved in the consideration of DMIs have varied on the basis of the nature of the DMI and any existing precedent for the actions appropriate to manage any associated concerns or risks to participants. In each case, the SOC, the PPDM, or both are informed about the incident, and in some cases, these groups meet to deliberate options for any NIH response within days of the DMI notification. Table 4 provides a brief summary of each GAIN DMI, the corresponding DUC term impacted, and the preventive measures implemented at either the approved user’s institution or the NIH for ensuring that such incidents did not recur. Preventive measures include additional data-security protocols, investigator education regarding the roles and responsibilities of an approved user, and dbGaP system checks prior to data release. Seven of these incidents occurred in the first few years in which GAIN data were available and, in every case, resolution of the DMI led to improved education of GAIN approved users and their colleagues, as well as improvements to dbGaP and GAIN DAC review processes.
Table 4.
Summary of GAIN DMIs and the Preventive Measures Implemented
| DMI Type | Number of Incidents | Brief Summary of the DMI | Corresponding DUC Agreement Term | Preventative Measures Implemented |
|---|---|---|---|---|
| Security breach | 1 | The computer system at an AU’s institution was determined to be vulnerable. The AU immediately contacted the institution’s IT department and captured key information from the affected machines. The GAIN DAC was contacted a few days after the vulnerability became apparent. After a thorough analysis by the IT department, there was no evidence that GAIN data were accessed. | Term 6. Data Security and Data Release Reporting. The AU agrees to notify the GAIN DAC of any unauthorized data sharing, breaches of data security, or inadvertent data releases that might compromise data confidentiality within 24 hr of when the incident is identified. | The AU’s institution implemented additional security measures to protect machines used for analyzing GAIN data. |
| Unapproved research use | 2 | AUs at two separate institutions submitted annual reports that described using GAIN data in a manner not described in the RUS. | Term 1. Research Use. New uses of these data outside those described in the PR require submission of a new PR. Modifications to the research project require submission of an amendment to this application. | At one of these two institutions, a memo in which dbGaP stressed the importance of the terms of access for NIH GWAS data was circulated to all investigators. As of December 2008, AUs have access to GAIN data for only 1 year instead of 3 years, after which they must renew their PRs and update the RUS if the research focus has changed. |
| Missing IT director | 1 | An AU was approved to access GAIN data sets despite the absence of an IT director on the PR. | Term 1. Research Use.The IT director, someone with authority to vouch for the IT capacities at an institution, should be listed as a “Senior/Key Person” on the PR. | GAIN DAC staff implemented additional checks in the PR review process, and the NCBI modified the PR system to make the requirement for an IT director more apparent. |
| Publication embargo violation | 1 | An abstract was submitted for a scientific meeting prior to the expiration of the publication embargo date. The GAIN DAC was notified when the AU called GAIN staff to clarify how to acknowledge GAIN in presentations. The abstract was withdrawn and did not appear in the meeting materials. | Term 8. Research Dissemination and Acknowledgment of NIH GWAS Data Sets. AUs acknowledge the NIH’s expectation that they will comply with the embargo date identified in dbGaP and will not present or publish findings generated from the GAIN data sets until the embargo date passes. | The AU has since improved education of trainees and staff associated with research using dbGaP data sets to prevent any further incidents. |
| IRB approval | 1 | An investigator from a GAIN study site was approved to access a GAIN data set before it was confirmed that he or she did not have access to personal identifying information and did not need IRB approval. This was an error on the part of the DAC staff. The AU’s access to the GAIN data set was suspended until the IRB memo was received. | Term 2. Institutional and AU Responsibilities. AUs who might have access to personal identifying information for research participants in the original study at their institution or through their collaborators might be required to have IRB approval. | Multiple checkpoints were implemented in the GAIN DAC review process. The DUC was updated to clarify when IRB approval is required for GAIN data sets. |
| Unapproved data access | 1 | The AU was able to access a GAIN data set for which he or she was not approved. This resulted from an error in how the files were configured in dbGaP. The NCBI identified the issue shortly after access was granted, and the AU submitted a revised PR to include the additional data set. | Term 1. Research Use. Modifications to the research project require submission of an amendment to this application. | NCBI staff reviewed the dbGaP system to verify the configuration files and implemented additional checks for each data set before it was released to an AU. |
| Unapproved data access | 1 | The AU changed institutions and downloaded a data set to which he or she was previously granted access. The issue was identified when the AU contacted the NCBI for help with the file that was just downloaded. | Term 5. Nontransferability. AUs agree that if they change institutions during the access period, they will submit a new PR and DUC in which the new institution agrees to the NIH GWAS policy before data access resumes. | When a PI changes institutions, a close-out report is submitted to verify that data were destroyed at the original institution. The NCBI developed an automated system for DACs to close the PR, ensuring that data files could no longer be accessed. If a PI submits a new request from a new institution and it is approved, then files can be accessed again. |
Discussion
Providing access to large-scale genomic data sets that include rich phenotypic information has allowed hundreds of authorized investigators to replicate results of their GWASs, explore new hypotheses regarding genetic contributions to complex disease, and develop statistical methods to analyze large-scale genomics data sets. For example, the GAIN psoriasis data sets were used for the development of a mathematical model for immune cell interactions via the specific dose-dependent cytokine production rates of cell populations,7 and the GAIN schizophrenia data sets were used for illustrating a new method for controlling confounding in case-control studies.8 Additional findings generated from GAIN data sets accessed through dbGaP were presented at the GAIN Analysis II and GAIN Analysis III workshops held in 2007 and 2008, respectively. The PIs of the GAIN psychiatric-disorder data sets went on to form the Psychiatric Genomics Consortium (PGC) in 2007. The PGC was founded to integrate and analyze genome-wide SNP array data for meta- and mega-analyses and to conduct cross-disorder and comorbidity analyses of GAIN and other data sets. This consortium has since grown to more than 200 investigators from 19 institutions in 60 countries and encompasses more than 100,000 participants.9 A total of 188 submitted or published manuscripts have been reported to the GAIN DAC (115 from GAIN PIs and co-PIs and 73 from secondary approved users) as a result of making just six GAIN studies available to the broader research community, and there has been a proportionately small number of DMIs. This experience demonstrates the benefits of the GAIN program, which we believe is indicative of the benefits to be realized through the NIH GWAS policy and the NIH controlled-access system.
Although the GAIN data-access experience has been positive overall, recent reports have highlighted the potential for privacy risks when genomic and other molecular data are broadly shared.10–12 Robust controlled-access data systems and strict adherence of approved users to the terms of access can minimize many of these risks; however, they do not reduce the risk to zero. It remains important that research participants be made aware of relevant privacy risks during the informed-consent process and be given ample time to decide whether to participate in a study.13 Most importantly, we must continue to view the controlled-access data-sharing model as but one approach to promoting participant privacy while still enabling advances in biomedical research to be pursued.
Limitations to the GAIN data-access process include the complexities of the request system, difficulty in receiving annual reports from approved users in a timely fashion, relying on annual reports and approved users to identify potential data misuses, and a diversity of approaches that are used for evaluating requests by other NIH DACs and that can sometimes lead to confusion for investigators requesting data sets managed by different DACs. If multiple data sets are requested under a single PR and some of the responsible DACs request specific revisions, the revised request must then be rereviewed by all the DACs involved. Modifications to satisfy one DAC might then run afoul of requirements by another DAC and so on. To minimize or avoid this, NIH DAC chairs are meeting regularly through the PPDM to increase inter-DAC communication regarding requests that span multiple DACs and standardize the PR review process to the extent possible.
Several improvements have been suggested or implemented on the basis of DAC experience and user comments. For example, as the number of genomic data sets in dbGaP continues to grow, the NIH PPDM is actively working in conjunction with dbGaP staff to automate and improve the current PR process to allow investigators to simultaneously submit annual reports and access renewal requests through an online system. An automated system directly tied to renewing access to dbGaP data sets might also increase compliance in submission of annual reports. In addition, given the 5 years of data showing that not a single requesting investigator or his or her collaborators have been identified on the sanctions lists for federal research, the SOC recently decided that NIH DACs may discontinue the review of public websites for determining whether a requesting investigator or his or her collaborators have been prohibited from conducting federal research. This decision was also based on the fact that the signing officials affirm that requesting investigators are in good standing at their institutions. This procedural change will afford the DACs more time to focus on other aspects of the review process. These changes, along with new services (developed by dbGaP staff), including providing genotype-data files in PLINK format, allowing PIs to assign an authorized downloader from their lab to mange file download, simplifying download management for data sets with thousands of files, summarizing the publication embargo properties of downloaded data, and quickly extracting subsets of data from the extremely large original genome-sequence submissions, can make the data-access system more straightforward and efficient for investigators, their institutions, and the NIH DACs while maintaining the system’s integrity.
GAIN and the NIH GWAS policy were founded on the principle that maximum public benefit can be achieved if innovative research is pursued through rich and readily accessible genomic data in a manner that promotes the utmost respect and protection for research-participant interests. The experience of the GAIN program illustrates that the data-access process has facilitated widespread data dissemination, measured by the nearly 1,000 requests for data from investigators from academic, nonprofit, and industry researchers. There are a growing number of presentations and publications from approved users, continued requests for GWAS data, and relatively few policy violations. Improvements, such as those described above, are still needed to enhance efficiency and ease of use and maintain vigilance in oversight practices. The availability of GAIN data has allowed for numerous advances in both the understanding of the genetic underpinnings of mental-health disorders, diabetes, and psoriasis and the development and refinement of statistical methods for identifying genetic and environmental factors related to complex common diseases.
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Library of Medicine, National Institutes of Health.
Supplemental Data
Document S1. Figure S1, Box S1, rosters for key GAIN governance groups, and a detailed description of the dbGaP request system and the GAIN data-access review process
Web Resources
The URLs for data presented herein are as follows:
- Bibliography of GAIN publications, http://genome.gov/19518664#al-2
- dbGaP, http://www.ncbi.nlm.nih.gov/
- eRA Commons, https://commons.era.nih.gov/commons/
- GAIN Analysis Workshop I, http://www.genome.gov/27532173
- GAIN Analysis Workshop II, http://www.genome.gov/27532174
- GAIN Analysis Workshop III, http://www.genome.gov/27532175
- NIH Advisory Committee to the Director, http://acd.od.nih.gov/index.htm
- NIH GWAS Policy, http://gwas.nih.gov/index.html
- Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
References
- 1.Manolio T.A., Rodriguez L.L., Brooks L., Abecasis G., Ballinger D., Daly M., Donnelly P., Faraone S.V., Frazer K., Gabriel S., GAIN Collaborative Research Group; Collaborative Association Study of Psoriasis; International Multi-Center ADHD Genetics Project; Molecular Genetics of Schizophrenia Collaboration; Bipolar Genome Study; Major Depression Stage 1 Genomewide Association in Population-Based Samples Study; Genetics of Kidneys in Diabetes (GoKinD) Study Nat. Genet. 2007;39:1045–1051. [Google Scholar]
- 2.Shurin S.B. Clin. Transl. Sci. 2008;1:91. doi: 10.1111/j.1752-8062.2008.00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mailman M.D., Feolo M., Jin Y., Kimura M., Tryka K., Bagoutdinov R., Hao L., Kiang A., Paschall J., Phan L. Nat. Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGuire A.L., Basford M., Dressler L.G., Fullerton S.M., Koenig B.A., Li R., McCarty C.A., Ramos E., Smith M.E., Somkin C.P. Genome Res. 2011;21:1001–1007. doi: 10.1101/gr.120329.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ludman E.J., Fullerton S.M., Spangler L., Trinidad S.B., Fujii M.M., Jarvik G.P., Larson E.B., Burke W. J. Empir. Res. Hum. Res. Ethics. 2010;5:9–16. doi: 10.1525/jer.2010.5.3.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker L., Starks H., West K.M., Fullerton S.M. Sci. Transl. Med. 2011;3:13cm34. doi: 10.1126/scitranslmed.3002788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valeyev N.V., Hundhausen C., Umezawa Y., Kotov N.V., Williams G., Clop A., Ainali C., Ouzounis C., Tsoka S., Nestle F.O. PLoS Comput. Biol. 2010;6:e1001024. doi: 10.1371/journal.pcbi.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen A.S., Satten G.A. Am. J. Epidemiol. 2011;173:752–760. doi: 10.1093/aje/kwq406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan P.F. Neuron. 2010;68:182–186. doi: 10.1016/j.neuron.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Homer N., Szelinger S., Redman M., Duggan D., Tembe W., Muehling J., Pearson J.V., Stephan D.A., Nelson S.F., Craig D.W. PLoS Genet. 2008;4:e1000167. doi: 10.1371/journal.pgen.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schadt E.E., Woo S., Hao K. Nat. Genet. 2012;44:603–608. doi: 10.1038/ng.2248. [DOI] [PubMed] [Google Scholar]
- 12.Gymrek M., McGuire A.L., Golan D., Halperin E., Erlich Y. Science. 2013;339:321–324. doi: 10.1126/science.1229566. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez L.L., Brooks L.D., Greenberg J.H., Green E.D. Science. 2013;339:275–276. doi: 10.1126/science.1234593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1. Figure S1, Box S1, rosters for key GAIN governance groups, and a detailed description of the dbGaP request system and the GAIN data-access review process