High-throughput sequencing of B- and T-lymphocyte antigen receptors in hematology (original) (raw)
Abstract
Application of high-throughput DNA sequencing to the analysis of B- and T-lymphocyte antigen receptors has great potential for improving the monitoring of lymphoid malignancies, assessing immune reconstitution after hematopoietic cell transplantation, and characterizing the composition of lymphocyte repertoires. Current technology can define the number and frequency of immunoglobulin heavy, T-cell receptor (TCR)α, TCRβ, or TCRγ chains expressed in a population of lymphocytes; techniques for determining the number of antigen receptor heterodimers, such as TCRαβ pairs, expressed in the population are under development.
Introduction
B and T lymphocytes are unique among somatic cells because much of their fundamental biology is determined by DNA sequences that are not encoded in the germline. The genes that encode B- and T-cell antigen receptors undergo rearrangement during lymphocyte development in a process that concatenates noncontiguous variable (V), diversity (D), and joining (J) gene segments to assemble sequences encoding functional receptors whose antigenic specificity is determined by 3 complementarity-determining regions (CDRs) (Figure 1; supplemental movies 1A and 1B).1,2,3 The V gene segments encode the CDR1 and CDR2 of both BCRs and TCRs, whereas the V, D, and J segments collectively encode CDR3, which is the most critical determinant of antigenic specificity. Extremely diverse repertoires of B- and T-cell antigen receptors are created by exploiting both combinatorial diversity, made possible by the existence of many distinct V, D, and J gene segments in the 4 T-cell and 3 B-cell antigen receptor loci, as well as junctional diversity. The latter is created by the unique molecular mechanism that mediates V-D-J rearrangement and involves template-independent insertion and deletion of nucleotides at the V-D, D-J, and V-J splice junctions. The diversity of the BCR repertoire is further increased by somatic hypermutation (SHM) of previously rearranged BCR genes during affinity maturation after initial antigen encounter. SHM is not limited to the V-D-J region that encodes the CDR3 and occurs throughout the V gene segment, potentially affecting the sequence of CDR1 and CDR2, as well as the intervening framework regions.
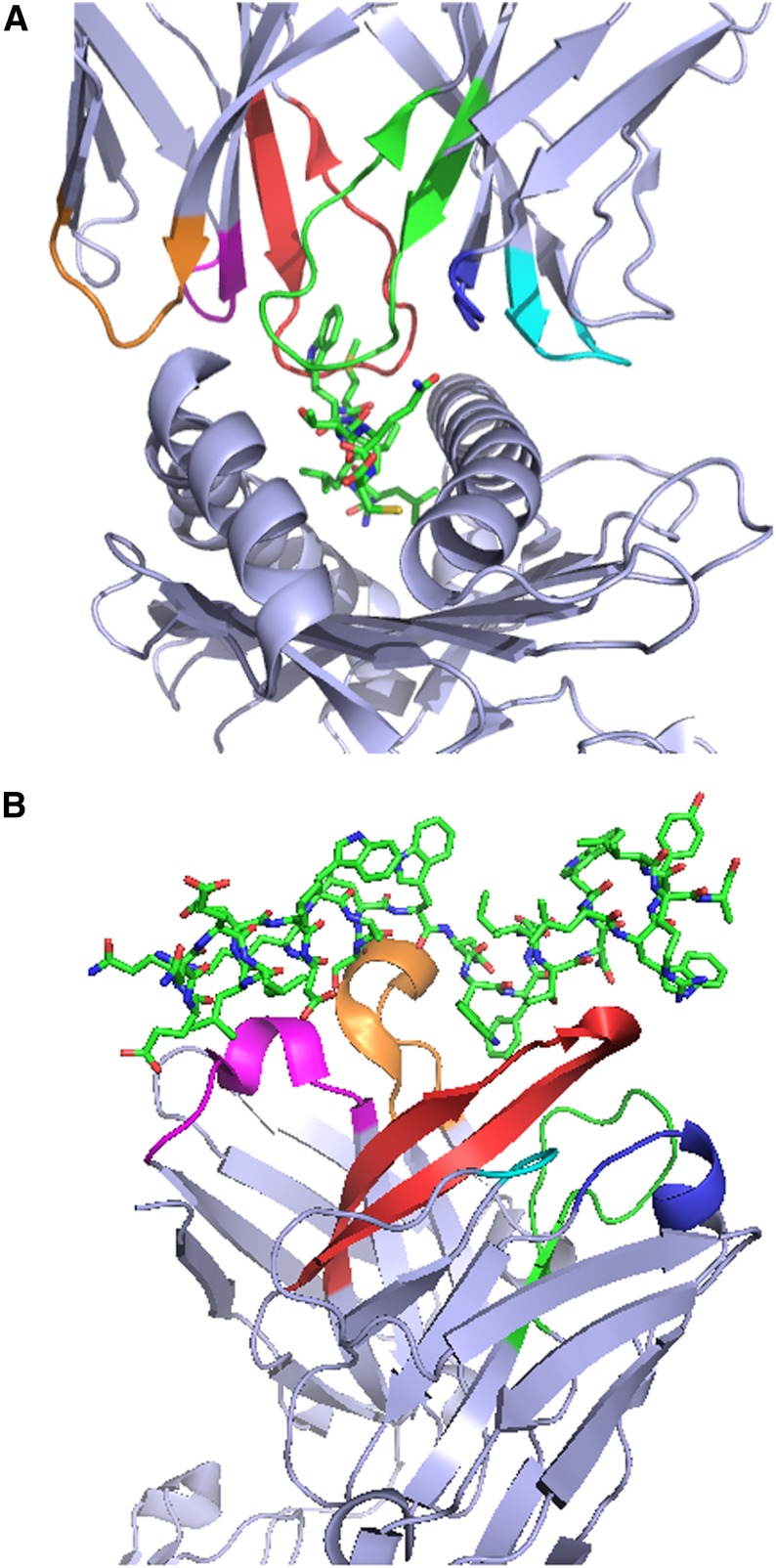
Figure 1.
Crystal structures of T-cell receptors (TCRs) and B-cell receptors (BCRs) in complex with their cognate antigens illustrate the critical role of CDRs in antigen recognition. (A) Structure of the 1G4 TCR in complex with the NY-ESO-1157-165 peptide and HLA-A*02:01.1 The first, second, and third CDRs (CDR1, CDR2, and CDR3) of the 1G4 α chain are colored magenta, orange, and red, respectively, to enable easier visualization, and the corresponding regions of the 1G4 β chain are colored blue, cyan, and green. (B) Structure of the broadly neutralizing 10E8 anti-HIV gp41 antibody antigen-binding fragment in complex with a gp41-derived peptide.2 The CDR1, CDR2, and CDR3 of the 10E8 heavy chain and the corresponding regions of the 10E8 light chain are colored magenta, orange, red, blue, cyan, and green, respectively. The structures in (A) and (B) were rendered with PyMOL.3
The antigen receptor repertoires created by the processes of V-D-J rearrangement and SHM are far too diverse to be amenable to comprehensive exploration and definition by conventional capillary-based DNA sequencing. Over the past 5 years, high-throughput DNA sequencing has been applied to the analysis of BCR4-7 and TCR8-11 CDR3 region sequence repertoires, enabling analysis of the repertoires realized in any given individual with unprecedented depth, resolution, and accuracy. The CDR3 region in the vast majority of successfully rearranged TCRβ chains and immunoglobulin heavy (IGH) chains comprises no more than 668,9 or 907 bp, encoding no more than 22 or 30 amino acids, respectively. This region is therefore ideally suited to comprehensive definition by current sequencing platforms that can generate ∼1 × 109 reads of the requisite length in a single run. Although these platforms cannot yet capture the entire V gene segment in addition to the V-D-J region, which limits their utility for analysis of SHM in BCR genes, SHM can be studied using other platforms that provide longer read lengths, albeit with less depth per sequencing run (Figure 2).5,12 Both genomic DNA and complementary DNA from lymphocytes have been used as the template for sequencing, but genomic DNA is preferred for studies in which the relative frequency of specific BCR or TCR sequences is to be assessed. Molecular and computational strategies based on high-throughput DNA sequencing have made it possible to explore BCR and TCR sequence repertoires of any degree of complexity, and thereby to address biological questions that were never before amenable to direct experimental analysis.
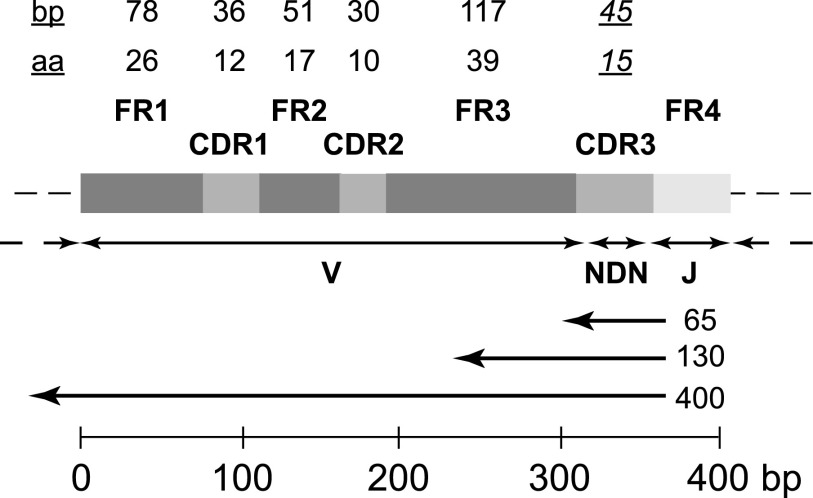
Figure 2.
Generic structure of a locus encoding the variable region of a TCRβ or IGH chain after V-D-J rearrangement, illustrating the intervals that encode the 4 framework (FR1-FR4) and 3 complementarity-determining (CDR1-CDR3) regions, the portions contributed by the V, D, and J gene segments, and the “N” nucleotides inserted at the V-D and D-J junctions. The maximum length of FR1-FR3, CDR1, and CDR2, and the modal length of CDR3, in base pairs (bp) and amino acids (aa), are indicated by the numbers at the top of the figure. The left-pointing arrows below the block structure indicate the approximate fraction of the locus that would be sequenced with reads of the indicated length (65, 130, or ≥400 bp) initiated by sequencing primers annealing to the 5′ region of the J gene segment, typical of current deep-sequencing protocols. The scale bar at the bottom of the figure indicates the length of the generic locus in base pairs.
Defining the characteristics of B- and T-cell repertoires
High-throughput sequencing of rearranged BCR and TCR genes in lymphocytes from peripheral blood has begun to define the essential characteristics of the B- and T-cell repertoires in healthy adults. Sequencing of the IGH locus in naive B cells from 4 individuals revealed an average of ∼86 000 unique CDR3 sequences per person.7 Up to 650 000 unique TCRβ CDR3 sequences were observed in naive CD8**+** T cells from single individuals,10 and exhaustive TCRβ sequencing of pooled naive and memory T cells from a single healthy adult revealed >1 × 106 unique sequences.13 Statistical methods developed for estimating total species diversity in ecological and microbial communities14,15 have been used to estimate total CDR3 sequence diversity, including the sequences that are directly observed as well as those that remain “unseen” due to the fact that such diverse repertoires cannot be sampled completely. Such efforts have suggested that the naive and memory CD8**+** and CD4**+** TCRβ chain CDR3 repertoires in healthy adults collectively contain at least 3 × 106 to 4 × 106 unique sequences,9 with at least as many unique TCRα chain CDR3 sequences.11 The IGH CDR3 repertoire is estimated to contain at least 106 distinct sequences.5 The IGH and TCRβ repertoires are both characterized by an extremely large number of CDR3 sequences that are present at low frequencies.5,8-10 It has therefore proven difficult, despite the application of methods for estimating unseen species diversity, to determine the true diversity in individual BCR and TCR repertoires with great accuracy, and the published estimates are best viewed as lower bounds for repertoire diversity.
Although most of the deep-sequencing studies published to date have focused on the TRB and IGH loci, the loci that encode the α chains of αβ T cells and the γ chains of γδ T cells have also been explored. Analysis of TCRα CDR3 sequences in several subsets of αβ T cells from a single individual revealed 1.2- to 2.4-fold more unique TCRα than TCRβ CDR3 sequences in each of the subsets studied.11 Sequencing of the TRG locus has provided valuable insight into the structure of the γδ T-cell repertoire. Although the theoretical diversity of γδ TCRs rivals that of αβ TCRs, the antigen receptors expressed by peripheral blood γδ T cells demonstrate very limited TCR CDR3 sequence diversity. Indeed, deep sequencing of the rearranged TRG genes expressed in peripheral blood γδ T cells from 3 healthy adults revealed that 45% of the TCRγ CDR3 sequences from the 3 individuals were identical to a previously described sequence found in a “public” γδ TCR, found in most individuals, that is specifically reactive with nonpeptide prenyl pyrophosphate antigens.16,19
Deep sequencing of the TRG locus in αβ and γδ T cells from peripheral blood of healthy adults has also provided valuable insight into the process by which T cells are committed to the αβ or γδ lineages during thymic development. The vast majority of both αβ and γδ T cells in peripheral blood contain rearranged TRG genes, suggesting that rearrangement of the TRG locus occurs before αβ/γδ lineage commitment.17-19 In contrast, only a very small fraction (<4%) of γδ T cells appear to have rearranged TRB loci, suggesting that rearrangement of the TRB locus only occurs in T cells that have committed to the αβ lineage.19
Comparison of the TCR repertoires carried in specific T-cell subsets within single individuals is the focus of several current studies and has great potential for defining the relationships between subsets as well as illuminating the developmental pathways that lead to each subset. Analysis of TCRα and TCRβ diversity in the CD4**+** regulatory T-cell (Treg) compartment in a single individual, for example, demonstrated higher TCR diversity in this subset than in any other major subset.11 This observation is consistent with the results of previous studies of both murine20 and human21 Tregs, performed before the advent of high-throughput sequencing, which demonstrated that CD4**+** Treg TCR diversity is at least comparable to, if not in fact greater than, that observed in the conventional CD4**+** compartment.
Reconstitution of B- and T-cell repertoires after HCT
Recent TCRβ and IGH sequencing studies have characterized reconstitution of the αβ T-cell and B-cell compartments in recipients of allogeneic hematopoietic cell transplantation (HCT). One study of αβ T-cell reconstitution in 10 allogeneic HCT recipients, in which a uniform sequencing depth of >1 × 106 reads per sample was achieved, demonstrated reduced TCRβ complexity at 100 days posttransplant, with modest improvement noted by 1 year posttransplant.22 A different study of 28 allogeneic HCT recipients, in which the sequencing depth was nonuniform and 50- to 200-fold lower (<1.5 × 104 reads per sample), evaluated reconstitution of TCRβ diversity in the CD8**+** and CD4**+** compartments of cord blood and T cell–depleted (TCD) HCT recipients.23 At 6 months posttransplant, the cord blood recipients demonstrated CD8**+** and CD4**+** TCRβ diversity comparable to healthy adults, but the TCD recipients had >10-fold lower diversity; by 1 year posttransplant, the CD4**+** but not the CD8**+** TCRβ diversity in the TCD recipients had improved. Serial sequencing of the IGH locus with a uniform number of 15 000 reads per sample in 6 CLL patients after allogeneic HCT revealed that reconstitution of IGH diversity to levels observed in the HCT donor typically required more than 1 year.12 Although repertoire diversity was a primary end point in all 3 of these studies, they did not address the important issues of sampling depth, the existence of “unseen” CDR3 sequence diversity, or possible confounding by intersample variation in absolute T- or B-lymphocyte count. Nonetheless, these 3 pilot studies have provided valuable initial data on the reconstitution of lymphocyte repertoire diversity after allogeneic HCT and have laid the foundation for more comprehensive future studies.
Tracking malignant or therapeutically administered lymphocytes
High-throughput sequencing can be exploited to detect the rearranged CDR3 sequences carried in malignant B and T cells with unprecedented sensitivity and specificity. IGH4,5,12,24-27 and TCRβ/γ28 sequencing is already being used to identify the malignant clone or clones in patients with B- and T-lymphoid malignancies, respectively, at the time of diagnosis, and to track them during and after therapy. Deep sequencing of antigen-receptor loci has multiple advantages when compared with multiparameter flow cytometry or custom-designed patient- and clone-specific polymerase chain reaction (PCR) assays for the monitoring of lymphoid malignancies. Deep sequencing demands less time and labor, has superior sensitivity,25,26,28 and can simultaneously track all of the clones that comprise the malignant population. Its utility for monitoring disease burden in patients with chronic lymphocytic leukemia,4,5,12,24 pediatric B-lineage acute lymphoblastic leukemia (ALL),25,26 and T-lineage ALL28 has been compellingly demonstrated, and it will undoubtedly have utility for monitoring other lymphoid malignancies. Serial sequencing of the IGH locus in pediatric B-ALL has revealed surprisingly dynamic evolution of the locus in some patients,25 demonstrating that this technology may also generate valuable insights into the biology of B- and T-cell cancers.
Deep sequencing of antigen receptor genes has numerous other potential applications. It will likely become the technique of choice for tracking the fate of therapeutically administered lymphocytes, as its sensitivity, precision, and capacity for monitoring multiple clones simultaneously are far superior to that which can be achieved with clone-specific PCR or tetramer-based assays. Comprehensive profiling of specific lymphocyte populations, such as tumor-infiltrating lymphocytes or lymphocytic infiltrates associated with infections, graft rejection, or autoimmune disease, is also readily achieved with deep sequencing.
Limitations of antigen receptor sequencing
Current strategies for analysis of BCR and TCR repertoires using high-throughput sequencing have notable limitations. All protocols use multiplex PCR to amplify the rearranged variable regions of antigen receptor loci to provide sufficient template DNA for sequencing, which introduces bias due to more efficient amplification of some templates compared with others.9 Such bias can be addressed molecularly and computationally if its magnitude can be accurately determined. Analysis and interpretation of antigen receptor sequence data also poses a number of lingering computational challenges, one of the most difficult of which is inferring the most likely sequence of molecular events that produced each antigen receptor sequence. Addressing these challenges has provided the rationale for the nascent field of computational immunology.
Conclusions and future research directions
Application of high-throughput sequencing to the analysis of B- and T-cell antigen receptor repertoires is elucidating the fundamental structure of lymphocyte repertoires and seems poised to transform the diagnosis and monitoring of B- and T-lymphoid malignancies. The scientific and clinical utility of this innovative technology will strongly depend on the accuracy, precision, and reproducibility with which it can define both the number of unique antigen receptor sequences in a given clinical specimen and their relative frequency. Future studies that aim to compare the relative frequency of specific BCR or TCR sequences in 2 or more samples must take into account the many potentially confounding factors that could influence such comparisons, prominent among which are the proportion of lymphocyte DNA in the samples, the sequencing depth, and the underlying clonal structure of the samples. Studies that aim to determine the diversity of antigen receptors expressed in lymphocyte populations should explicitly take into account the existence of “unseen” diversity and the possibility that its magnitude may differ significantly from sample to sample.
The antigen receptors in B cells, αβ T cells, and γδ T cells comprise heterodimers whose 2 constituent chains are generated by independent rearrangement events at distinct chromosomal loci. The true diversity of BCRs or TCRs expressed in an individual is thus defined by the number of distinct pairs of chains that occur, which is likely to be higher than the simple number of distinct IGH or TCRβ monomers that are generated. The development of molecular and computational strategies for estimating the number and frequency of distinct heavy/light, αβ, and γδ pairs in the B- and T-cell repertoires is therefore an important research priority. Techniques for simultaneous sequencing of the variable regions of the heavy/light or αβ gene pairs expressed in single lymphocytes have been developed,29,30 and extension of these techniques to the analysis of millions of lymphocytes is likely achievable.
Future applications of deep sequencing to antigen-receptor repertoires will hopefully address a broad range of critical questions in hematology and immunology. Perhaps most importantly, are specific characteristics or dimensions of the B- or T-cell repertoires significantly correlated with clinical immunity and pathogen resistance? Can one or more of these characteristics serve as a reliable surrogate for immune reconstitution after HCT? Can serial analysis of the B- or T-cell repertoire in an individual reliably identify changes that, in turn, are associated with specific clinical events or interventions, such as infections or immunizations? The current pace of research in this field suggests that answers to these questions will soon be at hand.
Supplementary Material
Supplemental Videos
Acknowledgments
This work was supported by grants from the National Institutes of Health (CA015704-37, AI033484, and HL110907).
Footnotes
The online version of this article contains a data supplement.
Authorship
Contribution: E.H.W. wrote the manuscript; and E.H.W., F.A.M., and J.C. read and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Edus H. Warren, Program in Immunology, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, D3-100, P.O. Box 19024, Seattle, WA 98109-1024; e-mail: ehwarren@u.washington.edu.
References
- 1.Chen JL, Stewart-Jones G, Bossi G, et al. Structural and kinetic basis for heightened immunogenicity of T cell vaccines. J Exp Med. 2005;201(8):1243–1255. doi: 10.1084/jem.20042323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang J, Ofek G, Laub L, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491(7424):406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, version 1.5.0.5. Schrödinger.
- 4.Campbell PJ, Pleasance ED, Stephens PJ, et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci USA. 2008;105(35):13081–13086. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd SD, Marshall EL, Merker JD, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med. 2009;1(12):12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu YC, Kipling D, Leong HS, Martin V, Ademokun AA, Dunn-Walters DK. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood. 2010;116(7):1070–1078. doi: 10.1182/blood-2010-03-275859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larimore K, McCormick MW, Robins HS, Greenberg PD. Shaping of human germline IgH repertoires revealed by deep sequencing. J Immunol. 2012;189(6):3221–3230. doi: 10.4049/jimmunol.1201303. [DOI] [PubMed] [Google Scholar]
- 8.Freeman JD, Warren RL, Webb JR, Nelson BH, Holt RA. Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing. Genome Res. 2009;19(10):1817–1824. doi: 10.1101/gr.092924.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robins HS, Campregher PV, Srivastava SK, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114(19):4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robins HS, Srivastava SK, Campregher PV, et al. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med. 2010;2(47):47ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C, Sanders CM, Yang Q, et al. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci USA. 2010;107(4):1518–1523. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Logan AC, Gao H, Wang C, et al. High-throughput VDJ sequencing for quantification of minimal residual disease in chronic lymphocytic leukemia and immune reconstitution assessment. Proc Natl Acad Sci USA. 2011;108(52):21194–21199. doi: 10.1073/pnas.1118357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warren RL, Freeman JD, Zeng T, et al. Exhaustive T-cell repertoire sequencing of human peripheral blood samples reveals signatures of antigen selection and a directly measured repertoire size of at least 1 million clonotypes. Genome Res. 2011;21(5):790–797. doi: 10.1101/gr.115428.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher RA, Corbet AS, Williams CB. The relation between the number of species and the number of individuals in a random sample of an animal population. J Anim Ecol. 1943;12(1):42–58. [Google Scholar]
- 15.Efron B, Thisted R. Estimating the number of unseen species: how many words did Shakespeare know? Biometrika. 1976;63(3):435–447. [Google Scholar]
- 16.Wang H, Fang Z, Morita CT. Vgamma2Vdelta2 T cell receptor recognition of prenyl pyrophosphates is dependent on all CDRs. J Immunol. 2010;184(11):6209–6222. doi: 10.4049/jimmunol.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blom B, Verschuren MC, Heemskerk MH, et al. TCR gene rearrangements and expression of the pre-T cell receptor complex during human T-cell differentiation. Blood. 1999;93(9):3033–3043. [PubMed] [Google Scholar]
- 18.van Dongen JJ, Langerak AW, Brüggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17(12):2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 19.Sherwood AM, Desmarais C, Livingston RJ, et al. Deep sequencing of the human TCRγ and TCRβ repertoires suggests that TCRβ rearranges after αβ and γδ T cell commitment. Sci Transl Med. 2011;3(90):90ra61. doi: 10.1126/scitranslmed.3002536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pacholczyk R, Ignatowicz H, Kraj P, Ignatowicz L. Origin and T cell receptor diversity of Foxp3+CD4+CD25+ T cells. Immunity. 2006;25(2):249–259. doi: 10.1016/j.immuni.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 21.Fazilleau N, Bachelez H, Gougeon ML, Viguier M. Cutting edge: size and diversity of CD4+CD25high Foxp3+ regulatory T cell repertoire in humans: evidence for similarities and partial overlapping with CD4+CD25- T cells. J Immunol. 2007;179(6):3412–3416. doi: 10.4049/jimmunol.179.6.3412. [DOI] [PubMed] [Google Scholar]
- 22.Meier J, Roberts C, Avent K, et al. Fractal organization of the human T cell repertoire in health and after stem cell transplantation. Biol Blood Marrow Transplant. 2013;19(3):366–377. doi: 10.1016/j.bbmt.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 23.van Heijst JW, Ceberio I, Lipuma LB, et al. Quantitative assessment of T cell repertoire recovery after hematopoietic stem cell transplantation. Nat Med. 2013;19(3):372–377. doi: 10.1038/nm.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalos M, Levine BL, Porter DL, et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. 2011;3(95):95ra73. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gawad C, Pepin F, Carlton VE, et al. Massive evolution of the immunoglobulin heavy chain locus in children with B precursor acute lymphoblastic leukemia. Blood. 2012;120(22):4407–4417. doi: 10.1182/blood-2012-05-429811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faham M, Zheng J, Moorhead M, et al. Deep-sequencing approach for minimal residual disease detection in acute lymphoblastic leukemia. Blood. 2012;120(26):5173–5180. doi: 10.1182/blood-2012-07-444042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brentjens RJ, Davila ML, Riviere I, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med. 2013;5(177):177ra38. [DOI] [PMC free article] [PubMed]
- 28.Wu D, Sherwood A, Fromm JR, et al. High-throughput sequencing detects minimal residual disease in acute T lymphoblastic leukemia. Sci Transl Med. 2012;4(134):134ra63. [DOI] [PubMed]
- 29.Kim SM, Bhonsle L, Besgen P, et al. Analysis of the paired TCR α- and β-chains of single human T cells. PLoS ONE. 2012;7(5):e37338. doi: 10.1371/journal.pone.0037338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeKosky BJ, Ippolito GC, Deschner RP, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol. 2013;31(2):166–169. doi: 10.1038/nbt.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Videos