Variant of TREM2 Associated with the Risk of Alzheimer’s Disease (original) (raw)
. Author manuscript; available in PMC: 2013 Jul 10.
Published in final edited form as: N Engl J Med. 2012 Nov 14;368(2):107–116. doi: 10.1056/NEJMoa1211103
Abstract
BACKGROUND
Sequence variants, including the _ε_4 allele of apolipoprotein E, have been associated with the risk of the common late-onset form of Alzheimer’s disease. Few rare variants affecting the risk of late-onset Alzheimer’s disease have been found.
METHODS
We obtained the genome sequences of 2261 Icelanders and identified sequence variants that were likely to affect protein function. We imputed these variants into the genomes of patients with Alzheimer’s disease and control participants and then tested for an association with Alzheimer’s disease. We performed replication tests using case–control series from the United States, Norway, the Netherlands, and Germany. We also tested for a genetic association with cognitive function in a population of unaffected elderly persons.
RESULTS
A rare missense mutation (rs75932628-T) in the gene encoding the triggering receptor expressed on myeloid cells 2 (TREM2), which was predicted to result in an R47H substitution, was found to confer a significant risk of Alzheimer’s disease in Iceland (odds ratio, 2.92; 95% confidence interval [CI], 2.09 to 4.09; P = 3.42×10−10). The mutation had a frequency of 0.46% in controls 85 years of age or older. We observed the association in additional sample sets (odds ratio, 2.90; 95% CI, 2.16 to 3.91; P = 2.1×10−12 in combined discovery and replication samples). We also found that carriers of rs75932628-T between the ages of 80 and 100 years without Alzheimer’s disease had poorer cognitive function than noncarriers (P = 0.003).
CONCLUSIONS
Our findings strongly implicate variant TREM2 in the pathogenesis of Alzheimer’s disease. Given the reported antiinflammatory role of TREM2 in the brain, the R47H substitution may lead to an increased predisposition to Alzheimer’s disease through impaired containment of inflammatory processes. (Funded by the National Institute on Aging and others.)
Alzheimer’s disease, the most common form of dementia in the elderly, is a neurodegenerative disorder that is characterized by a slow but progressive loss of cognitive function. Extracellular amyloid plaques, intracellular neurofibrillary tangles, and loss of neurons and synapses resulting in brain atrophy are the main pathological hallmarks of Alzheimer’s disease.1 Disease onset is usually after the age of 70 years, although the prevalence increases exponentially with age after the age of 65 years and exceeds 25% in those over the age of 90 years.2
The vast majority of variants in the sequence of the genome that have been shown to markedly affect the risk of Alzheimer’s disease are rare variants in APP, PSEN1, and PSEN2 (encoding amyloid precursor protein, presenilin 1, and presenilin 2, respectively). These variants appear to be fully penetrant and result in Alzheimer’s disease with an early onset, in most cases before the age of 60 years.3 However, these variants do not shed light on the most common, late-onset form of the disease. Although a number of common, low-risk variants have been associated with late-onset Alzheimer’s disease,4 the _ε_4 allele of apolipoprotein E (ApoE), originally discovered as a risk factor for Alzheimer’s disease in 1993,5,6 remains by far the most important sequence variant affecting the risk of late-onset Alzheimer’s disease because of its prevalence and the size of its effect on risk, with reported odds ratios ranging from 3 to 4 (a meta-analysis is available at www.alzgene.org/meta.asp?geneID = 83).
To search for sequence variants that influence the risk of Alzheimer’s disease, we performed a genomewide association analysis with variants (found by whole-genome sequencing of samples from 2261 Icelanders) that were likely to affect protein function. These variants were imputed in patients with Alzheimer’s disease and controls with the use of long-range haplotype phasing and chip-genotype information. Using this approach, we have recently reported variants that greatly influence the risk of the sick sinus syndrome,7 gout,8 gliomas,9 ovarian cancer,10 and Alzheimer’s disease.11
METHODS
STUDY PARTICIPANTS
Icelandic Population
Approval for these studies was obtained from the National Bioethics Committee and the Icelandic Data Protection Authority. Written informed consent was obtained from all participants or their guardians before blood samples were drawn, and all sample identifiers were encrypted in accordance with the regulations of the Icelandic Data Protection Authority.
In 1062 patients, the diagnosis of Alzheimer’s disease was established according to the criteria for definite, probable, or possible Alzheimer’s disease of the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association (NINCDS-ADRDA).12 In another 2697 patients, the diagnosis was established according to the criteria for code F00 of the International Classification of Diseases, 10th Revision (ICD-10). We assessed cognitive function using data from the Resident Assessment Instrument (RAI), with which assessment is performed on an individual basis and recorded in a Minimum Data Set (MDS 2.0) form. Data were primarily obtained through RAI 2.0 for Nursing Homes, which is a comprehensive and standardized instrument originally developed for residential facilities for the elderly,13 with additional information provided by the InterRAI Assessment for Home Care.14 We assessed cognitive function using the MDS Cognitive Performance Scale (CPS), which combines selected MDS 2.0 items expressing different measures of cognitive function on a seven-category scale, ranging from 0 (intact) to 6 (severe impairment).15 The CPS is hierarchical and based on an assessment of several measures of cognitive function; a 1-unit change is a reflection of distinct and measurable changes in at least one cognitive domain. A total of 1236 study participants with a score of 0 on the CPS scale were used as cognitively intact controls. We selected 110,050 population controls from among participants in various research projects at deCODE Genetics, excluding those in whom Alzheimer’s disease had been diagnosed.
Norwegian Population
The Regional Ethical Committee and the Norwegian Data Protection Agency approved the studies. The sample of patients with Alzheimer’s disease consisted of home-dwelling outpatients referred to three memory clinics in the Southeast Health Region of Norway for suspicious dementia. The patients underwent a standardized comprehensive assessment, which consisted of taking a medical history from the patient as well as a close family member, comprehensive neuropsychological testing, a physical and psychiatric examination with the use of standardized assessment scales, blood sample analyses, and brain imaging.16 Diagnoses of Alzheimer’s disease were established in accordance with the ICD-10 criteria for research. Controls were recruited as a part of the Thematically Organized Psychosis study (enrolling 700 patients and 291 controls)17 and two Norwegian studies of attention deficit–hyperactivity disorder (enrolling 626 patients and 898 controls).
Emory Population
All participants underwent a research evaluation in the Emory Alzheimer’s Disease Research Center in Atlanta. Participants were classified as controls or as having probable Alzheimer’s disease after a review of the history, physical examination, neuropsychological testing, and available clinical records in a consensus conference among neurologists, neuropsychologists, and other health care professionals. All controls underwent initial cognitive screening with a Mini–Mental State Examination (MMSE) and Clock Drawing Test (CDT), and those who were impaired (z score, −1.79 or less) after adjustment for age, sex, and education and all patients with Alzheimer’s disease underwent further neuropsychological testing consisting of a Brief Visuospatial Memory Test–Revised, Wechsler Memory Scale–Revised Logical Memory I and II, Wechsler Adult Intelligence Scale–Revised Similarities, Wechsler Adult Intelligence Scale III Digit Span, Wechsler Adult Intelligence Scale–Revised Digit Symbol, Judgment of Line Orientation, Trail Making Test A and B, Category Fluency (Animals, Vegetables), Phonemic Fluency, Boston Naming Test, Consortium to Establish a Registry for Alzheimer’s Disease (CERAD) Word List Memory, CDT evaluation, the Beck Depression Inventory (for participants <65 years of age), and the Geriatric Depression Scale (for participants ≥65 years of age). In interviews, controls had a negative response to questions about a personal history of a neurologic disease, and medical records that included imaging (computed tomography or magnetic resonance imaging), neuropsychiatric assessment, and ancillary testing were requested and reviewed when available.
Munich Population
Patients with Alzheimer’s disease were recruited at the memory clinic of the Department of Psychiatry, University of Munich, Germany. Participants in whom dementia associated with Alzheimer’s disease was diagnosed fulfilled the criteria for probable Alzheimer’s disease, according to the NINCDS-ADRDA criteria. The control group included participants who were randomly selected from the general population of Munich. Controls who had a disease of the central nervous system or a psychotic disorder or who had a first-degree relative with a psychotic disorder were excluded.
Rotterdam Population
The Rotterdam Study I is a prospective, population-based cohort study enrolling 7983 residents who are 55 years of age or older and live in Ommoord, a suburb of Rotterdam, the Netherlands.18 Baseline examinations took place between 1991 and 1993; follow-up examinations were performed between 1997 and 1999 and between 2002 and 2006; a final follow-up examination was performed between 2009 and 2011.19
Participants were screened for prevalent dementia with the use of a three-stage process; those free of dementia remained under surveillance for incident dementia, a determination that was made with the use of record linkage and assessment at three subsequent examinations. We included all patients in whom Alzheimer’s disease was diagnosed before December 31, 2011; a subset of those in whom Alzheimer’s disease was not diagnosed served as controls.
Screening was done with the MMSE and Geriatric Mental Schedule (GMS) for organic (i.e., medical or physical) mental illness for all participants. Participants who were deemed to be positive on screening (a score of <26 on the MMSE or >0 on the GMS organic level) underwent the Cambridge Mental Disorders of the Elderly Examination (CAMDEX) schedule. Participants in whom dementia was suspected underwent more extensive neuropsychological testing. When available, imaging data were used. In addition, all participants were continuously monitored for major events (including dementia) through automated linkage of the study database with digitized medical records from general practitioners, the Regional Institute for Outpatient Mental Health Care, and the municipality.
In addition, physicians’ files from nursing homes and general practitioners’ records for participants who moved out of the Ommoord district were reviewed twice a year. For suspected dementia events, additional information (including neuroimaging) was obtained from hospital records, and research physicians discussed available information with a neurologist experienced in dementia diagnosis and research to verify all diagnoses.
Dementia was diagnosed in accordance with internationally accepted criteria for dementia in the revised third edition of the Diagnostic and Statistical Manual of Mental Disorders, and Alzheimer’s disease was diagnosed on the basis of the NINCDS-ADRDA criteria for possible, probable, or definite disease. The criteria of the National Institute of Neurological Disorders and Stroke–Association Internationale pour la Recherche et l’Enseignement en Neurosciences (NINDS-AIREN) were used to diagnose vascular dementia. The final diagnosis was determined by a panel consisting of a neurologist, a neurophysiologist, and a research physician. The diagnoses of Alzheimer’s disease and vascular dementia were not mutually exclusive.
DATA GENERATION AND ANALYSIS
Whole-Genome Sequencing, SNP Calling, and Imputation
We performed whole-genome sequencing on samples obtained from 2261 Icelandic participants, followed by single-nucleotide-polymorphism (SNP) calling and genotype imputation, using methods that were described previously.11 The chip-genotype imputation was based on chip genotypes from 95,085 persons. Approximately 34 million markers (SNPs and insertion–deletion polymorphisms), including the 191,777 functional variants identified through whole-genome sequencing, were imputed in the Icelandic cases and controls. The information content for rs75932628 in the imputed data was 0.999 (as compared with 1.0 for perfect information).
Single-Track Assay SNP Genotyping
We performed single SNP genotyping of rs75932628 using the Centaurus (Nanogen) platform.20 No mismatches resulted from a comparison of genotypes determined through imputation and Centaurus genotyping of 964 participants, including 30 participants who were predicted to be heterozygous for the rare allele and 2 who were predicted to be homozygous for the rare allele. Samples from the United States, Germany, and Norway were also typed with the use of Centaurus assays. Before analysis, we excluded samples with a genotype yield of less than 90% and one member of each pair of duplicate samples. The genotyping yield was at least 95% in both cases and controls in samples from all study locations, and all genotypes were in Hardy–Weinberg equilibrium. Samples from the Netherlands were genotyped for rs75932628 with Taqman allelic discrimination Assays-by-Design (Applied Biosystems). All measurements were performed in accordance with the manufacturer’s protocols; primer and probe sequences are available from the manufacturer.
Imputation of Genomewide Data
We downloaded genotype and phenotype data from the Genetic Alzheimer’s Disease Associations (GenADA) study, the National Institute on Aging Late Onset Alzheimer’s Disease and National Cell Repository for Alzheimer’s Disease Family Study (NIA-LOAD), and the Electronic Medical Records and Genomics (eMERGE) genomewide association study of dementia from the controlled-access portal of the National Institutes of Health Genotype and Phenotype database (dbGAP, accession number phs000234.v1.p1). Two small components of NIA-LOAD, phs000168.v1.p1.c2 (involving 28 participants) and phs000168.v1.p1.c1 (involving 570 participants), could not be included because consent from participants was for nonprofit use only. In addition, rs75932628 could not be successfully imputed on the basis of the GenADA data (information content associated with the additive test, <0.3), which led to the exclusion of that study from further analyses.
In the NIA-LOAD and eMERGE studies, patients were classified as having definite, probable, or possible Alzheimer’s disease, according to NINCDS–ADRDA criteria. In the eMERGE study, patients in whom dementia was diagnosed according to electronic-medical-record criteria were excluded. In both studies, controls were free of dementia, and participants who were biologic relatives of patients with Alzheimer’s disease were not included as controls. Imputation was performed with the use of IMPUTE2 with the March 2012 haplotype release of the 1000 Genomes Project as a reference. Before imputation, participants with a genotyping yield of less than 98% were removed; SNPs with a yield of less than 98%, a minor allele frequency of less than 1%, or a deviation from Hardy–Weinberg equilibrium (P<1.0×10−5) were also removed. The information content of the imputed data associated with the additive test was 0.79 and 0.75 for the NIA–LOAD and eMERGE studies, respectively.
STATISTICAL ANALYSIS
For the Icelandic data, we performed case–control association testing of imputed genotypes using methods that were described previously.11 Odds ratios were calculated on the basis of a multiplicative model for the two chromosomes of each individual. The method of genomic control was used to correct for relatedness and potential population stratification.
We used logistic regression to perform association analysis that was based on the NIA-LOAD and eMERGE data sets, with the first three principal components included as covariates to adjust for population stratification. Before the analysis, we removed data for participants with genotyped sex inconsistent with reported sex, the lower-yield sample in each pair of duplicates, genetically related older cases and younger controls (to eliminate the inclusion of first- or second-degree relatives), and participants with an estimated fraction of less than 0.9 European ancestry in analysis with STRUCTURE software and using as a reference HapMap samples of Utah residents with ancestry from northern and western Europe (CEU), Han Chinese in Beijing and Japanese in Tokyo (CHB+JPT), and Yoruba in Ibadan, Nigeria (YRI).
We used Fisher’s exact test to perform other association analyses. We combined results from the various replication groups, and from the discovery group and the replication groups, using inverse-variance–weighted meta-analysis. The relationship between the age at onset and rs75932628-T was examined with the use of a linear model with the age at onset as the response and rs75932628-T and the ApoE _ε_4 count as predictors.
We analyzed the effect of age on the CPS score, using determinations made at several ages for each participant. The CPS score is based on the Resident Assessment Instrument for Nursing Homes, which is applied on average three times per year in Icelandic nursing homes. Since the residency time in nursing homes in Iceland is on average 3 to 4 years, many determinations of CPS that are performed at different times are available for most persons. We assessed the difference in CPS score between rs75932628-T carriers and noncarriers in the age range from 80 to 100 years using a mixed model with age and carrier status as fixed effects and the individual as a random effect. We used bootstrapping methods to calculate standard errors for the analysis of the CPS score versus age.
RESULTS
ASSOCIATION OF VARIANT WITH ALZHEIMER’S DISEASE
Through whole-genome sequencing of samples from 2261 Icelanders, we found 191,777 nonsynonymous SNPs, frameshift variants, splicing variants, and stop gain–loss variants and imputed these variants in patients with Alzheimer’s disease and controls. A total of 3550 patients with Alzheimer’s disease were included in the analysis. Our control group included persons who had reached the age of 85 without a diagnosis of Alzheimer’s disease. With the exclusion of the ApoE locus and the A673T variant in APP,11 only one marker, rs75932628, showed a genomewide association, on the basis of either the Bonferroni-adjusted threshold of P<2.60×10−7 for 191,777 tests or the conventional threshold for genome-wide association studies (5×10−8). The T allele of rs75932628, which encodes a substitution of histidine for arginine at position 47 (R47H) in the gene encoding the triggering receptor expressed on myeloid cells 2 (TREM2) on chromosome 6p21.1, with an allelic frequency of 0.63% in Iceland, was found to confer a significant risk of Alzheimer’s disease (odds ratio, 2.92; 95% confidence interval [CI], 2.09 to 4.09; P= 3.42×10−10) (Table 1). No other variant in TREM2 that is likely to affect protein function showed nominally significant association with Alzheimer’s disease (Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org).
Table 1.
Association between the rs75932628-T Variant and Alzheimer’s Disease in Comparisons with Three Control Groups.
| Control Group | No. of Participants | Frequency | Odds Ratio (95% CI) | P Value |
|---|---|---|---|---|
| % | ||||
| All population controls | 110,050 | 0.63 | 2.26 (1.71–2.98) | 1.13×10−8 |
| Population controls ≥85 yr of age | 8,888 | 0.46 | 2.92 (2.09–4.09) | 3.42×10−10 |
| Cognitively intact controls ≥85 yr of age* | 1,236 | 0.31 | 4.66 (2.38–9.14) | 7.39×10−6 |
Risk variants for a late-onset disorder such as Alzheimer’s disease are expected to be more common in the general population than in elderly controls without the disease. Thus, the use of elderly controls without a history of Alzheimer’s disease would in general be expected to result in an increased statistical power to detect risk variants for this disease. We therefore investigated the association of rs75932628-T using samples from cognitively intact elderly controls, as determined by their CPS scores. In an analysis of samples from such controls who were at least 85 years of age, the odds ratio for the association with rs75932628-T was 4.66 (95% CI, 2.38 to 9.14; P = 7.39×10−6). By contrast, we observed a smaller odds ratio in a comparison with samples from a general population controls of all ages (odds ratio, 2.26; 95% CI, 1.71 to 2.98; P = 1.13×10−8) (Table 1). The less significant P value that was observed for the comparison with cognitively intact controls was due to the substantially smaller size of this control group (1236, vs. 8888 elderly controls and 110,050 population controls). Furthermore, we found that the frequency of rs75932628-T in controls age 85 years or older without a history of Alzheimer’s disease (0.46%) was significantly less than in controls under the age of 85 years (0.64%; P = 0.007). This observation is expected for alleles associated with common, late-onset disorders such as Alzheimer’s disease and thus provides further support for the association between rs75932628-T and Alzheimer’s disease.
We identified four homozygous carriers of rs75932628-T in Iceland. Of these homozygotes, Alzheimer’s disease had been diagnosed in two but not in two others (ages 51 and 52).
REPLICATION SERIES
In an attempt to replicate the association between rs75932628-T and Alzheimer’s disease, we genotyped rs75932628 in cohorts from the United States (Emory), Germany, the Netherlands (Rotterdam Study), and Norway. We found that rs75932628-T conferred a risk of Alzheimer’s disease in all replication cohorts, with a combined odds ratio of 2.83 (95% CI, 1.45 to 5.40; P = 0.002 (Table 2). Combining results from Iceland (using population controls who were at least 85 years of age) and the replication cohorts, we found that the overall association between rs75932628-T and Alzheimer’s disease was highly significant (odds ratio, 2.90; 95% CI, 2.16 to 3.91; P = 2.1×10−12). We also estimated the effect of rs75932628-T on the risk of Alzheimer’s disease by imputing the variant in two publicly available data sets (NIA-LOAD and eMERGE). Association results that were based on imputed genotypes for rs75932628-T in these data sets were found to be consistent with the observed effect of rs75932628-T on disease risk in the genotyped cohorts (odds ratio, 2.66; 95% CI, 1.46 to 4.84; P = 0.001) (Table S2 in the Supplementary Appendix).
Table 2.
Replication Analysis of the Association between the rs75932628-T Variant and Alzheimer’s Disease.
| Group | No. of Cases | No. of Controls | Frequency* | Odds Ratio (95% CI) | P Value |
|---|---|---|---|---|---|
| % | |||||
| Emory | 399 | 402 | 0.12 | 3.03 (0.33–78.35) | 0.37 |
| Munich | 517 | 1891 | 0.19 | 3.15 (1.06–10.40) | 0.04 |
| Rotterdam | 944 | 4950 | 0.15 | 2.45 (0.94–6.35) | 0.07 |
| Norway | 177 | 2484 | 0.16 | 3.52 (0.54–17.21) | 0.14 |
| Combined | 2037 | 9727 | 2.83 (1.45–5.40) | 0.002 |
ASSOCIATION WITH APOE _ε_4
We investigated the effect of the _ε_4 allele of ApoE on the association between rs75932628-T and Alzheimer’s disease. We found a somewhat higher odds ratio in _ε_4 noncarriers (4.03) than in _ε_4 carriers (2.38) (Table S3 in the Supplementary Appendix). The difference in the frequency of rs75932628-T in ApoE _ε_4 carriers, as compared with noncarriers, had borderline significance (odds ratio, 0.60; 95% CI, 0.37 to 0.98; P = 0.04). However, in a logistic-regression model, the interaction between rs75932628-T and ApoE _ε_4 was not significant (P = 0.18), and the difference in frequency according to ApoE _ε_4 status in cases did not replicate in the additional data sets (odds ratio, 1.79; 95% CI, 0.73 to 4.43; P = 0.20) (Table S4 in the Supplementary Appendix).
Although the population frequency of rs75932628-T was low (0.63% in Iceland), it conferred a risk of Alzheimer’s disease that was similar to the risk the ApoE _ε_4 allele, which has a population frequency of 17.3% in Iceland. (As compared with controls 85 years of age or older, the odds ratio for Alzheimer’s disease was 2.92 for rs75932628-T and 3.08 for the ApoE _ε_4 allele.) We also found that in Iceland, each copy of rs75932628-T was associated with an age at onset of Alzheimer’s disease that was lower by 3.18 years than in controls without the variant (P = 0.20). Although this effect was similar to that of ApoE _ε_4 (3.22 years; P = 4.1×10−8), it was not significant, owing to the low frequency of the variant, which resulted in a reduced effective sample size and an elevated standard error (2.49 for rs75932628-T vs. 0.58 for ApoE _ε_4). We found a similar result in the Dutch data (3.65 years per allele, P = 0.13), and the combined effect was found to be 3.4 years per allele (P = 0.048).
ASSOCIATION ACCORDING TO AGE
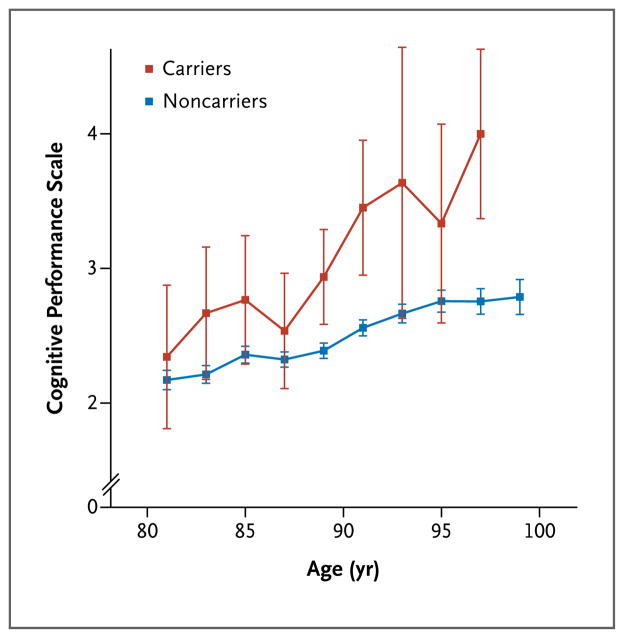
We also investigated how rs75932628-T affects cognitive function in elderly controls in whom Alzheimer’s disease had not been diagnosed. Cognitive function declined steadily with age in elderly persons between the ages of 80 and 100 (Fig. 1). We found that carriers of rs75932628-T showed worse cognition (a mean increase of 0.87 units on the CPS) than did noncarriers (P = 0.003). Clinical determination of Alzheimer’s disease is partially based on progressive loss of cognitive function, in particular memory, with time. Thus, the decline in cognitive function that we observed in rs75932628-T carriers may be due to early cognitive deficits that ultimately result in Alzheimer’s disease. Alternatively, the decline may at least partially be due to a loss of cognitive function in old age that is not associated with Alzheimer’s disease. The latter explanation is in keeping with the hypothesis that Alzheimer’s disease may be the extreme of the cognitive decline of the elderly and caused by the same biochemical mechanism.11
Figure 1. Cognition as a Function of Age in Controls Who Were Carriers or Noncarriers of the rs75932628-T Variant Associated with the Risk of Alzheimer’s Disease.
Shown are scores on the Cognitive Performance Scale (CPS) for carriers and noncarriers of the rs75932628-T variant associated with Alzheimer’s disease, according to age. Scores on the CPS range from 0 to 6, with higher scores indicating more severe impairment. Values are shown in 2-year bins (i.e., the data point for 81 years of age contains data for ages 80 and 81), except for the last bin, which represents ages of 98, 99, and 100 years. No CPS data were available for carriers in the last age bin. Each data point represents the average CPS score for participants in the respective age bin. The I bars represent standard errors. The graph is based on 307 measurements from 53 carriers and 24,152 measurements from 3699 noncarriers. Patients in whom Alzheimer’s disease had been diagnosed were not included in the analysis.
DISCUSSION
Inflammation is a well-established histologic feature in the brains of patients with Alzheimer’s disease. Complement factors were identified in amyloid plaques in the 1980s,21,22 followed by reports of clusters of activated microglia, complement-activation products, and cytokines in and near amyloid plaques.23–26 There is evidence that inflammation is an early event in the brains of patients with Alzheimer’s disease.27 It has also been noted that the expression of genes associated with inflammation in the brain is increased in aging and that this effect is accentuated in patients with Alzheimer’s disease.28 According to the amyloid hypothesis, which is the predominant theory about the pathogenesis of this disease, inflammation is a downstream effect of amyloidogenesis, which provides a trigger for the inflammatory response.
Genomewide association studies have also provided evidence of the importance of inflammation in Alzheimer’s disease. Thus, low-risk variants have been found in CR1,29 which belongs to the complement factor family of genes; in MS4A6A and MS4A4E,30 which are members of a cell-surface gene family expressed in lymphoid tissue; and in CD33,31 which encodes a myeloid cell-surface receptor.
TREM2 was originally identified as a DAP12-associated receptor that was expressed on macrophages and dendritic cells32 and was later shown to be expressed on osteoclasts and microglia.33 TREM2 is a transmembrane glycoprotein, consisting of an extracellular immunoglobulin-like domain, a transmembrane domain, and a cytoplasmic tail, which associates with DAP12 for its signaling function.32,34 TREM2 has both exogenous ligands on pathogens and endogenous ligands that remain largely unknown, although a recent study has shown that Hsp60 is an agonist of TREM2 in neuroblastoma cells and astrocytes.35 In addition, an endogenous ligand on dendritic cells has been found.36
In brain cells, TREM2 is primarily expressed on microglia, the resident histiocytes of the central nervous system.37 Activation of microglia may lead to phagocytosis of cell debris and amyloid, but microglia can also be activated to promote the production of proinflammatory cytokines, or they may differentiate into antigen-presenting cells.38 A recent study showed that TREM2 expression is induced concomitantly with the formation of amyloid plaques in APP transgenic mice expressing the Swedish mutation (K670N/M671L) in APP,39 and this expression was found to correlate positively with amyloid phagocytosis by unactivated microglia.
The expression of TREM2 also correlated positively with the ability of microglia to stimulate the proliferation of CD4+ T cells, as well as the secretion of tumor necrosis factor and CCL2, but not interferon-γ, into the extracellular milieu. This led the authors to speculate that TREM2-positive microglia on plaques capture and present self-antigens to lymphocytes infiltrating the central nervous system without promoting proinflammatory responses.39 Furthermore, knock-down of TREM2 or DAP12 in microglia resulted in reduced phagocytosis of apoptotic neurons, whereas the overexpression of TREM2 increased such phagocytosis,40 suggesting that microglia recognize and phagocytose apoptotic neurons through TREM2 ligation. TREM2 has an antiinflammatory function; it inhibits macrophage response to ligation of toll-like receptor (TLR),41 and it negatively regulates TLR-mediated maturation of dendritic cells, type I interferon responses, and the induction of antigen-specific T-cell proliferation.36 Furthermore, TREM2 stimulation of dendritic cells induces partial activation without any production of proinflammatory cytokines.34
Polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy, which produces increased signals from the deep white matter of the brain on T2-weighted magnetic resonance imaging, is called Nasu–Hakola disease. It is a rare recessively inherited disease that is characterized by painful bone cysts in wrists and ankles, psychotic symptoms, and progressive presenile dementia with onset in the fourth decade of life, usually leading to death in the fifth decade of life.42–44 Loss-of-function mutations in DAP12 and TREM2 were originally found in patients with Nasu–Hakola disease about a decade ago,45,46 suggesting that the TREM2–DAP12–mediated pathway may be important for human brain and bone tissue. Nasu–Hakola disease and Alzheimer’s disease are distinct from each other, and the clinical symptoms of Nasu–Hakola disease (early onset, painful bone cysts, fractures of bones of the limbs, and sclerosing leukoencephalopathy) are incompatible with the diagnosis of Alzheimer’s disease. Bearing in mind that it is possible that rare mutations accounting for a small proportion of cases of common diseases may define a clinical subgroup, we looked for but did not find clinical features (e.g., sex distribution, radiographic features, and rate of disease progression) that clearly separate carriers of the R47H mutation from noncarriers with Alzheimer’s disease, although the age at disease onset was on average 3.18 years earlier in the carriers than in the noncarriers.
A homozygous mutation in the 5′ consensus donor splice site in intron 1 of TREM2 in a Lebanese family, leading to early-onset dementia without bone cysts, has been reported.47 Furthermore, three homozygous mutations in TREM2 have recently been reported in three Turkish probands with frontotemporal dementia-like disease in the absence of bone cysts,48 and there is also a report of memory deficits in heterozygous carriers of a loss-of-function mutation in TREM2 in an Italian family.49 These findings suggest that TREM2 may be crucial for the integrity of cognitive function.
The R47H substitution encoded by rs75932628-T is located within the extracellular immunoglobulin-like domain of TREM2. The amino acid substitution may result in decreased affinity of TREM2 for its natural ligands and affect its signaling. It has recently been proposed that TREM2 may represent a proteolytic substrate for _γ_-secretase, although the exact cleavage site was not identified.50 If this proteolytic activity is confirmed, processing of TREM2 may be affected by the R47H substitution.
In conclusion, we have found a new risk variant, rs75932628-T, for Alzheimer’s disease. Although this variant occurs with less frequency than the ApoE _ε_4 allele, it confers a risk of Alzheimer’s disease with an effect size that is similar to that of ApoE _ε_4. Given the involvement of TREM2 in the phagocytic role of microglia on amyloid plaques, it is possible that reduced TREM2 activity caused by the R47H substitution may lead to brain damage through the inability of the brain to clear these toxic products.
Supplementary Material
Supplement1
Acknowledgments
Supported by grants from the National Institute on Aging (P50-AG025688, to Dr. Levey, for samples from the Alzheimer’s Disease Center at Emory University; U01AG006781 for the Alzheimer’s Disease Patient Registry and Adult Changes in Thought study; and the Division of Neuroscience for the NIA-LOAD study); a grant from the Research Council of Norway and South-Eastern Norway Health Authority for samples from Norway; a gift from 3M to expand the Adult Changes in Thought cohort; a grant from the National Institutes of Health (U01HG004438) for genotyping at Johns Hopkins University; cooperative agreements with the National Human Genome Research Institute (U01HG004610 for genomewide association analyses); by the eMERGE Administrative Coordinating Center (U01HG004603) for assistance with phenotype harmonization and genotype data cleaning; and by the National Center for Biotechnology Information. The Rotterdam Study was funded by Erasmus Medical Center and Erasmus University, Rotterdam; the Netherlands Organization for Health Research and Development; the Research Institute for Diseases in the Elderly; the Ministry of Education, Culture and Science; the Ministry for Health, Welfare and Sports; the European Commission; and the Municipality of Rotterdam; by a grant (014-93-015; RIDE2) from the Research Institute for Diseases in the Elderly, Stichting Alzheimer Onderzoek, Hersenstichting Nederland, the Netherlands Genomics Initiative–Netherlands Organization for Scientific Research (Center for Medical Systems Biology and the Netherlands Consortium for Healthy Aging), the Seventh Framework Program (FP7/2007-2013), and the ENGAGE project (grant agreement HEALTH-F4-2007-201413).
We thank Aaron Isaacs, Ben Oostra, and Jeannette Vergeer for genotyping and Renee de Bruijn for the clinical workup in the Rotterdam study; and Pål Zeiner and Heidi Aase for access to Norwegian control samples.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Castellani RJ, Rolston RK, Smith MA. Alzheimer disease. Dis Mon. 2010;56:484–546. doi: 10.1016/j.disamonth.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qiu C, Kivipelto M, von Strauss E. Epidemiology of Alzheimer’s disease: occurrence, determinants, and strategies toward intervention. Dialogues Clin Neurosci. 2009;11:111–28. doi: 10.31887/DCNS.2009.11.2/cqiu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cruts M, Theuns J, Van Broeckhoven C. Locus-specific mutation databases for neurodegenerative brain diseases. Hum Mutat. 2012;33:1340–4. doi: 10.1002/humu.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68:270–81. doi: 10.1016/j.neuron.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 5.Saunders AM, Strittmatter WJ, Schmechel D, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology. 1993;43:1467–72. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 6.Strittmatter WJ, Saunders AM, Schmechel D, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:1977–81. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holm H, Gudbjartsson DF, Sulem P, et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat Genet. 2011;43:316–20. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sulem P, Gudbjartsson DF, Walters GB, et al. Identification of low-frequency variants associated with gout and serum uric acid levels. Nat Genet. 2011;43:1127–30. doi: 10.1038/ng.972. [DOI] [PubMed] [Google Scholar]
- 9.Stacey SN, Sulem P, Jonasdottir A, et al. A germline variant in the TP53 poly-adenylation signal confers cancer susceptibility. Nat Genet. 2011;43:1098–103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rafnar T, Gudbjartsson DF, Sulem P, et al. Mutations in BRIP1 confer high risk of ovarian cancer. Nat Genet. 2011;43:1104–7. doi: 10.1038/ng.955. [DOI] [PubMed] [Google Scholar]
- 11.Jonsson T, Atwal JK, Steinberg S, et al. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012;488:96–9. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 12.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–44. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 13.Morris JN, Hawes C, Fries BE, et al. Designing the national resident assessment instrument for nursing homes. Gerontologist. 1990;30:293–307. doi: 10.1093/geront/30.3.293. [DOI] [PubMed] [Google Scholar]
- 14.Morris JN, Fries BE, Steel K, et al. Comprehensive clinical assessment in community setting: applicability of the MDS-HC. J Am Geriatr Soc. 1997;45:1017–24. doi: 10.1111/j.1532-5415.1997.tb02975.x. [DOI] [PubMed] [Google Scholar]
- 15.Morris JN, Fries BE, Mehr DR, et al. MDS Cognitive Performance Scale. J Gerontol. 1994;49:M174–M182. doi: 10.1093/geronj/49.4.m174. [DOI] [PubMed] [Google Scholar]
- 16.Brækhus A, Ulstein I, Wyller TB, Engedal K. The Memory Clinic — outpatient assessment when dementia is suspected. Tidsskr Nor Laegeforen. 2011;131:2254–7. doi: 10.4045/tidsskr.11.0786. [DOI] [PubMed] [Google Scholar]
- 17.Athanasiu L, Mattingsdal M, Kahler AK, et al. Gene variants associated with schizophrenia in a Norwegian genome-wide study are replicated in a large European cohort. J Psychiatr Res. 2010;44:748–53. doi: 10.1016/j.jpsychires.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofman A, Breteler MM, van Duijn CM, et al. The Rotterdam Study: objectives and design update. Eur J Epidemiol. 2007;22:819–29. doi: 10.1007/s10654-007-9199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofman A, van Duijn CM, Franco OH, et al. The Rotterdam Study: 2012 objectives and design update. Eur J Epidemiol. 2011;26:657–86. doi: 10.1007/s10654-011-9610-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kutyavin IV, Milesi D, Belousov Y, et al. A novel endonuclease IV post-PCR genotyping system. Nucleic Acids Res. 2006;34(19):e128. doi: 10.1093/nar/gkl679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eikelenboom P, Stam FC. Immunoglobulins and complement factors in senile plaques: an immunoperoxidase study. Acta Neuropathol. 1982;57:239–42. doi: 10.1007/BF00685397. [DOI] [PubMed] [Google Scholar]
- 22.Ishii T, Haga S. Immuno-electron-microscopic localization of complements in amyloid fibrils of senile plaques. Acta Neuropathol. 1984;63:296–300. doi: 10.1007/BF00687336. [DOI] [PubMed] [Google Scholar]
- 23.Akiyama H, Barger S, Barnum S, et al. Inflammation and Alzheimer’s disease. Neurobiol Aging. 2000;21:383–421. doi: 10.1016/s0197-4580(00)00124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffin WS, Stanley LC, Ling C, et al. Brain interleukin 1 and S-100 immunoreactivity are elevated in Down syndrome and Alzheimer disease. Proc Natl Acad Sci U S A. 1989;86:7611–5. doi: 10.1073/pnas.86.19.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers J, Luber-Narod J, Styren SD, Civin WH. Expression of immune system-associated antigens by cells of the human central nervous system: relationship to the pathology of Alzheimer’s disease. Neurobiol Aging. 1988;9:339–49. doi: 10.1016/s0197-4580(88)80079-4. [DOI] [PubMed] [Google Scholar]
- 26.Rozemuller JM, Eikelenboom P, Stam FC. Role of microglia in plaque formation in senile dementia of the Alzheimer type: an immunohistochemical study. Virchows Arch B Cell Pathol Incl Mol Pathol. 1986;51:247–54. doi: 10.1007/BF02899034. [DOI] [PubMed] [Google Scholar]
- 27.Eikelenboom P, van Exel E, Hoozemans JJ, Veerhuis R, Rozemuller AJ, van Gool WA. Neuroinflammation — an early event in both the history and pathogenesis of Alzheimer’s disease. Neurodegener Dis. 2010;7:38–41. doi: 10.1159/000283480. [DOI] [PubMed] [Google Scholar]
- 28.Blalock EM, Chen KC, Stromberg AJ, et al. Harnessing the power of gene microarrays for the study of brain aging and Alzheimer’s disease: statistical reliability and functional correlation. Ageing Res Rev. 2005;4:481–512. doi: 10.1016/j.arr.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Lambert JC, Heath S, Even G, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–9. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 30.Hollingworth P, Harold D, Sims R, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–35. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertram L, Lange C, Mullin K, et al. Genome-wide association analysis reveals putative Alzheimer’s disease susceptibility loci in addition to APOE. Am J Hum Genet. 2008;83:623–32. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouchon A, Dietrich J, Colonna M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol. 2000;164:4991–5. doi: 10.4049/jimmunol.164.10.4991. [DOI] [PubMed] [Google Scholar]
- 33.Colonna M. TREMs in the immune system and beyond. Nat Rev Immunol. 2003;3:445–53. doi: 10.1038/nri1106. [DOI] [PubMed] [Google Scholar]
- 34.Bouchon A, Hernández-Munain C, Cella M, Colonna MA. DAP12-mediated pathway regulates expression of CC chemokine receptor 7 and maturation of human dendritic cells. J Exp Med. 2001;194:1111–22. doi: 10.1084/jem.194.8.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stefano L, Racchetti G, Bianco F, et al. The surface-exposed chaperone, Hsp60, is an agonist of the microglial TREM2 receptor. J Neurochem. 2009;110:284–94. doi: 10.1111/j.1471-4159.2009.06130.x. [DOI] [PubMed] [Google Scholar]
- 36.Ito H, Hamerman JA. TREM-2, triggering receptor expressed on myeloid cell-2, negatively regulates TLR responses in dendritic cells. Eur J Immunol. 2012;42:176–85. doi: 10.1002/eji.201141679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sessa G, Podini P, Mariani M, et al. Distribution and signaling of TREM2/DAP12, the receptor system mutated in human polycystic lipomembraneous osteodysplasia with sclerosing leukoencephalopathy dementia. Eur J Neurosci. 2004;20:2617–28. doi: 10.1111/j.1460-9568.2004.03729.x. [DOI] [PubMed] [Google Scholar]
- 38.Carson MJ, Doose JM, Melchior B, Schmid CD, Ploix CC. CNS immune privilege: hiding in plain sight. Immunol Rev. 2006;213:48–65. doi: 10.1111/j.1600-065X.2006.00441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melchior B, Garcia AE, Hsiung BK, et al. Dual induction of TREM2 and tolerance-related transcript, Tmem176b, in amyloid transgenic mice: implications for vaccine-based therapies for Alzheimer’s disease. ASN Neuro. 2010;2(3):e00037. doi: 10.1042/AN20100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi K, Rochford CD, Neumann H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J Exp Med. 2005;201:647–57. doi: 10.1084/jem.20041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamerman JA, Jarjoura JR, Humphrey MB, Nakamura MC, Seaman WE, Lanier LL. Cutting edge: inhibition of TLR and FcR responses in macrophages by triggering receptor expressed on myeloid cells (TREM)-2 and DAP12. J Immunol. 2006;177:2051–5. doi: 10.4049/jimmunol.177.4.2051. [DOI] [PubMed] [Google Scholar]
- 42.Hakola HP. Neuropsychiatric and genetic aspects of a new hereditary disease characterized by progressive dementia and lipomembranous polycystic osteodysplasia. Acta Psychiatr Scand Suppl. 1972;232:1–173. [PubMed] [Google Scholar]
- 43.Madry H, Prudlo J, Grgic A, Freyschmidt J. Nasu-Hakola disease (PLOSL): report of five cases and review of the literature. Clin Orthop Relat Res. 2007;454:262–9. doi: 10.1097/01.blo.0000229364.57985.df. [DOI] [PubMed] [Google Scholar]
- 44.Nasu T, Tsukahara Y, Terayama K. A lipid metabolic disease — “membranous lipodystrophy” — an autopsy case demonstrating numerous peculiar membrane-structures composed of compound lipid in bone and bone marrow and various adipose tissues. Acta Pathol Jpn. 1973;23:539–58. doi: 10.1111/j.1440-1827.1973.tb01223.x. [DOI] [PubMed] [Google Scholar]
- 45.Paloneva J, Kestilä M, Wu J, et al. Loss-of-function mutations in TYROBP (DAP12) result in a presenile dementia with bone cysts. Nat Genet. 2000;25:357–61. doi: 10.1038/77153. [DOI] [PubMed] [Google Scholar]
- 46.Paloneva J, Manninen T, Christman G, et al. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet. 2002;71:656–62. doi: 10.1086/342259. [Erratum, Am J Hum Genet 2003;72:225.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chouery E, Delague V, Bergougnoux A, Koussa S, Serre JL, Mégarbané A. Mutations in TREM2 lead to pure early-onset dementia without bone cysts. Hum Mutat. 2008;29(9):E194–E204. doi: 10.1002/humu.20836. [DOI] [PubMed] [Google Scholar]
- 48.Guerreiro RJ, Lohmann E, Brás JM, et al. Using exome sequencing to reveal mutations in TREM2 presenting as a fronto-temporal dementia-like syndrome without bone involvement. Arch Neurol. 2012 Oct 8; doi: 10.1001/jamaneurol.2013.579. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montalbetti L, Ratti MT, Greco B, Aprile C, Moglia A, Soragna D. Neuropsychological tests and functional nuclear neuroimaging provide evidence of subclinical impairment in Nasu-Hakola disease heterozygotes. Funct Neurol. 2005;20:71–5. [PubMed] [Google Scholar]
- 50.Wunderlich P. γ-Secretase mediated proteolytic processing of the triggering receptor expressed on myeloid cells-2 — functional implications for intracellular signalling. Bonn, Germany: University of Bonn; 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement1