The Genetics of Mammalian Circadian Order and Disorder: Implications for Physiology and Disease (original) (raw)
. Author manuscript; available in PMC: 2013 Aug 30.
Published in final edited form as: Nat Rev Genet. 2008 Oct;9(10):764–775. doi: 10.1038/nrg2430
Abstract
Circadian cycles affect a variety of physiological processes, and disruptions of normal circadian biology therefore have the potential to influence a range of disease-related pathways. The genetic basis of circadian rhythms is well studied in model organisms and, more recently, studies of the genetic basis of circadian disorders has confirmed the conservation of key players in circadian biology from invertebrates to humans. In addition, important advances have been made in understanding how these molecules influence physiological functions in tissues throughout the body. Together, these studies set the scene for applying our knowledge of circadian biology to the understanding and treatment of a range of human diseases, including cancer, and metabolic and behavioural disorders.
Introduction
Circadian rhythms control a variety of biological processes in living systems, ranging from bacteria to humans 1, 2. Perhaps the most obvious function regulated by circadian rhythms is the daily sleep and wake cycle in animals; however, many other physiological processes are regulated by circadian rhythms, including body temperature, feeding behavior, hormone secretion, drug and xenobiotic metabolism, glucose homeostasis and cell cycle progression. When circadian cycles are disrupted, either by genetic or environmental insults, disorders of diverse physiological processes can occur 3. Links between circadian physiology and other physiological processes therefore have implications for human biology because it is likely that genetic variation in circadian clock genes can contribute significantly to physiological variation, and therefore potentially to variation in disease susceptibility. In addition, in modern societies, humans are increasingly ignoring natural circadian cues, so it is important to understand how the resulting perturbations of our circadian biology might affect our physiology and susceptibility to disease – both to understand the disease processes and to identify new targets for treatment.
In this Review, we bring together findings from recent studies that have begun to provide a molecular understanding of how circadian biology influences physiological processes that are relevant to human disease. We first briefly describe the generally accepted models of how our circadian rhythms are regulated at the neural, genetic and molecular levels, an understanding that has come from extensive work in model organisms. We then highlight recent findings from genetic studies of human circadian disorders, which illustrate the striking conservation of clock gene function in model organisms and humans and highlight implications of human clock gene variation for human circadian biology. We will also discuss recent studies that have revealed a number of surprisingly direct molecular links with the cell cycle, metabolism and behavioral disorders, providing a basis for understanding how the circadian system impinges on diverse aspects of mammalian physiology of relevance for human diseases and their treatment that include regenerative medicine and cancer, obesity and diabetes, and mental health.
Neural Control of Mammalian Circadian Rhythms
The ability of living systems to sustain ~24 hour rhythms in the absence of environmental cues shows that most daily oscillations are not responses to the diurnal cycle, but rather are generated by an internal clock (Text Box 1). The ‘master’ internal clock in mammals is located in the hypothalamic suprachiasmatic nuclei, or SCN — bilateral nuclei comprised of about 10,000 neurons each4, 5 (Figure 1). The primary environmental synchronizer of circadian rhythms in mammals is the daily light-dark cycle. A novel class of intrinsically photosensitive retinal ganglion cells that express the photopigment melanopsin integrates photic information for entrainment within the retina and projects to the core region of the SCN via the retinohypothalamic tract (RHT) 6, 7.
Text Box 1. Circadian Terminology and Activity Rhythms.
In the natural environment, an organism’s circadian system synchronizes a broad array of biological processes and behavior, such as sleep-wake cycle, periodic activity bouts, and blood hormone levels, to the prevailing light/dark cycle by phase advancing or phase delaying these daily rhythms. The time interval between phase reference points (e.g. two peaks) is called the period. Expression of endogenous circadian rhythm when subjects are isolated from the external cues, such as the light cycle, is described as free running. The timing of a reference point in the cycle (e.g. the peak) relative to a fixed event (e.g. beginning of the night phase) is known as the phase. The difference in the level between peak and trough values of the rhythm referred as amplitude (e.g. amount of activity or a circadian gene expression either in mRNA or protein levels).
To examine the effects of genetic disruption on behavioral circadian rhythms, long-term activity measurements are the most widely used method. When housed in a light:dark cycle, a normal mouse will exhibit nocturnal behavior by consolidating the majority of activity during the night, or dark, interval of the 24 hour day. When rodents are placed in constant darkness for extended periods of time, “free-running” circadian behavior persists and activity remains consolidated to the subjective night; however, the experimentally determined day length is usually different from 24 hours. For example, the internal clock in the inbred C57BL/6J strain of mice runs with an extremely precise free-running period of 23.6 hours. When individual genes involved in circadian rhythms are disrupted in mice, changes in free-running period, phase or amplitude of rhythms can be observed.
Figure 1. A schematic diagram of the SCN and its input and output pathways.
The mammalian circadian pacemaker in the hypothalamic suprachiasmatic nucleus (SCN) is organized into two major subdivisions, the core and the shell. The core region of the SCN receives information about the daily light cycle via the retinohypothalamic tract (RHT). Both neuronal and humoral signals act as output signals from the SCN to other regions of the brain and the periphery. The SCN output pathways are responsible for proper timing of diverse physiological functions, including hormone release, sleep-wake cycle, feeding behavior, and thermoregulation. The SCN output to the subparaventricular zone (sPVz) is relayed to the medial preoptic region (MPO) to control circadian rhythms of body temperature; and a separate projection through the dorsomedial nucleus of the hypothalamus (DMH) controls daily hormone secretion (via paraventricular nucleus, PVN) and sleep-wake cycles (via lateral hypothalamus, LH, and ventrolateral preoptic nucleus, VLPO). Figure modified from 8 [permission obtained].
Both neuronal and humoral signals originating from the SCN regulate output pathways, which control diverse physiological functions. SCN neurons project to many regions (Figure 1), and these output pathways are responsible for proper timing of hormone release, feeding behavior and body temperature fluctuations 8. In addition to these neuronal pathways, much evidence points to SCN-secreted circulating factors in mediating physiological rhythms in both the brain and the periphery 4. These factors are likely to include proteins such as TGFα 9, the cytokine CLC 10 and Prokineticin 2 11–13.
Molecular components of the mammalian circadian clock
The circadian clock mechanism involves a cell autonomous transcription-translation feedback loop comprised of a core set of genes that are highly conserved among animals 1, 2. In mammals, the circadian clock is composed of a primary negative feedback loop involving the genes, Clock and its paralog Npas2, Bmal1, Period1 (Per1), Per2, Cryptochrome1 (Cry1) and Cry2 (Figure 2). During the day, the basic helix-loop-helix (bHLH) PAS-domain containing transcription factors CLOCK/NPAS2 interact with BMAL1 to activate transcription of the Per and Cry genes, resulting in high levels of these transcripts. The resulting PER and CRY proteins heterodimerize, translocate to the nucleus and interact with the CLOCK:BMAL1 complex to inhibit their own transcription 14. During the night, the PER-CRY repressor complex is degraded, and CLOCK:BMAL1 can then activate a new cycle of transcription. The entire cycle takes approximately 24 hours to complete; however, little is known about the stoichiometry and kinetics of this feedback loop. In addition to the primary feedback loop, there is a second negative feedback loop involving the nuclear hormone receptor, Rev-erbα, which is a direct target of CLOCK:BMAL1 and which strongly represses Bmal1 transcription 15, 16. This secondary loop is not essential, but is thought to add robustness to the molecular clock. Finally, there are a number of other candidate clock components such as Timeless, Dec1, Dec2 and E4bp4 (reviewed in 2), whose roles remain to be more clearly defined.
Figure 2. The mammalian circadian clock is composed of a transcriptional–translational feedback network.
The circadian clock mechanism involves transcription-translation feedback loops comprised of a set of core clock genes. In mammals, the circadian clock is composed of a primary negative feedback loop involving the genes, Clock/Npas2, Bmal1, Period1 (Per1), Per2, Cryptochrome1 (Cry1) and Cry2. CLOCK/NPAS2 and BMAL1 are basic helix-loop-helix (bHLH) PAS-domain containing transcription factors that activate transcription of the Per and Cry genes. The resulting PER and CRY proteins heterodimerize, translocate to the nucleus and interact with the CLOCK:BMAL1 complex to inhibit their own transcription. With time, the PER-CRY repressor complex is degraded, and CLOCK:BMAL1 can then activate a new cycle of transcription. The secondary autoregulatory feedback loop is composed of REV-ERB_α_ that is a direct target of CLOCK:BMAL1 transcription activator complex. REV-ERB_α_ feeds back to repress Bmal1 transcription and competes with RORa to bind retinoic acid-related orphan receptor response elements (RREs) in the Bmal1 promoter. In addition to the transcriptional activators and repressors, post-translational modification and degradation of circadian clock proteins are critical steps for determining circadian periodicity. Key kinases for PER (and CRY) phosphorylation are Casein kinase 1δ (CK1δ) and CK1ε. One of the roles for phosphorylation of clock proteins is to target them for polyubiquitination and degradation by the 26S proteosomal pathway. Both β-TCRP1 and FBXL3 E3 ubiquitin ligase complexes have been implicated in targeting the PER and CRY proteins, respectively, for degradation. Figure modified from 140 [permission sought].
Recent work shows that post-translational modification and degradation of circadian clock proteins are critical steps for determining circadian periodicity of the clock 14, 17. As seen previously in Drosophila, mammalian PER1 and PER2 proteins are progressively phosphorylated as they accumulate during the late afternoon and night. Key kinases involved in PER (and CRY) phosphorylation are Casein kinase 1δ and 1ε. The hamster tau mutant has a short 20-hour circadian period, which is caused by an A178C missense mutation in CK1ε leading to a dominant negative mutant allele 18; this and further studies of this mutant have revealed a key role for CK1ε and protein phosphorylation in regulating circadian period in mammals 19. One of the roles for phosphorylation of clock proteins is to target them for polyubiquitination and degradation by the 26S proteosomal pathway. In vitro studies have implicated both β-TCRP1 and FBXL3 E3 ubiquitin ligase complexes in directly targeting the PER and CRY proteins, respectively, for degradation 20–24. Two recent studies have demonstrated that mutations in the Fbxl3 gene result in long period phenotypes in mice, due to a defect in targeting CRY1 and CRY2 for degradation 24, 25. Thus, the turnover of PER and CRY, respectively, are specifically regulated by two different SCF E3 ubiquitin ligase complexes.
Recent work has shown that circadian transcription of CLOCK-BMAL1 target genes is accompanied by circadian changes in histone H3 acetylation and chromatin remodeling 26. Circadian transcription of the mouse Dbp gene is accompanied by rhythmic binding of CLOCK:BMAL1 to E-box _cis_-regulatory elements, acetylation of Lys9 of histone H3, trimethylation of Lys4 of histone H3, and a reduction of histone density 27. Interestingly, the CLOCK protein itself has been shown to possess histone acetyltransferase (HAT) activity 28, and can acetylate its own partner, BMAL1, on Lys537 29. The histone deacetylase (HDAC), SIRT1, interacts with CLOCK and can deacetylate Lys537 of BMAL1 as well as Lys9/Lys14 of histone H3 30. SIRT1 is expressed in a circadian manner and regulates circadian gene expression of Bmal1, Rorγ, Per2 and Cry1 via interaction with CLOCK:BMAL1 and deacetylation and degradation of PER2 31. Thus, chromatin remodeling is intimately linked to CLOCK:BMAL1-driven transcription both within the core clock autoregulatory feedback loop as well as downstream target genes of the CLOCK:BMAL1 complex. Since SIRT1 is an NAD+-dependent HDAC, it will be interesting to see if this pathway provides a link between cellular metabolism and the circadian clock.
Genetics of human circadian disorders
Given the molecular insights of the circadian clock mechanism from model organisms, an obvious question concerns the conservation and possible role of clock genes in human biology. Perhaps the most obvious circadian rhythm in humans is the daily pattern of sleep and wakefulness 32, 33 (Text Box 2). According to the well-accepted model of sleep regulation, the timing of sleep and wakefulness is controlled by two processes: a homeostatic process that underlies the rise of sleep propensity during wakefulness and its dissipation during sleep, and a circadian process that determines the thresholds for switching between sleep and wake 34. Among the human sleep disorders, a subset of insomnias has been clearly linked to circadian alterations in the timing of sleep. These sleep disorders are known as: advanced sleep phase syndrome, delayed sleep phase syndrome, non-24-hour sleep-wake syndrome, and irregular sleep-wake pattern 35. In an effort to understand the genetic basis of human circadian sleep disorders, both genetic linkage and association studies have been undertaken.
Text Box 2. Phenotyping Methods to Ascertain Circadian Rhythm Defects in Humans.
Individual daily rest-activity rhythm parameters in humans can be measured with relative ease by a wrist actigraph, a small watch-like monitor that detects movements, used in conjunction with a sleep log/diary recorded by the subject 131. Other phenotypic measures have been valuable in identifying individuals with altered sleep-wake cycle. These include self-report questionnaires, such as the Horne-Östberg Morningness/Eveningness Questionnaire (MEQ) 132, designed to quantitatively associate individuals to morning type (M-type) or evening-type (E-type) tendency of preferred activity time. The Horne-Östberg MEQ score has been shown to correlate well with an individual’s intrinsic circadian period 133 and has been shown to be effective in identifying individuals/families with extreme diurnal preference 36, 37. Additional physiological measures such as core body temperature, plasma melatonin, and saliva dim-light melatonin-onset time measurements have proven useful in determining the phase of the circadian clock with respect to the rest-activity cycle. Another self-report questionnaire, the Munich ChronoType (MCTQ) has been recently developed to include additional parameters to refine the assessment of an individual’s diurnal preference and, thus, facilitate evaluation of individual’s genetic chronotype 32, 33.
Familial Advanced Sleep Phase Syndrome
To date, there is one clearly established Mendelian circadian rhythm disorder in humans, known as familial advanced sleep phase syndrome (FASPS) 36, 37. FASPS is inherited in an autosomal dominant fashion and characterized by persistent 3–4 hour advanced sleep onsets and awakening times relative to conventional and desired times 36, 37. When patients are allowed to choose their preferred sleep schedule, sleep quality and duration are normal for age and only under the constraints of a forced bedtime and wake-time schedule does one become aware of this disorder. Thus, FASPS is thought to result from an abnormal phase position of the circadian cycle relative to the desired sleep-wake schedule. FASPS was the first disorder to link known core clock genes directly with human circadian sleep disorders (Supplementary Table 1). Using linkage analysis, Toh and colleagues found that a missense mutation (S662G) in human PER2 (hPER2) cosegregated with ASPS 38. Interestingly, the S662G mutation occurs in a phosphorylation site within a casein kinase 1 (CK1)-binding domain of hPER2. Biochemical analysis revealed decreased phosphorylation by CK1ε of an hPER2 peptide containing this site. Based on these results from the S662G hPER2 mutation and from other results from the hamster tau mutation in CK1ε, it was thought that decreased phosphorylation of PER could stabilize it leading to a premature accumulation of PER and a shortening of period. However, a recent report by Xu et al. showed that transgenic mice expressing the S662G mutant hPER2 revealed an unexpected reduction of PER2 39. These authors propose that PER2 is normally a positive regulator of its own transcription and with the S662G mutation, the level of hPER2 transcription is decreased because the level of PER2 is decreased by increased turnover of PER2 39. However, independent work by Vanselow and colleagues suggests that the turnover of PER2 is not significantly affected by the S662G mutation and that nuclear retention of PER2 is reduced by the FASPS mutation 40. As alluded to earlier, the hamster tau mutation in CK1ε also does not lead to a stabilization of PER as initially thought, but rather leads to a destabilization of PER by enhancing the turnover and degradation of PER via the proteasome 41. Thus, the molecular basis of this form of FASPS appears to be caused by an increased turnover of nuclear PER2 either by enhanced degradation or by reduced nuclear retention of PER.
Given that expression of daily circadian rhythms involves a repertoire of circadian clock genes, it is not surprising to observe genetic heterogeneity in FASPS. Several independent studies of FASPS have failed to demonstrate mutations in PER2 37, 42. Consistent with these observations, Xu and colleagues have identified a mutation in casein kinase δ (CK1δ), a paralog of CK1ε, in FASPS using candidate gene approaches43. Interestingly, although a transgenic mouse model carrying the human mutation (T44A) transgene showed shortened circadian period as expected, a Drosophila model carrying the same mutant transgene showed lengthened circadian period, suggesting different regulatory mechanisms between these species. It will be of great interest to see whether additional mutations in core clock genes will be found to underlie the additional cases of FASPS. The fact that the first two cases of FASPS are caused by mutations in core circadian clock genes is remarkable and strongly suggests that the clock mechanism of humans is highly conserved with that seen in rodent animal models.
Delayed Sleep Phase Syndrome and circadian rhythm disorders with a complex genetic basis
In humans, delayed sleep phase syndrome (DSPS) represents the most common circadian rhythm sleep disorder in the general population 35. DSPS is characterized by a chronic inability to fall asleep or awaken at the desired “normal” times of day; the average onset of sleep in DSPS individuals occurs in the early morning (0300 to 0600 h) and their usual rise time occurs in the late morning to early afternoon (1100 to 1400h). Though rare, familial cases of DSPS have been described, suggesting that Mendelian inheritance of DSPS may exist 44, 45. Several association studies of DSPS with sequence variants of known circadian genes have been reported. Both significant association and lack of association with the T3111C polymorphism in the 3′-untranslated region of CLOCK have been reported for DSPS and for diurnal preference 45–48. One study reported that an amino acid substitution (S408N) in the CK1ε gene, which is a putative phosphoacceptor site, may play a protective role in the development of DSPS as well as Non-24-hour sleep-wake syndrome (in which loss of circadian regulation of sleep occurs) 49. A single nucleotide polymorphism in the 5′-untranslated region of PER2 has also been reported to be associated with morning preferences 50. In both these cases the functional consequences of these sequence variants have not been reported and thus we do not understand their mechanism.
One of the more interesting sequence variants is a variable-number tandem-repeat (VNTR) polymorphism in the PER3 gene, which encodes 18 amino acids repeating either four times (PER3–4) or five times (PER3–5); this coding region is predicted to harbor potential CK1 phosphorylation sites (which have not been analyzed) 51. The shorter VNTR allele has been shown to occur with higher frequency in a study of 48 Japanese DSPS patients 51. In a sample from an English population, one study showed that homozygosity for the shorter repeat allele (PER3–4/4) was strongly associated with DSPS 51. However, in contrast to this finding, a study in a Brazilian population found that the frequency of the longer allele was higher in DSPS patients 52. It has been proposed that the differences in the latitude of the two cities, which considerably influences temperature, the length of days throughout the year, and solar intensity, may have a role in the influence of PER3 polymorphisms in DSPS 52. Replication of this finding in equivalent latitudes is necessary to determine whether these associations are robust and if so whether there will be significant gene-environmental interactions for circadian traits in humans as seen in Drosophila 53.
The PER3 VNTR polymorphism may also contribute to sleep homeostasis; differences in sleep-wake structure, sleep propensity, and cognitive performance during sleep loss were noted between individuals who are homozygous for the shorter or longer allele in the general population 54. It is noteworthy that mouse models of other clock genes, such as Clock, Cry1, Cry2, Npas2, Dbp and Bmal1, also have alterations in sleep duration and/or homeostasis 55–59. Thus, in addition to the known phenotypic links between the circadian system and sleep, there are many examples in which circadian clock genes can clearly affect both the circadian control of sleep as well as the homeostatic sleep system. Given the interdependent relationships between the circadian and sleep systems, it will be important in future work to determine whether sleep and circadian phenotypes can be clearly distinguished at the genetic level. That is, can we find genes that selectively regulate sleep homeostasis independently of circadian rhythms and vice versa? Perhaps the only known genes definitively associated with sleep independent of the circadian system thus far are those genes causing narcolepsy, in which the loss of hypocretin/orexin ligand or hypocretin/orexin receptor 2, cause a defect in sleep stage entry into REM (rapid eye movement) sleep 60, 61. Inroads into the genetics of sleep are emerging in Drosophila 62, 63, but there is still a paucity of work on the genetics of sleep in mammalian animal models where a ripe opportunity for genetic analysis exists 64, 65.
In future work it will be important to integrate our knowledge of circadian mechanisms in humans so that the ‘chronotype’ of individuals can be more easily and reliably assessed 33 (Text Box 3). Because there is such wide diversity in diurnal preference among people and because our 24/7 society demands such temporal variability in schedules, it likely that the genetically programmed internal clock of humans will be in conflict (e.g., phase desynchrony) with modern society 33. Such desynchrony with the external world and the consequential internal desynchrony of the circadian system can contribute significantly to both the morbidity as well as the well-being of human health 3, 66. If the chronotype of an individual can be utilized to inform and optimize adaptation to the 24-hour society, then it is likely that an understanding of clock genetics in humans should have direct application: a clock gene allelic profile should be predictive of an individual’s genetically programmed clock, and this information could then be used to optimize an individual’s schedule and treatment regime for many disorders 66.
Text Box 3. In vitro methods to dissect human circadian rhythm disorders.
A major impediment in the study of human circadian rhythms is the difficulty and expense of assessing circadian rhythms in people. Because circadian clocks must be studied in temporal isolation, human circadian studies have demanded strict requirements of isolating subjects from environmental cues under constant conditions for prolonged period of time 134, 135. Unlike a mouse, it is difficult to convince a person to spend weeks living in temporal isolation. Thus, only a few investigators have had the resources and facilities necessary for such experiments. Therefore, surrogate measurements for assessing human circadian rhythms are essential. These have taken two forms: One has been to assess ‘diurnal preference’ or ‘chronotype’ of subjects using questionnaires, hormonal markers and actigraphy to measure phase of entrainment of circadian rhythms while living in the real world 32, 131, 132, 136. A second approach has been to develop cell-based and molecular approaches to assess circadian rhythms in humans by biopsy or blood sample 137. In animal models, precise period and phase estimates of circadian rhythms can be made by real-time measurements using circadian reporter genes in cultured cells 70, 71, 73. In humans, the challenge is to develop methods that are effective and robust in defining inter-individual phenotypic differences, but also that would be sufficiently cost-effective to screen large number of subjects.
Recently, Brown and colleagues 137 have developed a skin biopsy/culture method to measure inter-individual differences in circadian rhythms. By introducing a stable lentiviral-luciferase circadian reporter into cultured human fibroblasts, the authors showed that inter-individual differences of circadian period length were much more variable than different biopsies of the same individual. The same reporter system was implemented using known mouse circadian mutants and showed that circadian period measurements from fibroblast cultures were correlated with the behavioral rhythms of circadian clock mutants 137. More recently, Brown and colleagues have gone on to show that there are individual differences in circadian amplitude and phase-resetting responses in fibroblasts from human subjects that are correlated with their diurnal phase preferences 138. Thus cell-based assessments of circadian rhythms in fibroblast biopsies appear to be an excellent surrogate method to assess circadian periodicity in humans. It is important to note, however, that a recent study that examined different Per and Cry mutant mice has shown that SCN neuronal coupling can compensate for cell autonomous genetic defects 139. The authors show that while fibroblasts and isolated SCN neurons derived from Per1−/− and Cry1−/− mice exhibited defective Per2::luciferase expression, intact SCN explants from the same mice showed robust oscillation of Per2::luciferase expression. Therefore, in some cases, examination of primary fibroblasts from patients with suspected circadian disorders may overestimate the effect of genetic mutations on circadian rhythms at the behavioral level. Nonetheless, the technique should be valuable in identifying affected individuals and indeed may be a more sensitive indicator of genetic variants in human circadian disorders.
Peripheral clocks and tissue-specific control
With the initial discovery of circadian clock genes in mammals, it became clear that the expression of these genes was ubiquitous and that the majority of tissues throughout the body expressed circadian oscillations in the gene expression (Figure 3) 67–69. Indeed it is now well established that virtually every tissue in mammals has the capacity for generating circadian oscillations 70, 71. Using single-cell measurements of fibroblasts engineered with either circadian-driven bioluminescent or fluorescent reporter genes, cell-autonomous circadian rhythms can be observed that persist with undiminished amplitude for many days in vitro 72, 73. Interestingly, there is no evidence for coupling of these cell autonomous circadian oscillators in fibroblasts. The existence of self-sustained circadian oscillators in peripheral tissues and cells raises a number of organizational questions. What is the role of these peripheral oscillators and how are they coordinated in the intact organism? Evidence suggests that peripheral oscillators are normally entrained by the SCN pacemaker, but that they are only weakly coupled to the SCN. For example, experimentally inducing ‘jet lag’ in rodents by advancing or delaying the light cycle by 6 hours reveals that the SCN can reset rapidly to these shifts while at the same time resynchrony of peripheral oscillators can take over a week 70. Proximal cues such as feeding can override phase control of peripheral oscillators by the SCN under conditions of restricted feeding 74, 75. It is thought that circulating factors such as glucocorticoids and the circadian variation in body temperature may be acting as entraining signals for peripheral clocks 76, 77. Thus peripheral clocks are controlled at multiple levels. They receive entraining cues centrally from the SCN, and they have their own local cell autonomous oscillators that can operate independently from the SCN.
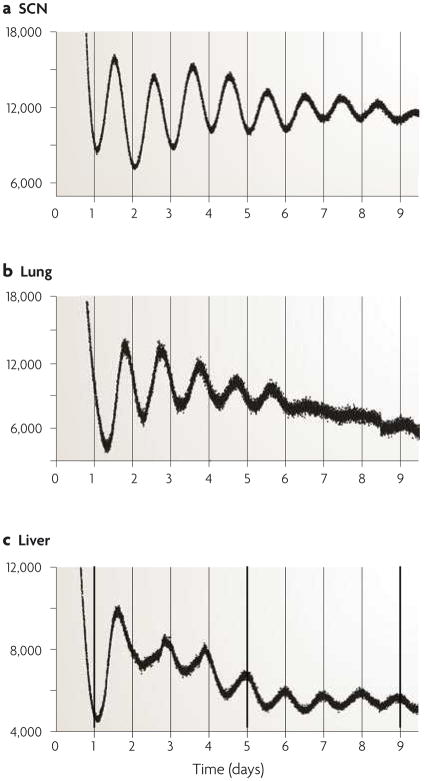
Figure 3. Central and peripheral oscillators.
The expression of the core circadian clock genes is ubiquitous and reflects the presence of circadian oscillators in virtually every tissue and cell in the body. The SCN expresses robust oscillations of PER2::Luciferase activity when isolated in explant culture ex vivo, as do virtually every tissue in the mouse, such as the lung and the liver. Data from 71.
The relative role of central vs. local control of circadian rhythms in peripheral tissues can be addressed by tissue-specific conditional regulation of circadian gene expression in transgenic mouse models. For the regulation of circadian behavioral rhythms, it is clear that central SCN control is sufficient to generate circadian rhythmicity. For example, in Bmal1 null mutant mice, circadian rhythms of behavior are abolished 78; however, transgenic expression of Bmal1 in the brain and SCN is sufficient to rescue circadian rhythms of behavior 79. Interestingly, other phenotypic effects of the Bmal1 null mutation (activity levels, morbidity, body weight loss) cannot be rescued by expression of Bmal1 in the brain, but can be rescued by transgenic expression in skeletal muscle 79. These experiments illustrate that the circadian behavioral effects of the Bmal1 mutation can be partitioned from other physiological effects by tissue-specific rescue and show that Bmal1 influences many other functions beyond circadian rhythmicity. Whether these pleiotropic effects of Bmal1 are a consequence of _Bmal1_’s peripheral circadian function or whether they reflect independent functions remains to be determined.
To address the role of local cell autonomous oscillators in the liver, Kornmann and colleagues 80 used conditional overexpression of Rev-erbα in the liver to knockdown Bmal1 in hepatocytes while maintaining normal circadian function in the rest of the animal. Disruption of cell autonomous clocks in the liver led to the abolition of cycling of the majority of genes in vivo; however, a subset of genes including Per2, Nocturnin and members of the heat shock protein family continue to oscillate suggesting that these genes may be responding to rhythmic systemic cues generated by SCN-driven rhythmic behavior of the animal 80. These experiments suggest that cell autonomous oscillators in the liver play a dominant role in the local control of circadian gene expression in these cells; however, systemic cues in vivo can still drive rhythms in some genes in hepatocytes presumably to entrain these peripheral oscillators.
Taken together, the discovery of cell autonomous circadian oscillators throughout the body of mammals has radically changed our view of the circadian system. Instead of a model in which a dominant pacemaker in the SCN alone generates and drives rhythms throughout the organism, we now appreciate that almost every cell and tissue in the body contains circadian oscillators and that there is a hierarchy of control in which the SCN acts as a coordinator or synchronizer by centrally regulating activity, feeding and body temperature rhythms, all of which can then influence the phase of cell autonomous oscillators in the periphery.
Interaction with physiological processes
In addition to their primary role in the generation of circadian rhythms, recent work has shown that circadian clock genes can affect a wide variety of other physiological processes (Supplementary Table 2) 81–83. The circadian system regulates these processes in two major ways; 1) outputs from the circadian clock originating either centrally from the SCN or locally from cell autonomous oscillators regulate key components in physiological pathways in a rhythmic fashion to drive circadian rhythms in these pathways; and 2) the core circadian clock components themselves appear to be elements within cellular pathways not traditionally thought to be under circadian regulation and may reflect additional roles for these genes. Emerging examples of circadian regulation of physiological pathways include diverse aspects of cellular metabolism, cell growth and DNA damage control, xenobiotic responses, as well as, the modulation of behavioral responses to drugs and alcohol.
Circadian Clocks and Metabolism
Perhaps the most striking discovery to be made from circadian gene profiling experiments was the observation that a significant fraction of the transcriptome (3–10%) is under circadian regulation and that the majority of the pathways regulated by the clock are imbedded in fundamental metabolic pathways 67. In the SCN, genes involved in protein biosynthesis and trafficking, including ribosomal synthesis, translation initiation, folding, targeting, post-translational modification, and transport, were under coordinated circadian control. In addition, genes involved in energy metabolism, the redox state of the cell, and cell signaling also showed circadian variation in their steady state message levels. In the liver, basic cellular pathways such as glycolysis, fatty acid metabolism, cholesterol biosynthesis, and xenobiotic and intermediate metabolism were under circadian regulation. Importantly, rate-limiting steps in these various pathways were key sites of circadian control, highlighting the fundamental role that circadian clocks play in cellular and organismal physiology. Recent gene expression analysis of the nuclear receptor family has shown that of the 49 nuclear receptors in mice, 28 of them display tissue-specific circadian rhythms in various tissues 84. Given the diverse roles nuclear receptors play in regulating metabolism, the circadian control of nuclear receptor expression provides a key nodal point for regulation of the overall physiology of the organism. Although there has been evidence for circadian modulation of metabolism previously, the pervasiveness of circadian control at a global level was not appreciated. These observations coupled with the cell autonomy of circadian oscillators in the majority of cells in the body suggest that circadian regulation may be a fundamental housekeeping function of the cell.
In addition to the circadian control of metabolism, circadian mutant animal models have been found to have metabolic disorders (Supplementary Table 2). For example, ClockΔ19 mutant mice are hyperphagic and obese, and not only lose rhythms of core clock genes, but also have lower levels of hypocretin/orexin and ghrelin neuropeptide gene expression in the arcuate nucleus 85. As a result, ClockΔ19 mice exhibit metabolic syndrome with hyperlipidemia, hepatic steatosis, hyperglycemia and hypoinsulinemia. A preliminary study in humans suggests that CLOCK polymorphisms may be associated with obesity and metabolic syndrome 86. In addition, both ClockΔ19 and _Bmal1_−/− mice are hypersensitive to insulin shock, suggesting a direct role for the molecular circadian clock in regulating glucose homeostasis 87. In humans, BMAL1 has been associated with susceptibility to hypertension and type 2 diabetes 88. In addition, BMAL1 appears to be involved in the control of adipogenesis and lipid metabolism in adipocytes 89. During adipose differentiation, Bmal1 is strongly induced in 3T3-L1 cells and both gain-of-function and loss-of-function experiments support a critical role for Bmal1 in this pathway. _Bmal1_−/− mice also exhibit numerous other phenotypes, including decreased adult body weight and activity levels, increased tendon calcification, increased sleep duration and fragmentation, and sarcopenia 58, 78, 90, 91. Some of these may reflect a general tendency to age quicker than wildtype mice 91, but also result from the loss of tissue-specific BMAL1 functions 79. Additional abnormalities have been characterized in other circadian mutants, for example the CK1ε (tau) mutant hamster exhibits reduced growth rate and elevated metabolic rate 92, 93. In addition, mice deficient in Nocturnin, a gene with robust circadian expression that encodes a polyA deadenylase, are resistant to diet-induced obesity 94. Thus, in these genetic models, we see a more direct link between circadian clock genes and metabolism. Mutations in a number of core clock genes are also associated with metabolic defects. Whether these effects are caused by alterations in circadian rhythmicity per se, or whether they are due to transcriptional targets of CLOCK and BMAL1 that are independent of the circadian clock system remains to be determined.
In addition to the circadian control of metabolism, it is also becoming clear that metabolism itself can have effects on circadian clocks 82. It has been known for some time that restricted feeding cycles can entrain circadian rhythms in rodents. Although the SCN is generally resistant to the entraining effects of restricted feeding, a food-entrainable oscillator (FEO) in the brain and circadian oscillators in the periphery are strongly reset by restricted feeding 74, 75. While it has not been established that restricted feeding entrains these oscillators by metabolic pathways, an hypothetical mechanism for how metabolism could have direct access to the clock mechanism has been proposed by McKnight and colleagues 82. For example, in biochemical experiments it can be shown that the reduced forms of the metabolic products NADH and NADPH modulate CLOCK:BMAL1 and NPAS2:BMAL1 heterodimers and enhance their DNA binding, while the oxidized forms NAD and NADP diminish DNA binding 95. In addition, the PAS domains of NPAS2 can bind heme, and carbon monoxide can regulate the DNA-binding of NPAS2:BMAL1 dimers 96. Recent work also shows that heme is the ligand for the circadian nuclear receptor, Rev-erbα 97, 98. To make things more complicated, heme biosynthesis is also under circadian regulation so these pathways appear to feedback on each other 99. It will be important to determine whether these redox state and heme interactions are physiological and operative in vivo. If so, then fluctuations in cellular metabolism could directly influence the transcriptional activity of circadian clock components, which in turn regulate the expression of many metabolic pathways.
Finally, additional evidence for effects of metabolism on circadian rhythms are experiments showing that alteration of metabolic state by genetic or environmental means can alter circadian rhythms in animals. For example, disruption of PGC1a, a transcriptional co-activator involved in energy metabolism, is also associated with abnormal circadian rhythms of activity, temperature and metabolic rate in mice 100. Interestingly, environmental modulators of metabolic state (high fat diet) can subtly influence circadian period and the expression of cycling genes in mice 101.
Taken together, it is clear that circadian clocks and metabolism are intimately linked and reflect adaptive responses of living systems to the cyclic environment (Figure 4). Because metabolic processes vary with the physiological demands of the organism and many of these physiological demands occur on a daily basis, it is not surprising that direct circadian coordination of metabolism might have adaptive significance. On the other hand, disruption of the optimal tuning of rhythmic metabolic pathways could then have suboptimal consequences and important implications for metabolic disorders.
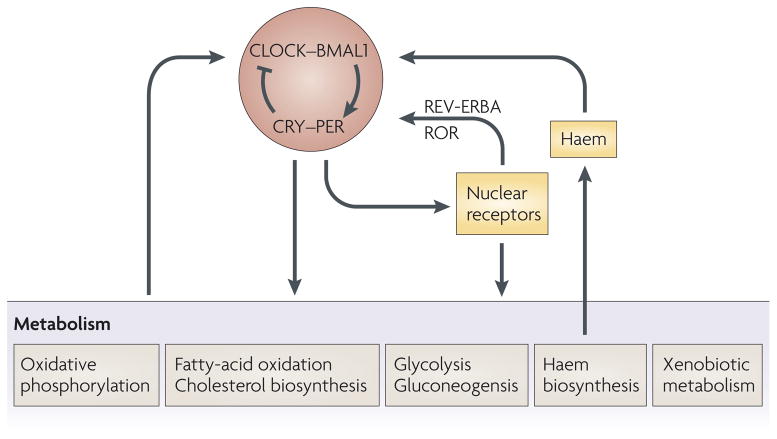
Figure 4. Interactions between circadian and metabolic systems.
Fundamental metabolic pathways such as glycolysis, fatty acid metabolism, cholesterol biosynthesis, and xenobiotic and intermediate metabolism are all under circadian regulation. Metabolism itself can also have effects on circadian clocks: these include NADH and NADPH which can modulate CLOCK:BMAL1 and NPAS2:BMAL1 DNA binding; the PAS domains of NPAS2 which can bind heme and can be modulated by carbon monoxide; and, heme can also be a ligand for the circadian nuclear receptor, Rev-erbα.
Circadian Clocks and the Cell Cycle
As alluded to above, the cell cycle has been observed to be under circadian modulation in many organisms and the selective factor for such modulation may have been to ‘escape from light’ and restrict DNA replication to the night 102 (Figure 5). Recently, it has been observed that some circadian clock mutants in mice can affect cell growth and proliferation, and in some cases can predispose mice to cancer. For example, in regenerating liver following partial hepatectomy in mice, Okamura and colleagues have shown that the circadian clock system controls cell-cycle related genes that modulate the activity of Cyclin B1-Cdc2 kinase, a key mitotic regulator 103. _Cry_-deficient mice exhibit delayed rates of liver regeneration after partial hepatectomy and the expression profiles of Cyclin D1 and Wee1 are deregulated. Importantly, the transcription of Wee1 is circadian and appears to be a direct target of the CLOCK:BMAL1 transcriptional complex. Thus, in regenerating liver the expression of Wee1 is co-regulated with Per1 and the entry of the cell cycle into M phase is suppressed during this time of day. This may provide a molecular mechanism coupling the circadian clock to the cell cycle. Similarly, the Clock mutant strongly affects the expression patterns of many cell-cycle related genes and leads to reduction in cell proliferation in Clock mutant fibroblasts 104.
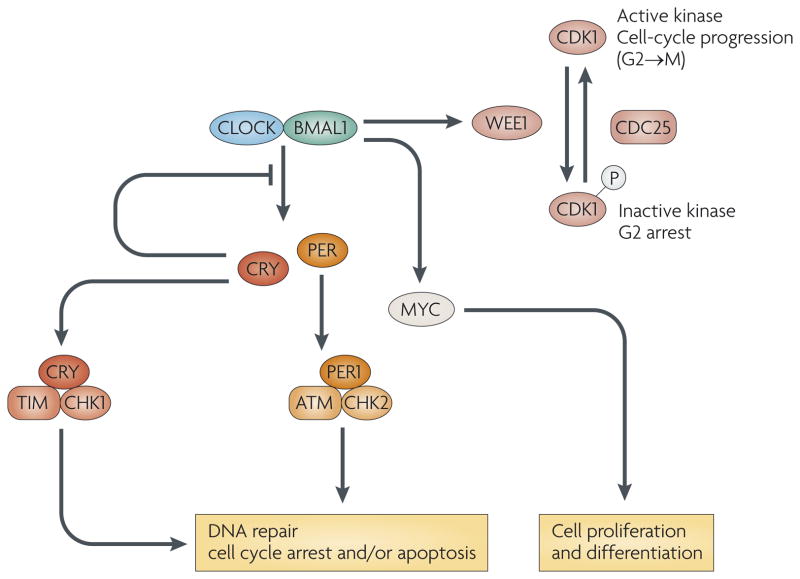
Figure 5. Circadian control of cell division cycles.
Circadian system is linked to the cell division cycle via circadian control of gene expression and posttranslational mechanisms. The transcription of c-Myc and Wee1 is circadian and appears to be a direct target of the CLOCK:BMAL1. The expression of Wee1 is co-regulated with Per and the entry of the cell cycle into M phase is suppressed during the day time when the transcription of Per (and Wee1) is high. In addition, the PER1 protein interacts with the check-point proteins, ATM and Chk2; while related work has linked the TIMELESS (TIM) and CRY proteins with Chk1. Activation of the DNA damage pathway can also reset the phase of the circadian clock.
In other work, Fu and Lee 105 have shown that Per2 mutant mice are predisposed to cancer following γ radiation treatment and that the expression of c-myc and Cyclin D1 are regulated by the CLOCK/NPAS2:BMAL1 pathway. The Per2 pathway involving c-myc and Cyclin D1 also appears to be operative in leptin-dependent osteoblast proliferation in bone 106. The Per1 gene also has been linked to cell growth and DNA damage control 107, and overexpression of Per1 sensitized cancer cells to DNA damage-induced apoptosis and caused significant growth reduction. The PER1 protein interacted with the check-point proteins, ATM and Chk2 107, while related work has linked the TIMELESS protein with Chk1 and ATR-ATRIP 108. Unexpectedly, activation of the DNA damage pathway itself has been shown to be able to reset the phase of the circadian clock in cells and in vivo 109, 110. Thus, the circadian clock not only regulates key cell cycle regulatory proteins, but can also be reset by DNA damaging agents. While these links between the circadian clock and the cell cycle occur at the molecular level, there is also evidence that abolition of circadian rhythms in the host by SCN lesions can accelerate the growth rates of implanted tumors 111. Thus, the circadian gene pathway can both interact directly with cell cycle pathways and influence cell growth through physiological system level interactions (Figure 5).
Circadian Sensitivity to Chemotherapeutic Agents
It has been know for many decades that the morbidity and mortality (drug-induced toxicity) of anticancer agents vary strikingly with time of day 66. Using the chemotherapeutic agent, cyclophosphamide, we have recently reinvestigated this phenomenon and found that a treatment that causes 20% mortality when administered at dusk (ZT10-ZT14) results in 100% mortality when administered at dawn (ZT22-ZT02) 112. To test the role of clock genes in this response, we used mouse mutants that have disrupted circadian rhythms. Both the Clock and Bmal1 mouse mutants were hypersensitive to cyclophosphamide treatment, while _Cry_-deficient mice were resistant to the drug’s toxicity 112. These unexpected results point to a CLOCK:BMAL1 target underlying the sensitivity to cyclophosphamide. Contrary to conventional wisdom (changes in pharmacokinetics with time of day), there were no significant differences in the metabolism and turnover of cyclophosphamide that could explain the dramatic differences in circadian sensitivity. Instead, evidence suggests that the pharmacodynamic effects of cyclophosphamide-induced toxicity are best correlated with CLOCK:BMAL1-dependent modulation of B cell survival. The elucidation of the molecular mechanisms underlying the circadian control of B cell survival will provide a rationale not only for adjusting the timing of chemotherapeutic treatment to be less toxic but also for providing a basis for a search for pharmacological modulators of drug toxicity acting through circadian system regulators. This result may significantly increase the therapeutic index and reduce morbidity associated with anticancer treatment.
Thus, circadian rhythms and core clock components play crucial roles in many aspects of cell cycle regulation and morbidity of chemotherapeutic agents. These results, combined with epidemiological studies examining the link between rotating shift work and cancer risk, suggest a significant association between circadian rhythm disruptions and abnormal cell growth 113–115. While some human studies also have shown the beneficial effects of timed chemotherapy delivery on both drug effectiveness and side effects, translation to recommended clinical practices has not been generally adopted 116, 117. Further evidence that timed drug administration is beneficial will be needed to initiate this course of therapy for cancer treatment as well as therapies for a variety of other disorders. Indeed recent work demonstrates that even hematopoietic stem cell release is regulated by the circadian clock and that the efficacy of stem cell transplantation can vary by as much as two- to three-fold depending on the time of day 118. Thus, chemotherapy and stem cell transplantation are two important targets for circadian optimization in clinical treatments in the future.
Circadian rhythms and affective disorders
In addition to physiological effects, circadian mutants can also exhibit behavioral dysfunctions associated with alcohol consumption, addiction and human mood disorders. The Per2 gene affects the glutamatergic system in mice and Per2 mutations enhance alcohol consumption 119. Interestingly, Clock Δ19 mutant mice display an overall behavioral profile that is strikingly similar to the manic state in human bipolar patients, including hyperactivity in a novel environment, decreased sleep, lower risk aversion, lower anxiety and an increase in susceptibility to reward by cocaine and alcohol 120, 121. In humans, mood disorders are often associated with sleep disturbances and it has been suggested that circadian rhythms may influence susceptibility to depression (reviewed in 122, 123). In support of this, NPAS2, a CLOCK paralog, has been implicated in seasonal affective disorder 124. One study showed suggestive evidence for association of PER3 and ARNTL (human ortholog of mouse Bmal1) with bipolar disorder 125; however, association studies with CLOCK, CRY1 and PER2 genes in affective or bipolar disorders have so far been negative 126–128. Whether circadian clock genes contribute to pathogenesis in non-seasonal affective/mood disorders in humans remains to be further investigated in additional patient populations. Thus far all of the association studies performed on circadian genes have been performed on small samples and few have been replicated. Clearly these small-scale, underpowered, candidate-gene studies need to be replaced by unbiased genome-wide association studies that are sufficiently powered and replicated. Thus, much caution must be taken in interpreting the existing reports on association of circadian genes with phenotypes in humans.
Perspectives
The elucidation of the molecular mechanism of circadian clocks in model organisms has revealed striking conservation of genes regulating human circadian rhythms. Emerging genetic studies on human clock genes suggest that genetic variation in these genes may be associated with metabolic, sleep and mood disorders. Given the strong links between circadian components and diverse physiological processes in animal models, genetic variation in human clock genes could also contribute to phenotypic variation relevant to disease pathways such as metabolic and mood disorders. As such, it will be important in future studies to consider the impact of circadian variation on the design and assessment of human phenotypes whether normal or disease-related. In addition, the clear role for circadian genes in sleep disorders such as FASPS provides new opportunities for addressing whether human circadian and sleep disorders, as seen in model organisms, are also associated with metabolic, cell cycle, or mood disorders.
An important issue to resolve concerning the mechanism of action of circadian alterations on physiological and disease processes is whether they reflect a primary effect on circadian (i.e., cyclic) function or modulation in pathways, or whether there may be functions of circadian clock components that are independent of the circadian system. These two scenarios could have important implications for therapeutic strategies because on the one hand treatments or lifestyles that perturb the normal patterns of circadian behavior could then have detrimental effects on metabolic and physiological pathways that are coordinated by the circadian system. On the other hand, if circadian clock proteins have other non-circadian functions, then therapeutic strategies that target them would need to consider whether the non-circadian functions are involved in the physiological processes linked to circadian genes, or whether they are off-target sites for side effects. In either case, delineating the mechanisms in which circadian clock gene products are linked to disease processes will be critical for therapeutic interventions targeting the circadian system.
Finally, with the knowledge that circadian clocks are cell autonomous and distributed throughout the body, it is important to target both central and peripheral circadian oscillators for therapeutic intervention. Because the circadian clock is so intimately linked to cellular metabolism and cell proliferation, new therapeutic indications such as the predisposition to type 2 diabetes and obesity may become amenable to treatments that modulate the circadian and sleep systems 129, 130. Future exploration in these research areas, along with increased public awareness, may allow us to appropriately diagnose and treat human circadian disorders as well as improve lifestyle choices to best align our physiological systems with the daily clock.
Supplementary Material
S. Table 1
S. Table 2
References
- 1.Bell-Pedersen D, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–56. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–41. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–61. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 4.Antle MC, Silver R. Orchestrating time: arrangements of the brain circadian clock. Trends Neurosci. 2005;28:145–51. doi: 10.1016/j.tins.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Herzog ED. Neurons and networks in daily rhythms. Nat Rev Neurosci. 2007;8:790–802. doi: 10.1038/nrn2215. [DOI] [PubMed] [Google Scholar]
- 6.Berson DM. Phototransduction in ganglion-cell photoreceptors. Pflugers Arch. 2007;454:849–55. doi: 10.1007/s00424-007-0242-2. [DOI] [PubMed] [Google Scholar]
- 7.Hankins MW, Peirson SN, Foster RG. Melanopsin: an exciting photopigment. Trends in Neurosciences. 2008 doi: 10.1016/j.tins.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005;437:1257–63. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- 9.Kramer A, et al. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science. 2001;294:2511–5. doi: 10.1126/science.1067716. [DOI] [PubMed] [Google Scholar]
- 10.Kraves S, Weitz CJ. A role for cardiotrophin-like cytokine in the circadian control of mammalian locomotor activity. Nat Neurosci. 2006;9:212–9. doi: 10.1038/nn1633. [DOI] [PubMed] [Google Scholar]
- 11.Cheng MY, et al. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature. 2002;417:405–10. doi: 10.1038/417405a. [DOI] [PubMed] [Google Scholar]
- 12.Li JD, et al. Attenuated circadian rhythms in mice lacking the prokineticin 2 gene. J Neurosci. 2006;26:11615–23. doi: 10.1523/JNEUROSCI.3679-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prosser HM, et al. Prokineticin receptor 2 (Prokr2) is essential for the regulation of circadian behavior by the suprachiasmatic nuclei. Proc Natl Acad Sci U S A. 2007;104:648–53. doi: 10.1073/pnas.0606884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–67. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 15.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–60. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 16.Sato TK, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–37. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 17.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–48. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 18.Lowrey PL, et al. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–92. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng QJ, et al. Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58:78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eide EJ, et al. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirogane T, Jin J, Ang XL, Harper JW. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem. 2005;280:26863–72. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- 22.Reischl S, et al. Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms. 2007;22:375–86. doi: 10.1177/0748730407303926. [DOI] [PubMed] [Google Scholar]
- 23.Busino L, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–4. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 24.Siepka SM, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–23. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Godinho SI, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. References 23–25 identify a novel SCF E3 ubiquitin ligase complex in which the F-box protein, FBXL3, interacts specifically with the Cryptochrome proteins to target them for degradation by the protesome. [DOI] [PubMed] [Google Scholar]
- 26.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–82. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 27.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–74. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 28.Doi M, Hirayama J, Sassone-Corsi P. Circadian Regulator CLOCK Is a Histone Acetyltransferase. Cell. 2006 doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 29.Hirayama J, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 30.Nakahata Y, et al. The NAD+-Dependent Deacetylase SIRT1 Modulates CLOCK-Mediated Chromatin Remodeling and Circadian Control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asher G, et al. SIRT1 Regulates Circadian Clock Gene Expression through PER2 Deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. References 30–31 show that the NAD+ dependent deacetylase, SIRT1, interacts with the CLOCK protein and plays a role in the regulation of BMAL1 and PER2 deacetylation. [DOI] [PubMed] [Google Scholar]
- 32.Roenneberg T, Wirz-Justice A, Merrow M. Life between clocks: daily temporal patterns of human chronotypes. J Biol Rhythms. 2003;18:80–90. doi: 10.1177/0748730402239679. [DOI] [PubMed] [Google Scholar]
- 33.Roenneberg T, et al. Epidemiology of the human circadian clock. Sleep Med Rev. 2007;11:429–38. doi: 10.1016/j.smrv.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 34.Borbely AA, Achermann P. Sleep homeostasis and models of sleep regulation. J Biol Rhythms. 1999;14:557–68. doi: 10.1177/074873099129000894. [DOI] [PubMed] [Google Scholar]
- 35.Sack RL, et al. Circadian rhythm sleep disorders: part II, advanced sleep phase disorder, delayed sleep phase disorder, free-running disorder, and irregular sleep-wake rhythm. An American Academy of Sleep Medicine review. Sleep. 2007;30:1484–501. doi: 10.1093/sleep/30.11.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones CR, et al. Familial advanced sleep-phase syndrome: A short-period circadian rhythm variant in humans. Nat Med. 1999;5:1062–5. doi: 10.1038/12502. [DOI] [PubMed] [Google Scholar]
- 37.Reid KJ, et al. Familial advanced sleep phase syndrome. Arch Neurol. 2001;58:1089–94. doi: 10.1001/archneur.58.7.1089. [DOI] [PubMed] [Google Scholar]
- 38.Toh KL, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–3. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 39.Xu Y, et al. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanselow K, et al. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–72. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallego M, Eide EJ, Woolf MF, Virshup DM, Forger DB. An opposite role for tau in circadian rhythms revealed by mathematical modeling. Proc Natl Acad Sci U S A. 2006;103:10618–23. doi: 10.1073/pnas.0604511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satoh K, Mishima K, Inoue Y, Ebisawa T, Shimizu T. Two pedigrees of familial advanced sleep phase syndrome in Japan. Sleep. 2003;26:416–7. doi: 10.1093/sleep/26.4.416. [DOI] [PubMed] [Google Scholar]
- 43.Xu Y, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–4. doi: 10.1038/nature03453. References 38–40 and 43 provide the first evidence that a human disorder, FASPS, involves mutations in the core circadian genes, PER2 and CKIdelta. [DOI] [PubMed] [Google Scholar]
- 44.Ancoli-Israel S, Schnierow B, Kelsoe J, Fink R. A pedigree of one family with delayed sleep phase syndrome. Chronobiol Int. 2001;18:831–40. doi: 10.1081/cbi-100107518. [DOI] [PubMed] [Google Scholar]
- 45.Iwase T, et al. Mutation screening of the human Clock gene in circadian rhythm sleep disorders. Psychiatry Res. 2002;109:121–8. doi: 10.1016/s0165-1781(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 46.Katzenberg D, et al. A CLOCK polymorphism associated with human diurnal preference. Sleep. 1998;21:569–76. doi: 10.1093/sleep/21.6.569. [DOI] [PubMed] [Google Scholar]
- 47.Robilliard DL, et al. The 3111 Clock gene polymorphism is not associated with sleep and circadian rhythmicity in phenotypically characterized human subjects. J Sleep Res. 2002;11:305–12. doi: 10.1046/j.1365-2869.2002.00320.x. [DOI] [PubMed] [Google Scholar]
- 48.Mishima K, Tozawa T, Satoh K, Saitoh H, Mishima Y. The 3111T/C polymorphism of hClock is associated with evening preference and delayed sleep timing in a Japanese population sample. Am J Med Genet B Neuropsychiatr Genet. 2005;133:101–4. doi: 10.1002/ajmg.b.30110. [DOI] [PubMed] [Google Scholar]
- 49.Takano A, et al. A missense variation in human casein kinase I epsilon gene that induces functional alteration and shows an inverse association with circadian rhythm sleep disorders. Neuropsychopharmacology. 2004;29:1901–9. doi: 10.1038/sj.npp.1300503. [DOI] [PubMed] [Google Scholar]
- 50.Carpen JD, Archer SN, Skene DJ, Smits M, von Schantz M. A single-nucleotide polymorphism in the 5′-untranslated region of the hPER2 gene is associated with diurnal preference. J Sleep Res. 2005;14:293–7. doi: 10.1111/j.1365-2869.2005.00471.x. [DOI] [PubMed] [Google Scholar]
- 51.Archer SN, et al. A length polymorphism in the circadian clock gene Per3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep. 2003;26:413–5. doi: 10.1093/sleep/26.4.413. [DOI] [PubMed] [Google Scholar]
- 52.Pereira DS, et al. Association of the length polymorphism in the human Per3 gene with the delayed sleep-phase syndrome: does latitude have an influence upon it? Sleep. 2005;28:29–32. [PubMed] [Google Scholar]
- 53.Sandrelli F, et al. A molecular basis for natural selection at the timeless locus in Drosophila melanogaster. Science. 2007;316:1898–900. doi: 10.1126/science.1138426. [DOI] [PubMed] [Google Scholar]
- 54.Viola AU, et al. PER3 polymorphism predicts sleep structure and waking performance. Curr Biol. 2007;17:613–8. doi: 10.1016/j.cub.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 55.Naylor E, et al. The circadian clock mutation alters sleep homeostasis in the mouse. J Neurosci. 2000;20:8138–43. doi: 10.1523/JNEUROSCI.20-21-08138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franken P, Lopez-Molina L, Marcacci L, Schibler U, Tafti M. The transcription factor DBP affects circadian sleep consolidation and rhythmic EEG activity. J Neurosci. 2000;20:617–25. doi: 10.1523/JNEUROSCI.20-02-00617.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wisor JP, et al. A role for cryptochromes in sleep regulation. BMC Neurosci. 2002;3:20. doi: 10.1186/1471-2202-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laposky A, et al. Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep. 2005;28:395–409. doi: 10.1093/sleep/28.4.395. [DOI] [PubMed] [Google Scholar]
- 59.Franken P, et al. NPAS2 as a transcriptional regulator of non-rapid eye movement sleep: genotype and sex interactions. Proc Natl Acad Sci U S A. 2006;103:7118–23. doi: 10.1073/pnas.0602006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taheri S, Zeitzer JM, Mignot E. The role of hypocretins (orexins) in sleep regulation and narcolepsy. Annu Rev Neurosci. 2002;25:283–313. doi: 10.1146/annurev.neuro.25.112701.142826. [DOI] [PubMed] [Google Scholar]
- 61.Sakurai T. The neural circuit of orexin (hypocretin): maintaining sleep and wakefulness. Nat Rev Neurosci. 2007;8:171–81. doi: 10.1038/nrn2092. [DOI] [PubMed] [Google Scholar]
- 62.Hendricks JC, Sehgal A. Why a fly? Using Drosophila to understand the genetics of circadian rhythms and sleep. Sleep. 2004;27:334–42. doi: 10.1093/sleep/27.2.334. [DOI] [PubMed] [Google Scholar]
- 63.Cirelli C, et al. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–92. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 64.Dauvilliers Y, Maret S, Tafti M. Genetics of normal and pathological sleep in humans. Sleep medicine reviews. 2005;9:91–100. doi: 10.1016/j.smrv.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 65.Mignot E. Why We Sleep: The Temporal Organization of Recovery. PLoS Biol. 2008;6:e106. doi: 10.1371/journal.pbio.0060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Levi F, Schibler U. Circadian rhythms: mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007 doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 67.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 68.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 69.Ueda HR, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–9. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 70.Yamazaki S, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–5. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 71.Yoo SH, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–46. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nagoshi E, et al. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 73.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14:2289–95. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Damiola F, Le Minh N, Preitner N, Kornmann B. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker …. Genes & Development. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–3. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 76.Balsalobre A, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–7. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 77.Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N. Rhythms of Mammalian Body Temperature Can Sustain Peripheral Circadian Clocks. Current Biology. 2002 doi: 10.1016/s0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- 78.Bunger MK, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–17. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McDearmon EL, et al. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314:1304–8. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. Conditional inactivation of the cell autonomous clock in the liver reveals the role of local control vs. systemic cues for regulating cycling liver gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramsey KM, Marcheva B, Kohsaka A, Bass J. The clockwork of metabolism. Annu Rev Nutr. 2007;27:219–40. doi: 10.1146/annurev.nutr.27.061406.093546. [DOI] [PubMed] [Google Scholar]
- 82.Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Annu Rev Biochem. 2002;71:307–31. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- 83.Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–48. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- 84.Yang X, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–10. doi: 10.1016/j.cell.2006.06.050. A comprehensive analysis of nuclear receptor gene expression reveals extensive circadian regulation of this superfamily of proteins. [DOI] [PubMed] [Google Scholar]
- 85.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–5. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Scott EM, Carter AM, Grant PJ. Association between polymorphisms in the Clock gene, obesity and the metabolic syndrome in man. Int J Obes (Lond) 2007 doi: 10.1038/sj.ijo.0803778. [DOI] [PubMed] [Google Scholar]
- 87.Rudic RD, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Woon PY, et al. Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc Natl Acad Sci USA. 2007;104:14412–7. doi: 10.1073/pnas.0703247104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shimba S, et al. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci USA. 2005;102:12071–6. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bunger MK, et al. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis. 2005;41:122–32. doi: 10.1002/gene.20102. [DOI] [PubMed] [Google Scholar]
- 91.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–73. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lucas RJ, Stirland JA, Mohammad YN, Loudon AS. Postnatal growth rate and gonadal development in circadian tau mutant hamsters reared in constant dim red light. J Reprod Fertil. 2000;118:327–30. doi: 10.1530/jrf.0.1180327. [DOI] [PubMed] [Google Scholar]
- 93.Oklejewicz M, Hut RA, Daan S, Loudon AS, Stirland AJ. Metabolic rate changes proportionally to circadian frequency in tau mutant Syrian hamsters. J Biol Rhythms. 1997;12:413–22. doi: 10.1177/074873049701200503. [DOI] [PubMed] [Google Scholar]
- 94.Green CB, et al. From the Cover: Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci U S A. 2007;104:9888–93. doi: 10.1073/pnas.0702448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–4. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 96.Dioum EM, et al. NPAS2: a gas-responsive transcription factor. Science. 2002;298:2385–7. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- 97.Raghuram S, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007 doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yin L, et al. Rev-erb{alpha}, a Heme Sensor That Coordinates Metabolic and Circadian Pathways. Science. 2007 doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 99.Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–71. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- 100.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–81. doi: 10.1038/nature05767. A key link between the circadian and metabolic systems. [DOI] [PubMed] [Google Scholar]
- 101.Kohsaka A, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–21. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 102.Pittendrigh CS. Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol. 1993;55:16–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 103.Matsuo T, et al. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–9. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 104.Miller BH, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A. 2007;104:3342–7. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 106.Fu L, Patel MS, Bradley A, Wagner EF, Karsenty G. The Molecular Clock Mediates Leptin-Regulated Bone Formation. Cell. 2005 doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 107.Gery S, et al. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–82. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 108.Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005;25:3109–16. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oklejewicz M, et al. Phase resetting of the Mammalian circadian clock by DNA damage. Curr Biol. 2008;18:286–91. doi: 10.1016/j.cub.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 110.Pregueiro AM, Liu Q, Baker CL, Dunlap JC, Loros JJ. The Neurospora checkpoint kinase 2: a regulatory link between the circadian and cell cycles. Science. 2006;313:644–9. doi: 10.1126/science.1121716. [DOI] [PubMed] [Google Scholar]
- 111.Filipski E, et al. Host circadian clock as a control point in tumor progression. J Natl Cancer Inst. 2002;94:690–7. doi: 10.1093/jnci/94.9.690. [DOI] [PubMed] [Google Scholar]
- 112.Gorbacheva VY, et al. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proc Natl Acad Sci U S A. 2005;102:3407–12. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schernhammer ES, et al. Rotating night shifts and risk of breast cancer in women participating in the nurses’ health study. J Natl Cancer Inst. 2001;93:1563–8. doi: 10.1093/jnci/93.20.1563. [DOI] [PubMed] [Google Scholar]
- 114.Megdal SP, Kroenke CH, Laden F, Pukkala E, Schernhammer ES. Night work and breast cancer risk: a systematic review and meta-analysis. Eur J Cancer. 2005;41:2023–32. doi: 10.1016/j.ejca.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 115.Kubo T, et al. Prospective cohort study of the risk of prostate cancer among rotating-shift workers: findings from the Japan collaborative cohort study. Am J Epidemiol. 2006;164:549–55. doi: 10.1093/aje/kwj232. [DOI] [PubMed] [Google Scholar]
- 116.Antoch MP, Kondratov RV, Takahashi JS. Circadian clock genes as modulators of sensitivity to genotoxic stress. Cell Cycle. 2005;4:901–7. doi: 10.4161/cc.4.7.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lis CG, et al. Circadian timing in cancer treatment: the biological foundation for an integrative approach. Integr Cancer Ther. 2003;2:105–11. doi: 10.1177/1534735403002002002. [DOI] [PubMed] [Google Scholar]
- 118.Méndez-Ferrer S, Lucas D, Battista M, Frenette PS. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008 doi: 10.1038/nature06685. Surprising observation of circadian regulation of haematopoietic stem cell release and transplantation efficiency. [DOI] [PubMed] [Google Scholar]
- 119.Spanagel R, et al. The clock gene Per2 influences the glutamatergic system and modulates alcohol consumption. Nat Med. 2005;11:35–42. doi: 10.1038/nm1163. [DOI] [PubMed] [Google Scholar]
- 120.McClung CA, et al. Regulation of dopaminergic transmission and cocaine reward by the Clock gene. Proc Natl Acad Sci U S A. 2005;102:9377–81. doi: 10.1073/pnas.0503584102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roybal K, et al. Mania-like behavior induced by disruption of CLOCK. Proc Natl Acad Sci U S A. 2007;104:6406–11. doi: 10.1073/pnas.0609625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wehr TA, Sack D, Rosenthal N, Duncan W, Gillin JC. Circadian rhythm disturbances in manic-depressive illness. Fed Proc. 1983;42:2809–14. [PubMed] [Google Scholar]
- 123.Jones SH. Circadian rhythms, multilevel models of emotion and bipolar disorder--an initial step towards integration? Clin Psychol Rev. 2001;21:1193–209. doi: 10.1016/s0272-7358(01)00111-8. [DOI] [PubMed] [Google Scholar]
- 124.Johansson C, et al. Circadian clock-related polymorphisms in seasonal affective disorder and their relevance to diurnal preference. Neuropsychopharmacology. 2003;28:734–9. doi: 10.1038/sj.npp.1300121. [DOI] [PubMed] [Google Scholar]
- 125.Nievergelt CM, et al. Suggestive evidence for association of the circadian genes PERIOD3 and ARNTL with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2006;141:234–41. doi: 10.1002/ajmg.b.30252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Desan PH, et al. Genetic polymorphism at the CLOCK gene locus and major depression. Am J Med Genet. 2000;96:418–21. doi: 10.1002/1096-8628(20000612)96:3<418::aid-ajmg34>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 127.Bailer U, et al. No association of clock gene T3111C polymorphism and affective disorders. Eur Neuropsychopharmacol. 2005;15:51–5. doi: 10.1016/j.euroneuro.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 128.Nievergelt CM, et al. Examination of the clock gene Cryptochrome 1 in bipolar disorder: mutational analysis and absence of evidence for linkage or association. Psychiatr Genet. 2005;15:45–52. doi: 10.1097/00041444-200503000-00008. [DOI] [PubMed] [Google Scholar]
- 129.Taheri S, Lin L, Austin D, Young T, Mignot E. Short sleep duration is associated with reduced leptin, elevated ghrelin, and increased body mass index. PLoS Med. 2004;1:e62. doi: 10.1371/journal.pmed.0010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Spiegel K, Tasali E, Penev P, Van Cauter E. Brief communication: Sleep curtailment in healthy young men is associated with decreased leptin levels, elevated ghrelin levels, and increased hunger and appetite. Ann Intern Med. 2004;141:846–50. doi: 10.7326/0003-4819-141-11-200412070-00008. [DOI] [PubMed] [Google Scholar]
- 131.Ancoli-Israel S, et al. The role of actigraphy in the study of sleep and circadian rhythms. Sleep. 2003;26:342–92. doi: 10.1093/sleep/26.3.342. [DOI] [PubMed] [Google Scholar]
- 132.Horne JA, Ostberg O. A self-assessment questionnaire to determine morningness-eveningness in human circadian rhythms. Int J Chronobiol. 1976;4:97–110. [PubMed] [Google Scholar]
- 133.Duffy JF, Rimmer DW, Czeisler CA. Association of intrinsic circadian period with morningness-eveningness, usual wake time, and circadian phase. Behav Neurosci. 2001;115:895–9. doi: 10.1037//0735-7044.115.4.895. [DOI] [PubMed] [Google Scholar]
- 134.Aschoff J. Circadian rhythms in man. Science. 1965;148:1427–32. doi: 10.1126/science.148.3676.1427. [DOI] [PubMed] [Google Scholar]
- 135.Czeisler CA, et al. Stability, precision, and near-24-hour period of the human circadian pacemaker. Science. 1999;284:2177–81. doi: 10.1126/science.284.5423.2177. [DOI] [PubMed] [Google Scholar]
- 136.Sack RL, et al. Circadian rhythm sleep disorders: part I, basic principles, shift work and jet lag disorders. An American Academy of Sleep Medicine review. Sleep. 2007;30:1460–83. doi: 10.1093/sleep/30.11.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Brown SA, et al. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol. 2005;3:e338. doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Brown SA, Kunz D, Dumas A, Westermark PO. Molecular insights into human daily behavior. Proceedings of the National Academy of Sciences. 2008 doi: 10.1073/pnas.0707772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liu AC, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–16. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15 (Suppl 2):R271–7. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S. Table 1
S. Table 2