BAP1 loss defines a new class of renal cell carcinoma (original) (raw)
. Author manuscript; available in PMC: 2013 Oct 2.
Published in final edited form as: Nat Genet. 2012 Jun 10;44(7):751–759. doi: 10.1038/ng.2323
Abstract
The molecular pathogenesis of renal cell carcinoma (RCC) is poorly understood. Whole-genome and exome sequencing followed by innovative tumorgraft analyses (to accurately determine mutant allele ratios) identified several putative two-hit tumor suppressor genes including BAP1. BAP1, a nuclear deubiquitinase, is inactivated in 15% of clear-cell RCCs. BAP1 cofractionates with and binds to HCF-1 in tumorgrafts. Mutations disrupting the HCF-1 binding motif impair BAP1-mediated suppression of cell proliferation, but not H2AK119ub1 deubiquitination. BAP1 loss sensitizes RCC cells in vitro to genotoxic stress. Interestingly, BAP1 and_PBRM1_ mutations anticorrelate in tumors (_P_=3×10−5), and combined loss of BAP1 and PBRM1 in a few RCCs was associated with rhabdoid features (_q_=0.0007). BAP1 and PBRM1 regulate seemingly different gene expression programs, and BAP1 loss was associated with high tumor grade (_q_=0.0005). Our results establish the foundation for an integrated pathological and molecular genetic classification of RCC, paving the way for subtype-specific treatments exploiting genetic vulnerabilities.
INTRODUCTION
Kidney cancer is estimated to have been diagnosed in over 60,000 individuals in the US in 20111. Most kidney tumors are renal cell carcinomas (RCCs) and 70% are of clear-cell type (ccRCC)2. Despite recent advances3, when metastatic, ccRCC remains largely incurable.
ccRCC is characterized by von Hippel-Lindau (VHL) gene inactivation4–6. VHL, which is on 3p25, is a two-hit tumor suppressor gene. One allele is typically inactivated through a point mutation (or indel) and the other through a large deletion resulting in loss-of-heterozygosity (LOH)7,8. Also on 3p is Polybromo 1 (PBRM1), which is frequently mutated in ccRCC9. Other genes implicated in ccRCC development include SETD210, KDM5C10, and KDM6A11, but their mutation frequency is estimated at <5%10,11.
ccRCC are classified into low and high grade tumors12, and nuclear grade is an important prognostic factor13,14. High-grade tumors exhibit mammalian target of rapamycin (mTOR) complex 1 (mTORC1) activation15. mTORC1 is a critical regulator of cell growth and is negatively regulated by a complex formed by the tuberous sclerosis complex 1 (TSC1) and 2 (TSC2) proteins16.mTOR9,10,17 and TSC118 are both mutated in sporadic ccRCC, however, mutations are infrequent, and the genetic determinants of tumor grade remain largely unknown19.
RESULTS
Identification of candidate two-hit tumor suppressor genes
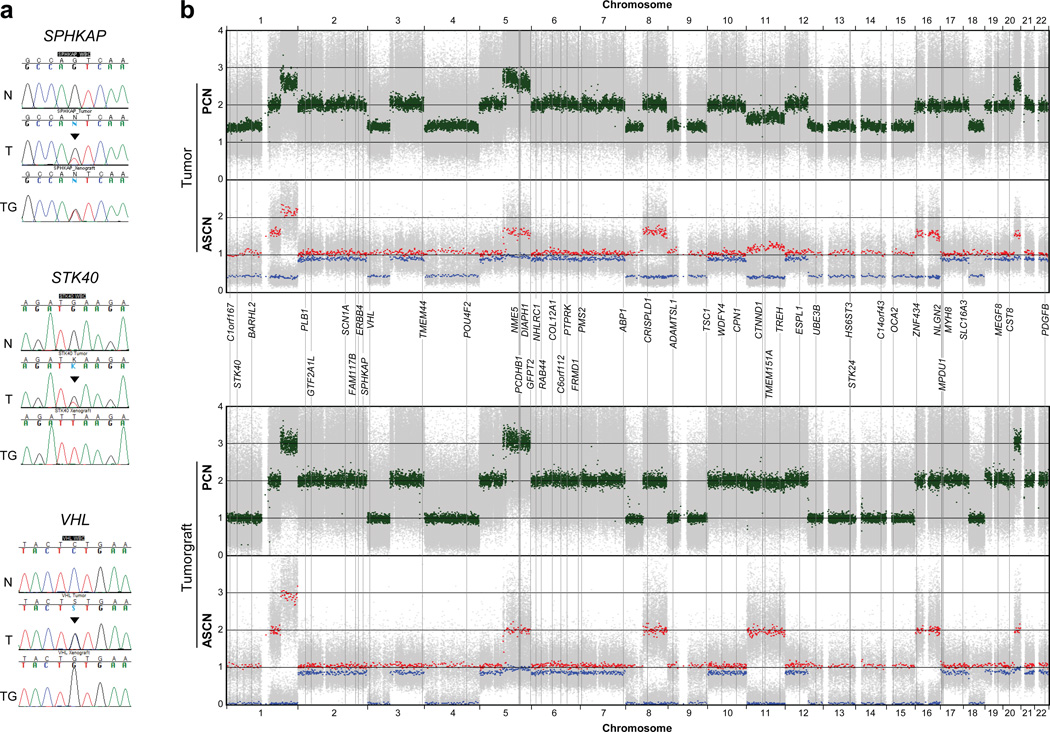
We sequenced the genome of a sporadic high-grade ccRCC and paired normal sample to >94% coverage and a mean depth ≥35× (Supplementary Figures 1 and 2). We found 6,571 somatically-acquired single-nucleotide mutations or indels including 59 in protein-coding regions (Supplementary Table 1). Every mutation evaluated was confirmed by Sanger sequencing (Table 1 and Supplementary Table 2). However, mutant allele ratios (MARs) - the fraction of mutant over mutant and wild type alleles for each mutation – were low; few mutations reached 0.5 (expected for heterozygous mutations), and no mutations reached 1 (expected for mutations accompanied by LOH) (Table 1 and Supplementary Table 2). In the case of VHL, the MAR was 0.52 (Table 1; Fig. 1a), which suggested a heterozygous mutation. However, these results conflicted with DNA copy number analyses showing that one copy of 3p was lost (Fig. 1b). We attributed the low MARs to tumor contamination by normal stroma. Contamination occurred despite careful sample selection (Supplementary Figure 2c).
Table 1.
Integrated analysis of a subset of somatic mutations and DNA copy-number alterations in the index subject
| Gene | Chr | Position§ | Nucleotidechange | Mutant Allele Ratios | T | TG | Change | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sanger Seq. | ASCN | ASCN | |||||||||||
| Illumina | T | TG | PCN | Min | Max | PCN | Min | Max | |||||
| C1orf167 | 1 | 11,767,238 | G>T | 0.38 | 0.36 | 1.00 | 1.39 | 0.43 | 1.00 | 1.02 | 0.003 | 1.04 | Splice site |
| STK40 | 1 | 36,593,565 | C>A | 0.30 | 0.32 | 1.00 | 1.39 | 0.43 | 1.00 | 1.00 | 0.003 | 1.04 | p.Met133Ile |
| VHL | 3 | 10,166,479 | C>G | 0.37 | 0.52 | 1.00 | 1.39 | 0.43 | 1.07 | 0.98 | 0.003 | 1.05 | p.Leu158Val |
| DIAPH1 | 5 | 140,885,872 | C>T | 0.26 | 0.20 | 0.31 | 2.53 | 0.97 | 1.55 | 3.05 | 0.97 | 2.00 | p.Arg1164Gln |
| GFPT2 | 5 | 179,662,025 | G>A | 0.43 | 0.38 | 0.65 | 2.51 | 0.97 | 1.54 | 3.05 | 0.97 | 2.00 | Splice site |
| CRISPLD1 | 8 | 76,088,864 | G>A | 0.57 | 0.56 | 1.00 | 2.02 | 0.41 | 1.63 | 1.98 | 0.003 | 2.00 | p.Val200Ile |
| ADAMTSL1 | 9 | 18,767,566 | del9 | 0.25 | 0.34 | 1.00 | 1.42 | 0.44 | 1.12 | 1.01 | 0.004 | 1.06 | p.Glu1114_Gln1116del |
| CTNND1 | 11 | 57,333,402 | delG | 0.36 | 0.38 | 1.00 | 1.74 | 0.41 | 1.16 | 1.95 | 0.003 | 1.96 | p.Val769Serfs*5 |
| TMEM151A | 11 | 65,818,643 | G>T | 0.36 | 0.18 | 1.00 | 1.62 | 0.41 | 1.16 | 1.93 | 0.003 | 1.96 | p.Cys117Phe |
| TREH | 11 | 118,035,289 | C>A | 0.54 | 0.50 | 1.00 | 1.62 | 0.41 | 1.19 | 1.89 | 0.003 | 1.96 | p.Gly478Cys |
| UBE3B | 12 | 108,456,960 | A>T | 0.37 | 0.40 | 1.00 | 1.39 | 0.43 | 1.01 | 1.00 | 0.003 | 1.06 | p.Glu1066Tyr |
| HS6ST3 | 13 | 96,283,428 | A>T | 0.38 | 0.38 | 1.00 | 1.39 | 0.43 | 1.06 | 0.98 | 0.004 | 1.04 | p.Tyr464Phe |
| STK24 | 13 | 97,907,504 | C>T | 0.28 | 0.38 | 1.00 | 1.39 | 0.43 | 1.06 | 0.98 | 0.004 | 1.04 | p.Arg405Gln |
| C14orf43 | 14 | 73,275,194 | del50 | 0.32 | 0.35 | 1.00 | 1.39 | 0.44 | 1.07 | 0.99 | 0.004 | 1.05 | p.Gln408Glyfs*65 |
| ZNF434 | 16 | 3,373,160 | T>C | 0.29 | 0.30 | 0.55 | 1.95 | 0.41 | 1.57 | 1.98 | 0.003 | 1.99 | p.Gln384Arg |
Fig. 1. Integrative mutation and DNA copy-number analyses in a tumor and tumorgraft from the index subject.
(a) Representative capillary sequencing chromatograms of normal (N), patient’s tumor (T), and tumorgraft (TG) illustrating different examples of mutant allele enrichment in the tumorgraft. Arrowheads indicate mutations. (b) Allele specific (ASCN) and paired (PCN) copy number representation of high-density SNP array data incorporating the estimated position of mutated genes. Green, paired copy numbers. Red and blue, maximum and minimum copy numbers for each heterozygous SNP.
Tumor implantation in mice expands the neoplastic compartment while human stroma is replaced by the host20 and therefore, tumorgrafts may serve to calculate MARs with accuracy. RCC tumors implanted orthotopically in mice preserve the characteristics of patient tumors21. We performed Sanger sequencing of mutated genes in a tumorgraft derived from the index subject’s tumor using human-specific primers. By comparison to tumor MAR (MART) values, tumorgraft MAR (MARTG) values often increased to ~0.5, and for several genes, including VHL, they reached 1 (Table 1, Supplementary Figure 3 and Supplementary Table 2).
To determine whether MARTG reflected those expected in the subject’s tumor, we asked whether a correlation existed between MARTG and the corresponding regional DNA copy numbers in the patient’s tumor (Table 1, Supplementary Table 2 andFig. 1b). A correlation was found with MARTG (_p_=0.000013), but not with MART(_p_=0.054). These data suggest that MARs in tumors are more accurately determined by evaluating tumorgrafts. Consistent with the notion that tumorgrafts represent largely pure populations of human tumor cells, paired copy numbers (PCNs) and allele-specific copy numbers (ASCNs) in tumorgrafts more closely approached integer values (Table 1 and Fig. 1b and Supplementary Table 2).
To identify putative two-hit tumor suppressor genes, we searched for genes with MARTG~1. Some genes (STK40,UBE3B, HS6ST3, STK24,C1orf167, ADAMTSL1 and C14orf43) were in regions of deletion (PCNTG~1), whereas others (CRISPLD1, TMEM151A, _TREH_and CTNND1) were in areas of copy-neutral LOH (PCNTG~2 and tumorgraft ASCNmin~0 and ASCNmax~2) (Table 1, Fig. 1b). Because mutations in copy-neutral LOH regions could be either homozygous (e.g.CRISPLD1) or heterozygous (such as ZNF434) (Table 1, Fig. 1b), accurate MARs were essential to establish whether mutated genes were putative two-hit tumor suppressors.
Accurate MARs were also helpful in inferring whether in areas of duplication (PCNTG~3) the allele amplified was mutant (e.g.GFPT2; MARTG = 0.65 (expected 0.66)) or wild-type (e.g. DIAPH1; MARTG = 0.31 (expected 0.33)) (Table 1). In the case of_GFPT2_, the mutation may have preceded the duplication, whereas in the case of DIAPH1, the mutation is likely to have followed the duplication. Thus, analyses in tumorgrafts identified candidate tumor suppressor genes and shed light into the temporal sequence of mutation acquisition.
Evaluation of somatically mutated genes in a discovery cohort
Twenty-one genes mutated in the sequenced ccRCC and not previously examined by the Sanger Institute10 were sequenced in a discovery set of 76 ccRCCs, and mutations were examined in the corresponding normal samples (Supplementary Table 3). As determined by VHL sequencing, which revealed somatically acquired mutations in 79% of tumors (see Supplementary Data 1), sensitivity for mutation detection was excellent. Several putative two-hit tumor suppressor genes were mutated at higher than expected frequencies including_CRISPLD1_, which was mutated in two additional tumors (_q_=0.044) and TMEM151A, mutated in three (q_=0.005) (Supplementary Table 4). In addition, several other genes were recurrently mutated including OCA2 and_MT-ND1 (Supplementary Table 4). Germline mutations in OCA2_cause autosomal recessive oculocutaneous albinism type 2 and the two somatic mutations we identified (p.Pro211Leu and p.Val443Ile; Supplementary Table 4) are known disease-causing mutations22,23. Two additional somatically-acquired mutations were found in MT-ND1(Supplementary Table 4), a gene mutated in oncocytomas, a benign tumor type24. Their presence in ccRCC suggests that oncocytomas could transform into malignant tumors. Transformation may result from VHL inactivation, which was observed in all the tumors with somatic ND1 mutations (Supplementary Data 1). VHL inactivation could change the morphological appearance of the tumor by affecting cellular metabolism and angiogenesis. In addition, 3 additional mutations were identified in_TSC1, which we previously reported elsewhere18.
Exome sequencing identifies BAP1 as a candidate two-hit tumor suppressor gene
We performed exome sequencing on 7 ccRCC primary tumors, including 6 of high grade (and corresponding normal samples). A metastasis from one patient was also sequenced. We found 345 somatically acquired mutations (Supplementary Table 5). In the tumor/metastasis pair we observed 37 and 39 mutations respectively and 32 were shared.
To determine concordance of the mutations called, we performed Sanger sequencing. If concordance was >95%, Sanger sequencing of 82 mutations would predict for >90% accuracy for the whole cohort. Among 82 randomly selected mutations, 78 were confirmed (see methods) with an accuracy >95% (Supplementary Table 6 and Supplementary Data 2). For 7 samples, there were tumorgrafts, and sequencing analyses of mutated genes therein, uncovered 16 potential two-hit tumor suppressor genes (Supplementary Table 6).
We focused on 10 genes mutated in at least two tumors (Supplementary Table 7). All the mutations validated by Sanger sequencing. Whereas MARTanalysis failed to identify any putative two-hit tumor suppressors, another gene besides VHL and PBRM1 exhibited MARTG~1, BAP1 (Supplementary Table 7).
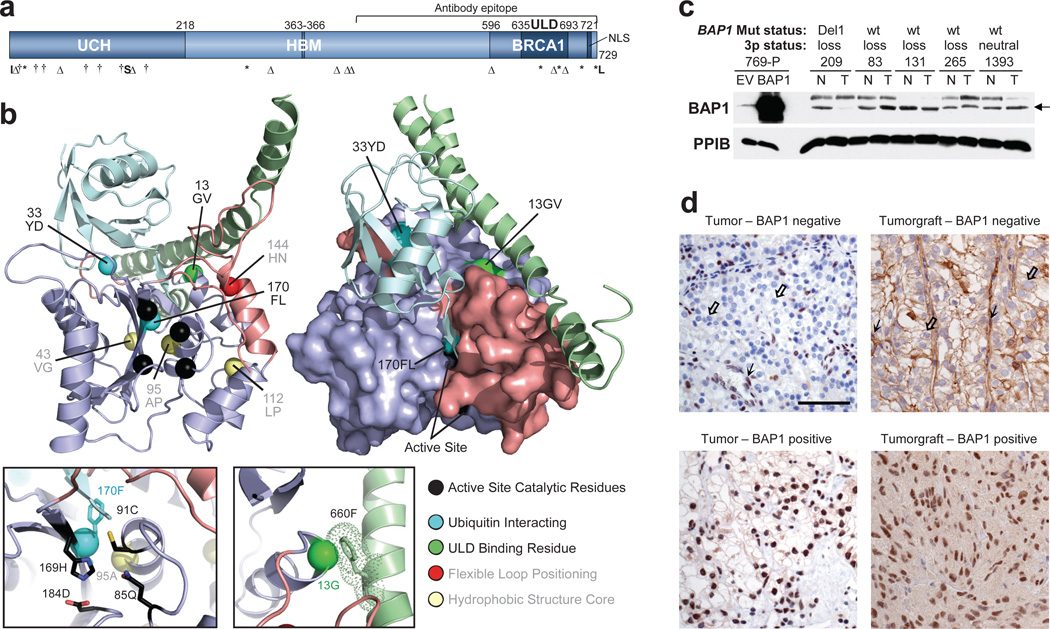
BAP1 sequencing in the discovery set of 76 ccRCCs identified 11 non-synonymous mutations, including 10 confirmed to be somatically acquired (Table 2). Examination of a validation ccRCC set (_n_=92), with corresponding normal samples, uncovered 11 additional nonsynonymous mutations, including 10 that were somatically acquired (Table 2). Two mutations without matching normal samples were truncating and likely deleterious. Altogether, the BAP1 mutation rate was 14% (24/176 tumors). BAP1 encodes a nuclear deubiquitinatinase (DUB) of the ubiquitin C-terminal hydrolase (UCH)-domain containing family25–27 that is mutated in both uveal28 and cutaneous melanoma29 as well as in mesothelioma30. In ccRCC, most mutations were predicted to truncate the protein and mutations were enriched in the UCH domain (Fig. 2a and b).
Table 2.
List of BAP1 mutations in ccRCC and cell lines
| ID | CDS | Protein |
|---|---|---|
| 3575 | c.5_6dupAT | p.Lys3Ilefs*33 |
| 63 | c.21_32del12 | p.Glu7Asp,Leu8_Asp11del |
| T145 | c.38G>T | p.Gly13Val |
| T211 | c.58G>T | p.Glu20* |
| T16 | c.128T>G | p.Val43Gly |
| T114 | c.193delT | p.Leu65Trpfs*7 |
| T166 | c.283G>C | p.Ala95Pro |
| T69 | c.335T>C | p.Leu112Pro |
| T115 | c.430C>A | p.His144Asn |
| 3397 | c.IVS438-1G>A | Splice site |
| T55 | c.458delC | p.Pro153Leufs*34 |
| T212 | c.510T>A | p.Phe170Leu |
| T184 | c.889G>T | p.Glu297* |
| 209 | c.971delC | p.Pro324Hisfs*11 |
| 162 | c.1219delG | p.Asp407Metfs*23 |
| T149 | c.1256delA | p.Lys419Argfs*11 |
| T26 | c.1271_1274delGGAA | p.Lys425Glnfs*4 |
| 78 | c.1793delC | p.Pro598Glnfs*19 |
| 9575 | c.1981A>T | p.Lys661* |
| T163 | c.2028_2046del19 | p.Cys676Trpfs*10 |
| T70 | c.2050C>T | p.Gln684* |
| T25 | c.2051delA | p.Gln684Argfs*8 |
| 40 | c.2134C>T | p.Gln712* |
| 9145 | c.2188T>G | p.*730Glyext*206 |
| 769-P | c.97T>G | p.Tyr33Asp |
| UMRC6 | c.430delC | p.His144Metfs*43 |
Fig. 2. BAP1 is a tumor suppressor in ccRCC.
(a) Schematic of BAP1 with mutations (UCH, ubiquitin C-terminal hydrolase domain; HBM, HCF-1 binding motif; BRCA1, [putative] BRCA1 interacting domain; ULD, Uch37-like domain; NLS, nuclear localization signal; I, insertion; Δ, deletion; †, missense; *, non-sense; S, splice site; *L, stop codon loss. (b) Structural model of BAP1 UCH domain (purple) and ULD tail (green) superimposed on a template DUB (not shown) bound to ubiquitin (cyan); structural elements that alter upon ubiquitin binding are colored salmon. Left, cartoon of BAP1 model. Right, surface representation of BAP1 highlighting the positions of RCC alterations on interaction surfaces. Left inset, enlarged view of the DUB active site. Right inset, enlarged view of the Gly13, interaction with an aromatic residue in the ULD tail (dots indicate interaction radius). (c) Western blot of extracts from tumors with defined BAP1 mutation and chromosome 3p status. 769-P cells transfected with either an empty vector (EV) or wild-type (WT) BAP1 were used as controls. PPIB, cyclophilin B is shown as a loading control. Arrow indicates BAP1. (d) Representative IHC of tumors and tumorgrafts positive or negative for nuclear BAP1. Scale bar, 50 μm. Open arrow, tumor cells; simple arrow, endothelial cells and lymphocytes, which express BAP1 and serve as internal controls.
Development of a clinical IHC assay for BAP1 detection
As most mutations were truncating, we developed, in a CLIA-certified laboratory, an immunohistochemistry (IHC) test for the presence/absence of BAP1 protein. Genetically characterized ccRCC samples validated by western blot were used as controls (Fig. 2c and d). Scoring was performed by a clinical pathologist (P.K.) masked to the_BAP1_ genotype. IHC was interpretable in 175/176 tumors. Nuclear BAP1 was detected in 150 tumors, and 148 were wild-type (Supplementary Figure 4). The 2 discordant samples had missense mutations (p.Gly13Val and p.Phe170Leu). Twenty-five samples were negative by IHC and 22 had _BAP1_mutations. Analysis of an IHC-negative but BAP1 wild type sample by western blot failed to reveal detectable BAP1 protein suggesting that other mechanisms exist to inactivate BAP1. Overall, the positive and negative predictive values of the IHC test were ~100% and 98.6%, respectively.
Structural analyses of BAP1 missense mutations
To evaluate missense mutations in a structural context, we generated a BAP1 protein model on the basis of the related family members Uch-L3 and Uch37 (Fig. 2b). Since ubiquitin binding orders a significant portion of the protein, the UCH domain of BAP1 was modeled after Uch-L3 bound to ubiquitin (PDB: 1xd3). The interaction with the ULD domain was built by superimposing Uch37 (PDB: 3ihr). Four mutations abrogated protein expression; 3 were predicted to destabilize the protein (p.Val43Gly and p.Leu112Pro removed side chains that contribute to the hydrophobic core, and p.Ala95Pro disrupted the backbone of a central α-helix) and the fourth (p.His144Asn) disrupted the position of a flexible loop (Fig. 2b). Two mutations did not abrogate protein expression (p.Gly13Val and p.Phe170Leu). These mutations disrupted side chains implicated in either an intramolecular interaction with the ULD domain (Gly13) or ubiquitin binding (Phe170), and highlight the importance of these interactions for tumor suppressor function.
BAP1 suppresses RCC cell proliferation and deubiquitinates H2Aub1 in renal cancer cells
Studies of the role of BAP1 in cell proliferation have led to conflicting results25–27,30–33. To examine BAP1 in an appropriate context, ccRCC cell lines were sought in which natural selection had led to BAP1 inactivation. Among 12 RCC cell lines examined initially, only 769-P had a BAP1 mutation (Supplementary Table 8). The mutation (c.97T>G; p.Tyr33Asp) disrupted a residue binding ubiquitin and did not abrogate protein expression (Fig. 2b and Fig. 3a).
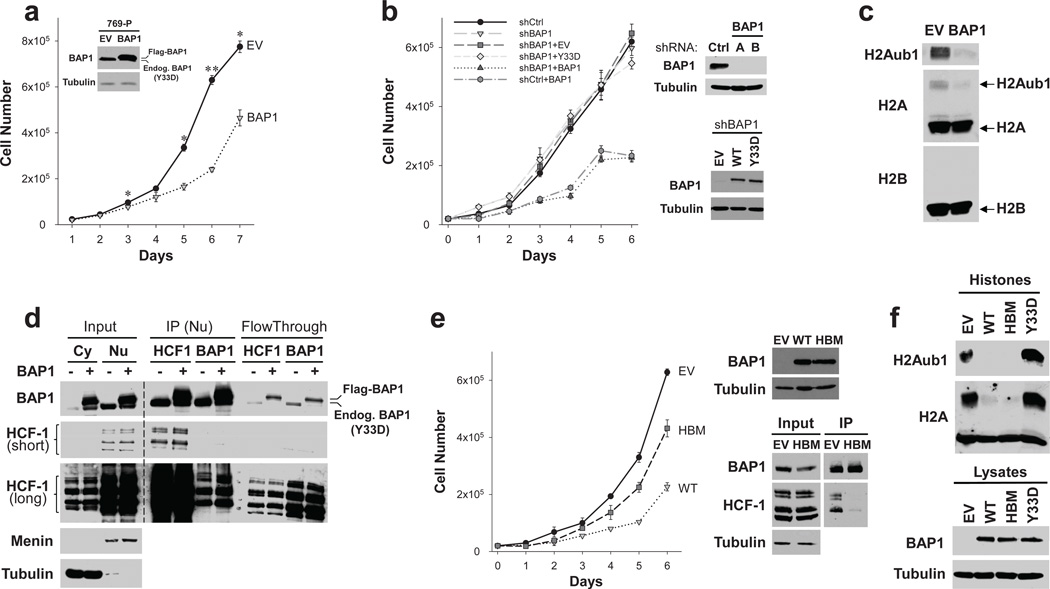
Fig. 3. HCF-1-dependent suppression of cell proliferation by BAP1.
(a) Proliferation curves of empty vector (EV) and BAP1 reconstituted 769-P cells with inset showing BAP1 western blot. (b) Proliferation curves of 769-P cells stably expressing an shRNA targeting endogenous (mutant) BAP1 (shBAP1) or a vector control (shCtrl) and in addition wild-type BAP1 (BAP1), BAP1Y33D (Y33D) or empty vector (EV). Western blot of cells transduced with shRNA targeting endogenous mutant BAP1 (A and B) or vector control (shCtrl) and transduced with expression vectors as indicated. (c) Western blot of partially purified histone fractions of 769-P cells reconstituted with an empty vector (EV) or wild-type BAP1 (BAP1). (d) Western blot of input (cytosolic [Cy] or nuclear [Nu] fractions) as well as immunoprecipitates (from nuclear fractions) and corresponding flow-through from empty vector (−) or wild-type BAP1 (+) expressing cells. Short and long exposures are indicated. Both ectopically expressed epitope tagged wild-type as well as endogenous mutant BAP1 bind HCF-1. (e) Proliferation curves of 769-P cells depleted of endogenous BAP1 shRNA and reconstituted with an empty vector (EV), wild-type BAP1 (WT) or HCF-1 binding motif mutant (HBM). Western blot from input as well as BAP1 immunoprecipitates. (f) Western blot of partially purified histone fractions or cell lysates from 769-P cells depleted of endogenous_BAP1_ and transduced with an empty vector (EV), wild-type BAP1 (WT), an HCF-1 binding motif mutant (HBM), or BAP1Y33D (Y33D). Error bars represent SEM (_n_=3). *,p<0.05; **, p<0.01.
To determine the role of BAP1, 769-P cells were reconstituted with epitope-tagged wild-type BAP1 (or an empty vector control). BAP1 repressed cell proliferation without causing apoptosis (Fig. 3a and data not shown). However, BAP1 did not completely abrogate cell proliferation. To determine whether endogenous mutant BAP1 acted as a dominant negative, endogenous BAP1 was depleted using shRNA. However, mutant BAP1 depletion did not increase the effects of ectopically expressed wild-type BAP1 indicating that mutant BAP1 does not function in a dominant negative fashion (Fig. 3b).
The BAP1 orthologue in Drosophila, Calypso, targets monoubiquitinated histone H2A (H2Aub1)34. An examination of H2AK119ub1 levels in 769-P cells reconstituted with wild-type BAP1 showed downregulation of basal H2Aub1 levels indicating that mammalian BAP1 similarly deubiquitinates H2A in renal cancer cells (Fig. 3c).
BAP1 binds HCF-1 and HCF-1 binding is required for suppression of cell proliferation
BAP1 interacts with host cell factor-1 (HCF-1)31,33,35, which serves as a scaffold for several chromatin remodeling complexes36. HCF-1 binds to multiple transcription factors including several E2Fs37,38, and recruits histone modifying enzymes such as Set1/MLL1 histone methyltransferases39–41, LSD1 histone demethylase42, Sin3 histone deacetylase39, and MOF histone acetyltransferase43.
We asked whether BAP1 interacted with HCF-1 in 769-P cells. An interaction was confirmed by reciprocal immunoprecipitation experiments (Fig. 3d). Interestingly, anti-HCF-1 antibodies depleted BAP1 from cell extracts to the same extent as anti-BAP1 antibodies, suggesting that, as in other cell types35, the majority of BAP1 in renal cancer cells is bound to HCF-1 (Fig. 3d). BAP1 has been proposed to deubiquitinate HCF-131,33 and regulate HCF-1 levels31, but consistent with other reports33, HCF-1 levels were similar in BAP1-deficient and reconstituted 769-P cells (Fig. 3d).
We mutated sequences in BAP1 encoding the HCF-1 binding motif and evaluated this mutant (HBM) in cell proliferation assays. HBM suppressed HCF-1 binding and compromised the inhibitory effect of BAP1 on cell proliferation (Fig. 3e). However, the HBM mutant did not differ from wild-type BAP1 in its ability to deubiquitinate H2A (Fig. 3f). Thus, BAP1 binds HCF-1, and binding to HCF-1, but not H2Aub1 deubiquitination, is important for the inhibition of cell proliferation.
Next, we performed gel-filtration chromatography. Extracts from 769-P cells expressing either an empty vector or wild-type BAP1 were fractionated using a size-exclusion column and subjected to western blotting. As shown inSupplementary Figure 5, most BAP1 was found in complexes >1 MDa and eluted with HCF-1.
BAP1 loss sensitizes renal cancer cells to radiation and PARP inhibitors
BAP1 is phosphorylated following DNA damage44,45 and we asked whether BAP1 loss affected the response to γ-irradiation. Empty vector and BAP1 reconstituted 769-P cells exhibited a similar pattern of Rad51 and γH2AX foci (Supplementary Figure 6a). However, BAP1-deficient cells were more sensitive to ionizing radiation (Supplementary Figure 6b) and fewer colonies formed in clonogenic assays (Supplementary Figure 6c). In addition, BAP1 loss sensitized cells to the PARP inhibitor olaparib (Supplementary Figure 6d and e).
We examined 4 additional ccRCC cell lines (Supplementary Table 8). UMRC6 lacked BAP1 protein and had a frameshift mutation (c.430delC) (Supplementary Figure 7a and b). As in 769-P cells, (i) cell proliferation was inhibited by wild-type BAP1 and substantially less so by an HBM mutant, (ii) the HBM mutant reduced H2Aub1 levels, (iii) BAP1 cofractionated with HCF-1, and (iv) restoration of BAP1 protected UMRC6 cells against genotoxic death (Supplementary Figure 7).
BAP1 binds HCF-1 and elutes with HCF-1 in tumorgrafts
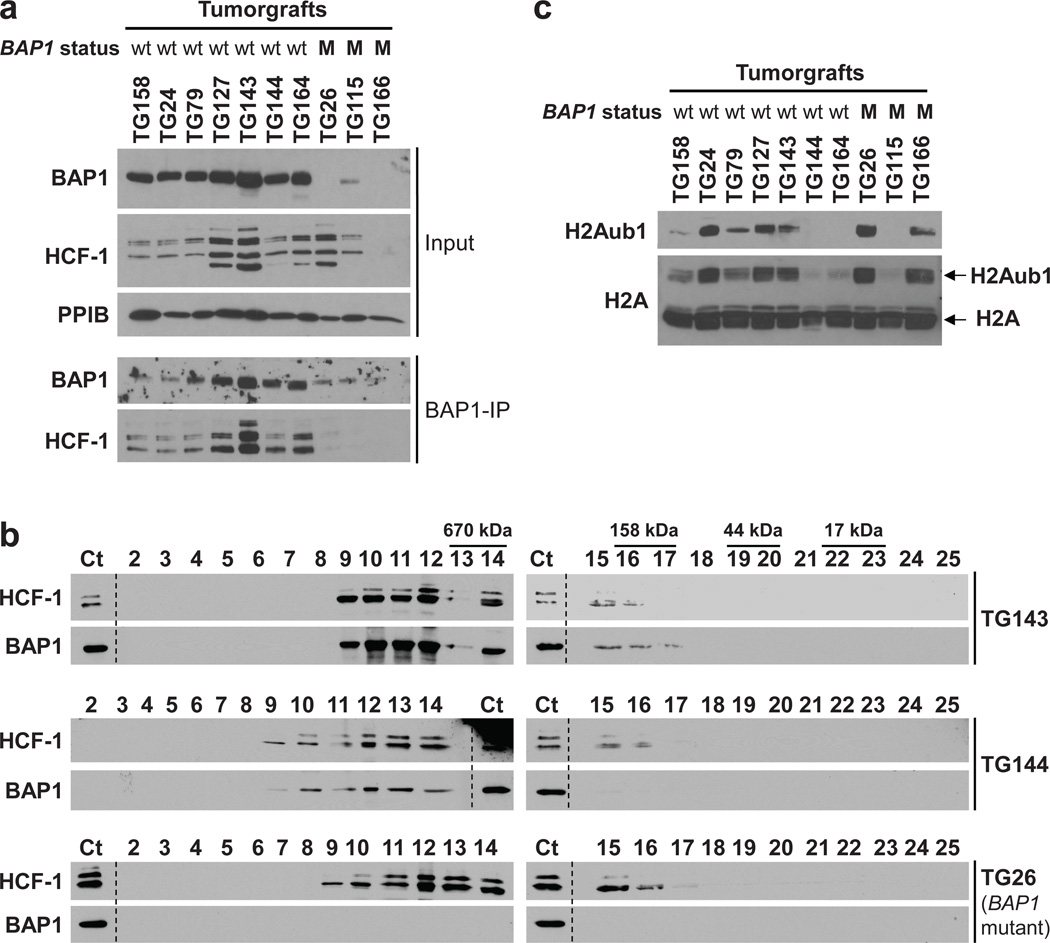
The usefulness of RCC cell lines is limited by the development of mutations and copy number alterations as tumor cells adapt to growth in culture7,46. Divergence from tumors may be particularly striking with respect to epigenetic regulation, as growth conditions of cell lines and tumors are very different. In contrast, the pattern of gene expression is reproduced in tumorgrafts growing orthotopically in mice21 and tumorgrafts, like cell lines, represent a renewable source of tumor material. To determine whether the interaction of BAP1 with HCF-1 was physiologically significant, we analyzed tumorgrafts. As in cell lines, BAP1 bound to and co-fractionated with HCF-1 (Fig. 4a and b). In addition, we examined whether there was a correlation between_BAP1_ mutation and H2Aub1 levels in tumorgrafts, but no correlation was observed (Fig. 4c). Taken together these data show that BAP1 binding to HCF-1 is likely to be important for BAP1 suppression of RCC development.
Fig. 4. BAP1 binds to and elutes with HCF-1 in tumorgrafts.
(a) Western blot from inputs as well as BAP1 immunoprecipitates (BAP1-IP) from the indicated tumorgrafts. wt, wild type; M, mutant. (b) Western blot of TCA precipitated gel-filtration fractions of tumorgrafts either wild type for BAP1 (TG143 and TG144) or mutant (TG26). Ct, Control lysate from 769-P cells. (c) Western blot of partially purified histone fractions from tumorgrafts with the indicated BAP1 status.
BAP1 loss is associated with high grade
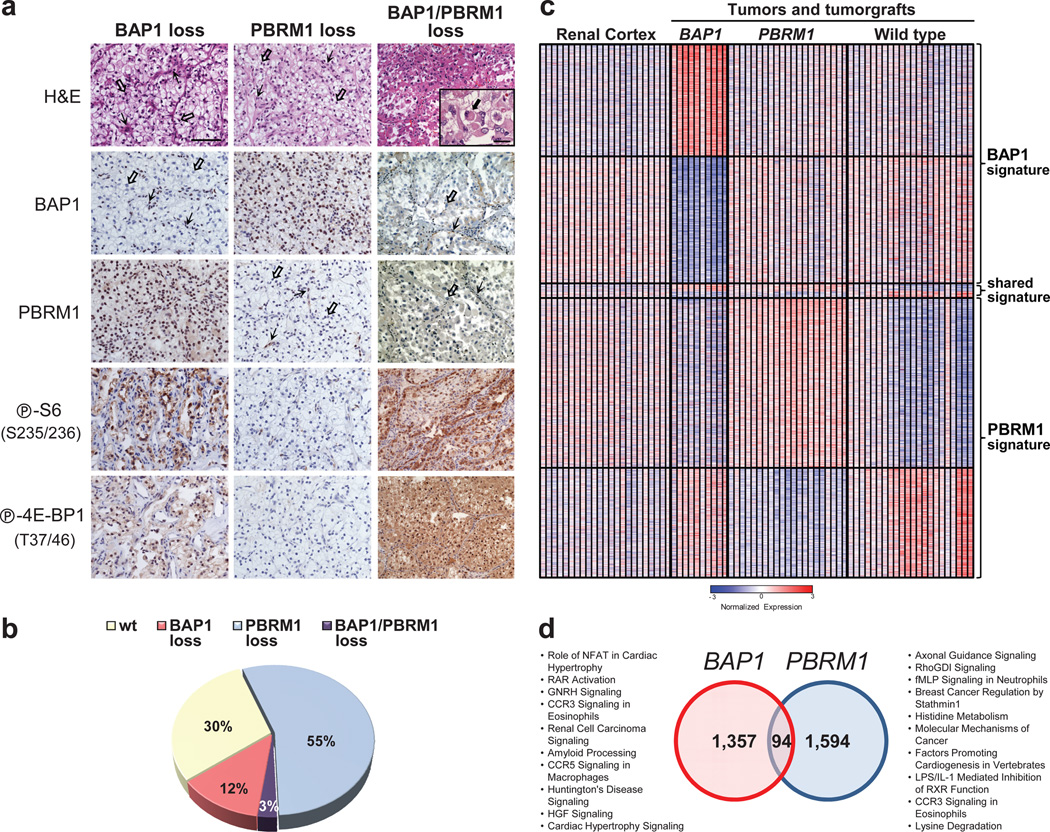
Deep sequencing studies largely focused on high-grade tumors. An analysis of all 176 tumors examined showed that BAP1 loss correlated with high Fuhrman nuclear grade (_q_=0.0005) (Supplementary Data 1). Because nuclear grade is associated with mTORC1 activation15, we tested whether a correlation existed with between BAP1 loss and mTORC1 activity. As determined by the phosphorylation of both S6 and 4E-BP1, BAP1 loss correlated with mTORC1 activation (_q_=3·10−4 and 0.029, respectively) (Fig. 5a and Supplementary Data 1). This association did not appear to be direct, however, and similar levels of mTORC1 activation were observed in BAP1-deficient and wild-type BAP1-reconstituted cells (Supplementary Figure 8).
Fig. 5. Loss of BAP1 and PBRM1 sets foundation for molecular genetic classification of ccRCC.
(a) Representative H&E and immunohistochemistry (IHC) images of tumors with loss of BAP1, PBRM1, or both. Scale bar, 50 µm; 10 µm for inset. Open arrows, tumor cells; simple arrows, stroma/inflammatory cells; filled arrow, rhabdoid tumor cell. (b) Pie chart of the distribution of ccRCC subtypes. (c) Heatmap of statistically significant probes distinguishing _BAP1_- and_PBRM1_-deficient tumors/tumorgrafts vs. wild type. Expression of the same probes in renal cortex included as a reference. The full data set is provided in Supplementary Data 4. (d) Venn diagram illustrating the overlap in BAP1 and PBRM1 gene expression signatures with associated global pathway analyses.
BAP1 and PBRM1 mutations anticorrelate in ccRCC
To explore whether a relationship existed between the loss of BAP1 and PBRM1, we first developed an IHC assay for PBRM1 (also known as BAF180) (Fig. 5a). Evaluation of the 176 tumors showed confident PBRM1 staining for 146 samples, and 53% were negative for PBRM1 (Supplementary Data 1). As PBRM1 was lost in ~50% of tumors, BAP1 loss should distribute equally between PBRM1-expressing and -deficient tumors. However, only 4 of 21 BAP1-IHC-deficient tumors were also deficient for PBRM1 (Supplementary Figure 9a). These results suggest that PBRM1 and BAP1 loss anticorrelated in tumors (_p_=7·10−4).
To explore this further, we sequenced PBRM1 in the 176 ccRCCs. We identified 92 somatic mutations, including 6 missense mutations (Supplementary Data 1). Structural analyses are shown in Supplementary Figure 10. We correlated sequencing data with the results from IHC; ~90% in of samples that were negative for PBRM1 by IHC had a mutation, and ~90% of the samples that were positive were wild type (_p_=4·10−23; Supplementary Figure 9b). An analysis of BAP1 and PBRM1 mutations in tumors revealed that only 3 of 24 samples with BAP1 mutations had a somatically-acquired PBRM1 mutation (Supplementary Figure 9c). Once again, an anticorrelation was found (_p_=3·10−5).
As a reference, we evaluated the distribution of mutations in_SETD2_ and KDM5C respect to_PBRM1_ in ccRCCs from the Sanger Institute9,10. Among 348 ccRCCs genotyped for PBRM1, 15 mutations in SETD2 were observed, and these mutations distributed similarly between PBRM1 mutant and wild-type tumors (8 vs. 7; Supplementary Data 3). KDM5C mutations were also similarly distributed (5 vs. 4; Supplementary Data 3).
Combining the IHC and mutation data, 5 out of 27 BAP1-deficient tumors were also deficient for PBRM1. Assuming a binomial distribution of BAP1 loss, these data indicate that simultaneous inactivation of BAP1 and PBRM1 is negatively selected for in tumors (_p_=0.0008). Notably, however, loss of BAP1 or PBRM1 was observed in 70% of ccRCC (Fig. 5b).
To obtain further insight into the relationship between BAP1 and PBRM1, we performed gene expression analyses. We grouped tumors and tumorgrafts according to their BAP1 and PBRM1 status and evaluated differences with respect to wild-type tumors and tumorgrafts (Fig. 5c). Probesets (probes) that we had previously determined using tumorgrafts to be driven by non-neoplastic cells21 were excluded from the analysis. We identified 1,451 probes that were deregulated in BAP1-deficient tumors by comparison to those that were wild type (for both BAP1 and_PBRM1_) (q<0.05) (Supplementary Data 4). A similar number of probes distinguished PBRM1-deficient tumors (Supplementary Data 4). These two datasets shared 94 probes in common (Fig. 5d). However, the overlap expected at random was 67. Similarly, pathway analyses of the two signatures showed little overlap. These results suggest that BAP1 and PBRM1 do not function in the same pathway, and that the tumorigenic advantage to mutating BAP1 and_PBRM1_ is context dependent.
Further supporting the notion that loss of BAP1 and PBRM1 in tumors is not equivalent, analyses of the 176 tumors showed that PBRM1 loss was not associated with high grade (_q_=0.26) (Supplementary Data 1). In the 348 ccRCC tumors sequenced by the Sanger Institute9,10 (Supplementary Data 3), we found a non-significant correlation between PBRM1 loss and low grade (_p_=0.074). Furthermore, when focusing the analyses of 176 tumors on those that had exclusively lost PBRM1, a statistically significant correlation with low tumor grade was found (_q_=0.025).
Tumors with simultaneous inactivation of BAP1 and_PBRM1_ exhibit rhabdoid features
A few tumors had loss of both BAP1 and_PBRM1_ (n_=5) (Supplementary Data 1). While co-ocurrence of mutations in tumors may not indicate their occurrence together in the same cell and there is substantial mutation heterogeneity in RCC17,47, in two tumors for which tumorgrafts were available, MARTG for both BAP1 and_PBRM1 were ~1 and no wild-type alleles were detected (data not shown). These data suggest that the two mutations were indeed present in the same tumor cells and highlight another application of tumorgrafts.
Tumors deficient for both BAP1 and PBRM1 were uniformly of high grade and exhibited characteristic features: abundant acidophilic cytoplasm, eccentric nuclei and prominent macronucleoli (Fig. 5a). These features were consistent with rhabdoid morphology48, a form of dedifferentiation portending aggressive tumor behavior49. They were present in all tumors for which there was sufficient material for analysis (4/5), and while not unique to tumors deficient for both BAP1 and PBRM1, the association was significant (_q_=0.0007; Supplementary Data 1).
DISCUSSION
These results implicate BAP1 as a tumor suppressor in ccRCC and establish the foundation for a molecular genetic classification of RCC. We show that 70% of ccRCCs lose either BAP1 or PBRM1, that tumors tend to segregate into BAP1 or PBRM1-deficient subtypes, and that BAP1 loss but not PBRM1 loss is associated with high tumor grade.
BAP1 functions as a two-hit tumor suppressor in ccRCC and consistent with this, mutant BAP1 does not act as dominant negative. Both copies of_BAP1_ are also lost in melanoma28,29,50 and mesothelioma30,51. While the number of RCC samples with _BAP1_mutations is small, it is interesting that no second-hit point mutations or indels were observed. In contrast, both BAP1 alleles may be inactivated through a point mutation (or indel) in mesothelioma51. We speculate that the different modes of inactivation of the “second” BAP1 allele reflect tissue-specific tumor suppressor gene cooperativity. Indeed, in ccRCC, 3p loss may simultaneously inactivate several genes suppressing renal tumorigenesis including, most importantly, VHL, which is rarely mutated in other tumor types. In metastatic uveal melanoma, whole chromosome 3 losses are frequent, and other melanoma metastasis suppressors may exist on 3q. Thus, the deletion architecture of tumors may reflect tissue-specific cooperativity of tumor suppressor genes.
We propose that following a VHL mutation, which likely represents an early event17, the loss of 3p leaves cells vulnerable to the loss of the remaining_PBRM1_ or BAP1 allele. The acquisition of a_PBRM1_ or BAP1 mutation may set the course for ccRCCs with different properties. PBRM1 and BAP1 likely affect different epigenetic programs, and BAP1 loss is associated with high grade and mTORC1 activation. Interestingly, whereas mutations in SETD2, also on 3p, appear to distribute equally between PBRM1_-deficient and wild-type tumors, this is not the case for BAP1 mutations. PBRM1 and_BAP1 mutations anticorrelate in ccRCC. These data suggest that there is a genetic context of tumor suppressor function and that simultaneous loss of BAP1 and PBRM1 in most tumors may be disadvantageous.
The clinical implications of BAP1 loss remain to be explored. Inasmuch as BAP1 loss was associated with high tumor grade and correlated with metastasis development in uveal melanoma28, BAP1 loss in ccRCC may be associated with poor prognosis. From a therapeutic standpoint, while RCC is considered radioresistant, BAP1-deficient tumors may be more sensitive. Evaluating the prognostic and therapeutic implications of BAP1 loss will be greatly facilitated by the development in a clinical laboratory of a highly sensitive and specific IHC assay.
Interestingly, BAP1 is mutated in the germline, where it predisposes to melanoma and mesothelioma28,29,50,51. Given the role of BAP1 in sporadic ccRCC, germline BAP1 mutations may similarly predispose to RCC. In fact, a germline variant (c.121G>A; p.Gly41Ser) was identified in one individual who had two first and one second degree relatives with RCC and who had been previously evaluated for a germline_VHL_ mutation, which he did not have. In addition, a recently reported pedigree had one individual with a germline BAP1 mutation who had RCC51. Thus, BAP1 mutation in the germline may predispose to RCC, in which case, RCC development may also be initiated by loss of BAP1.
Multiple lines of evidence implicate HCF-1 in BAP1-mediated RCC tumor suppression function. First, BAP1 binds to and cofractionates with HCF-1. Second, as determined by immunodepletion experiments, the majority of BAP1 is bound to HCF-1. HCF-1 is a very abundant protein52and this may explain why mutant BAP1 does not function as a dominant negative. Third, the growth inhibitory effect of BAP1 is compromised by a mutation that, while not disrupting protein structure (as determined by deubiquitinating activity), disrupts HCF-1 binding. Finally, the interaction with HCF-1 is unlikely to reflect an abnormal epigenetic state of tumor cell lines in culture, as BAP1 binds to and cofractionates with HCF-1 also in tumorgrafts. Tantalizingly however, the HCF-1 binding motif in BAP1 is not conserved in the_Drosophila_ Calypso protein.
The role of H2Aub1 in ccRCC requires further study. BAP1 binding to HCF-1 was required for the suppression of cell proliferation but dispensable for H2Aub1 deubiquitination. Thus, these two functions of BAP1, HCF-1 binding and H2Aub1 deubiquitination, can be separated. We did not find a correlation between BAP1 inactivation and global H2Aub1 levels in tumors. Nevertheless, the levels of H2Aub1 were not uniform across tumors and we cannot rule out that BAP1 may affect the levels of H2Aub1 at specific sites.
Our studies were greatly aided by the availability of tumorgrafts. Tumorgrafts were instrumental in determining mutant allele ratios with accuracy and for the identification of putative two-hit tumor suppressor genes. They made possible determining the co-occurrence of mutations in tumor cells and when mutations occurred in regions of amplification, they shed light on the temporal sequence of mutation acquisition. Finally, tumorgrafts provided a renewable source of tumor material allowing us to evaluate the significance of biochemical observations made in cell lines in culture.
While this manuscript was in preparation, a brief communication reported a list of 12 genes mutated in ccRCC53, including TSC1, which we previously showed to be mutated in sporadic ccRCC18, and BAP1. The mutation frequency reported for _BAP1_was 8%, but a VHL mutation frequency of 27% suggests low sensitivity.
ONLINE METHODS
Regulatory
Patients provided written informed consent of an Institutional Review Board (IRB)-approved protocol for tissue collection for genetic studies. Whole-genome and exome sequences were released in dbGaP for those patients giving explicit authorization in the consent form.
Annotation
Patient tumor samples are labeled with a number or a number preceded by a T if those samples were also used for tumorgraft generation. Tumorgrafts are labeled with the same number as patient tumors prefixed by “TG” and followed by the cohort “c” number (when applicable), referring to the tumor passage (e.g. c0 for primary tumorgraft).
Staging was based on the TNM classification from the American Joint Committee on Cancer. Samples were annotated according to the corresponding edition based on the date of surgery. Per the seventh edition, all tumors with lymph node metastases were referred to as pN1. All Fuhrman grade 3 and 4 samples were reviewed by P.K. for the presence of rhabdoid features.
Tissue selection
ccRCC and adjacent normal kidney samples were frozen fresh in liquid nitrogen and stored at −80°C. Tumor content and quality was inferred by a pathologist from perpendicular sections immediately flanking 1–3 mm thick fragments that were oriented using pathology dyes (Supplementary Fig. 2c). For whole-genome sequencing a sample was selected with ~90% tumor content in both sections. For the Discovery Set, 76 ccRCC samples with ≥80% tumor cellularity were selected among 431 fresh-frozen tumor samples from 133 patients. Seven tumor samples and a metastasis with ≥85% tumor cellularity were selected for exome sequencing among 16 patients with tumorgrafts growing in mice21. For the Validation Set, 92 ccRCC samples with ≥70% tumor cellularity were selected among 535 fresh-frozen tumor samples from 165 RCC patients. Genomic DNA and RNA were simultaneous extracted from each tissue (detailed in the Supplementary Note). Reference DNA, extracted from either adjacent normal kidney or peripheral blood mononuclear cells (PBMCs), was available for 71/76 tumors in the Discovery Set and 82/92 tumors of the Validation Set.
Whole-genome sequencing of paired-end libraries from tumor and matched normal genomes
Tumor and PBMCs samples were processed in a CLIA-certified and CAP-accredited laboratory. The preparation of short-insert (212–263 bp) Illumina paired-end sequencing libraries, flow cells and clusters have been described previously54. Paired-end sequence reads of 100 bases were generated using the Genome Analyzer IIx. Image analysis, base calling and Phred quality scoring were performed using the Illumina analysis pipeline (RTA, v1.5). Sequence reads were filtered out from clusters whose proximity to others resulted in mixed sequence data.
Whole-genome somatic substitution and indel detection
Single-nucleotide variants (SNVs) from the reference sequence (human NCBI36.1) were determined separately for the tumor and normal genomes using CASAVA, v1.6. Prediction of a homozygous SNV required a minimum allele score of 10 (equivalent to at least three high quality (Q33) base calls). Additionally, for heterozygous calls, the second allele was required to have a score of at least 6 (equivalent to two Q30 base calls) and the ratio of the two allele scores had to be ≤3, so that allele ratios did not deviate from the expected 1:1 for heterozygous calls. Indels relative to human NCBI36.1 were predicted using GROUPER. SNVs and indels in the tumor were only considered as candidate somatic events if the read depth at the equivalent site in the normal genome build was at least 10. SNVs and indels observed in both genomes were subtracted from the tumor calls. Previously known SNPs (dbSNP130) were also removed. Indels in the tumor overlapping a contig of assembled shadow reads in the normal genome, were removed. The impact of somatic changes on protein coding and non-coding genes was annotated using Ensembl version 54.
Exome capture and sequencing
Exome capture was performed by Illumina FastTrack using Illumina Truseq exome target enrichment. For more details, please see Supplementary Note.
Exome mutation detection and validation
The sequences of tumor, metastasis, and normal samples were compared to NCBI reference sequence and SNVs and indels were determined independently using CASAVA v1.8.0a4 without any filtering. Mutations predicted in tumors also present in the corresponding normal samples were eliminated (Supplementary Table 5). Synonymous mutations were removed and the resulting mutations were inspected visually using the Integrative Genomics Viewer (IGV, see URLs) to confirm that their presence in tumor but not normal sequence reads (see Supplementary Table 5). All genes with recurrent mutations were validated by Sanger sequencing (Supplementary Table 7). For the rest, a mutation calling accuracy of >95% for 82 mutations would signify >90% accuracy for the whole cohort, according to a cumulative hypergeometric distribution. Sanger sequencing of 82 randomly-selected mutations (proportional to the number of mutations in seven tumors and one metastasis) showed 4 false positives (MAR in tumor or metastasis <0.1 and MARTG<0.2; <5%), all of which had been scored based on two mutant reads (Supplementary Table 6). Among the remaining genes, those with just two mutant reads (3) were inferred to represent false positives (see Supplementary Data 2).
Mutation Analyses and Mutant Allele Ratios (MARs)
Single-nucleotide variants and indels in chromatograms were scored with Mutation Surveyor v3.30 and v3.98 (Softgenetics) using an overlapping factor of 0.2 and a dropping factor of 0.1. Reference sequences were obtained from NCBI. Only bidirectionally-observed somatic mutations are reported. Mutations within seven nucleotides before or after an exon were considered to be splice-site mutations. A somatically-acquired PBRM1 variant outside this range (15 nt upstream of exon 8) was not included in the analyses (sample ID 78).
Mutant allele ratios (MARs) refer to the fraction of mutant allele for a particular mutation over the sum of mutant plus wild-type alleles (MAR of 1, only mutant allele detected; MAR of 0.5, mutant and wild-type alelles detected at similar frequencies). MARs were calculated by measuring the nucleotide intensities of chromatograms using ImageJ (see URLs) (whole-genome data) or the Mutation Quantifier function of Mutation Surveyor v3.98 (exome data). For indels, MARs were calculated taking the average of the measurements of at least five nucleotides. MARs were scored as <0.10 if they accounted for <10% of all alleles, but were clearly present in tumorgrafts. For Illumina tracings, MARs were based on the number of mutant over total reads.
Copy Number Analyses
Genomic DNA was hybridized to Affymetrix SNP Arrays 6.0 at the Genome Science Resource (Vanderbilt University) using standard procedures. Several tumor and tumorgraft SNP arrays were previously evaluated for other purposes and have been reported elsewhere21. CEL files were quantile normalized with Partek Genomics Suite 6.5 (Partek Inc., St. Louis, MO) adjusting for fragment length and probe sequence without background correction. Paired copy numbers for tumors and tumorgrafts were calculated from the intensities of the corresponding normal samples. Genotypes were estimated using birdseed v2 algorithm in Affymetrix Genotyping Console 4.0. Regions of allelic imbalance were identified by determining the allele-specific copy number for the primary tumor or tumorgraft respect to normal DNA using Partek Genomics Suite. Copy numbers were adjusted for local GC content and were segmented using Circulary Binary Segmentation (CBS)55, where log2 ratios were analyzed with the _DNAcopy_package of R/Bioconductor (see URLs) considering a type I error (α=0.001) and a minimum segment size of 5 markers. Maximum and minimum allele-specific copy numbers were segmented independently by CBS.
Establishment and maintenance of tumorgrafts
Tumorgraft studies were approved by the IACUC. Fresh tumor fragments (~2 mm in diameter) were implanted in the kidney of NOD/SCID mice as described elsewhere21.
Gene Expression Analyses
RNA samples were labeled with biotin and hybridized to Affymetrix Human Genome U133 Plus 2.0 arrays by the UTSW Microarray Core. Gene expression arrays on 13 out of 29 tumors and tumorgrafts were previously evaluated to identify a tumor-specific signature and have been previously reported21. CEL intensity files were analyzed as described elsewhere56. Probesets with non-specific hybridization were removed (8,696, 16%). 2,443 probesets representing signal attributed to stromal/immune signature21were similarly removed. Tumors and tumorgrafts with mutations in either_BAP1_ or PBRM1 (but not both) were compared to tumors and tumorgrafts wild type for both BAP1 and_PBRM1_ using t tests and a Benjamini and Hochberg false discovery rate (FDR) correction57. Probesets with FDR_q_<0.05 were analyzed with Ingenuity Pathways Analysis (IPA).
Statistics
To determine whether a correlation (inverse correlation) existed between regional DNA copy numbers and MARTG (or MART), a two-tailed Spearman correlation test was utilized (data not normally distributed according to a Shapiro-Wilk test). Correlations were compared as previously described58. The_p_ values for the identification of 2 or 3 additional gene mutations among the 76 patients of the Discovery Set were calculated using a binomial distribution assuming, based on the index patient, that the probability of identifying a non-synonymous mutation in a gene was 0.0022 (47 mutations among the 21,099 protein-coding genes annotated in GRCh37.p6 assembly). For the Sanger Institute dataset9,10, the highest tumor grade was used for each tumor. Throughout, a Fisher's exact test was used to determine if there were nonrandom associations between two binary variables. A Benjamini and Hochberg FDR correction of the p values (q values) was calculated to account for multiple comparisons57. SPSS Statistics 17.0 and SAS 9.0 were used to analyze data.
Primer sequences and antibody information are provided in Supplementary Tables 9 and 10, respectively. Further details and description of other experimental methods and materials are available in the Supplementary Note.
Supplementary Material
Suppl. Data 1
Suppl. Data 2
Suppl. Data 3
Suppl. Data 4
Supp. Note, Figures, and Tables
ACKNOWLEDGEMENTS
We recognize the patients who participated in the study and donated samples. We thank Orlando Sepulveda, Aneesa Husain, and Amulya Yadlapalli for technical support, Shawn Cohenour and Diane Sheppard for contracts and regulatory assistance, Dr. Yuichi Machida (Mayo Clinic) for providing plasmids, Dr. Bart Grossman (MDACC) for providing the UMRC cells, Dr. Cristel Camacho and Dr. Nozomi Tomimatsu for irradiating cells, and the UT Southwestern tissue resource staff. This work was supported by a fellowship of excellence from Generalitat Valenciana (BPOSTDOC06/004) to S.P.-L., and the following grants to J.B.: Cancer Prevention and Research Institute of Texas (RP101075), a Clinical Scientist Development Award from the Doris Duke Charitable Foundation, American Cancer Society Research Scholar Grant (115739), and RO1CA129387. The tissue management shared resource is supported in part by NCI (1P30CA142543). J.B. is a Virginia Murchison Linthicum Scholar in Medical Research at UT Southwestern. The content is solely the responsibility of the authors and does not represent official views from any of the granting agencies.
Footnotes
ACCESSION NUMBERS
SNP and gene expression microarray data have been deposited in GEO (GSE25540 and GSE36895, respectively). Whole-genome and exome sequences for patients consenting to the deposit of their information are in dbGaP (phs000491).
AUTHOR CONTRIBUTIONS
S.P.-L. processed, managed, and extracted nucleic acids from tissues, evaluated and validated mutations, performed bioinformatic analyses on the exome data as well as copy-number, gene-expression, and statistical analyses. S.V.-R,-d,-C. was responsible for most biochemical studies using cell lines and tumorgrafts. A.L., T.H., S.J., and M.L. supervised the whole-genome sequencing process, performed quality control measures, and were responsible for the primary SNV analysis in the clinical lab. N.L. analyzed exome sequences under the supervision of C.D.H. A.P.-J. and P.S. helped with tissue processing and histology. S.W. helped with functional studies in UMRC6 cells. T.Y. assisted in mutation validation and mouse studies. L.Z. reviewed patient’s records. L.K. and N.G. performed in silico structural analyses for BAP1 and PBRM1. S.S. maintained the tumorgrafts and processed tissues. Y.L., V.M., and A.I.S. assisted with sample procurement. P.B.S. was the index patient’s genetic counselor. W.K. and P.K. evaluated the pathology slides and P.K. was responsible for the IHC assays. X-J.X. performed statistical analyses and revised statistics. S.W.W.W. performed the indel analysis. M.T.R. and D.R.B. supervised and managed the genome sequencing and annotation process. J.B. conceived the study, designed experiments, analyzed the data, and wrote the manuscript with input from S.P.-L. and other authors.
COMPETING FINANCIAL INTERESTS
None.
REFERENCES
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Baldewijns MM, et al. Genetics and epigenetics of renal cell cancer. Biochim Biophys Acta. 2008;1785:133–155. doi: 10.1016/j.bbcan.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Brugarolas J. Renal-cell carcinoma--molecular pathways and therapies. N Engl J Med. 2007;356:185–187. doi: 10.1056/NEJMe068263. [DOI] [PubMed] [Google Scholar]
- 4.Latif F, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 5.Gnarra JR, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 6.Nickerson ML, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res. 2008;14:4726–4734. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beroukhim R, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen M, et al. Genome-wide profiling of chromosomal alterations in renal cell carcinoma using high-density single nucleotide polymorphism arrays. Int J Cancer. 2009;125:2342–2348. doi: 10.1002/ijc.24642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuhrman SA, Lasky LC, Limas C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol. 1982;6:655–663. doi: 10.1097/00000478-198210000-00007. [DOI] [PubMed] [Google Scholar]
- 13.Bretheau D, et al. Prognostic value of nuclear grade of renal cell carcinoma. Cancer. 1995;76:2543–2549. doi: 10.1002/1097-0142(19951215)76:12<2543::aid-cncr2820761221>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 14.Ficarra V, et al. Prognostic and therapeutic impact of the histopathologic definition of parenchymal epithelial renal tumors. Eur Urol. 2010;58:655–668. doi: 10.1016/j.eururo.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Pantuck AJ, et al. Prognostic relevance of the mTOR pathway in renal cell carcinoma: implications for molecular patient selection for targeted therapy. Cancer. 2007;109:2257–2267. doi: 10.1002/cncr.22677. [DOI] [PubMed] [Google Scholar]
- 16.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 17.Gerlinger M, et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. New England Journal of Medicine. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kucejova B, et al. Interplay Between pVHL and mTORC1 Pathways in Clear-Cell Renal Cell Carcinoma. Mol Cancer Res. 2011;9:1255–1265. doi: 10.1158/1541-7786.MCR-11-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brugarolas J. Research Translation and Personalized Medicine. In: Figlin RA, Rathmell WK, Rini BI, editors. Renal Cell Carcinoma. New York: Springer; 2012. p. 161.p. 191. [Google Scholar]
- 20.Hahn SA, et al. Allelotype of pancreatic adenocarcinoma using xenograft enrichment. Cancer Res. 1995;55:4670–4675. [PubMed] [Google Scholar]
- 21.Sivanand S, et al. A Validated Tumorgraft Model Reveals Activity of Dovitinib Against Renal Cell Carcinoma. Science Translational Medicine. 2012;4:137ra75. doi: 10.1126/scitranslmed.3003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee ST, et al. Mutations of the P gene in oculocutaneous albinism, ocular albinism, and Prader-Willi syndrome plus albinism. N Engl J Med. 1994;330:529–534. doi: 10.1056/NEJM199402243300803. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki T, et al. Six novel P gene mutations and oculocutaneous albinism type 2 frequency in Japanese albino patients. J Invest Dermatol. 2003;120:781–783. doi: 10.1046/j.1523-1747.2003.12127.x. [DOI] [PubMed] [Google Scholar]
- 24.Gasparre G, Romeo G, Rugolo M, Porcelli AM. Learning from oncocytic tumors: Why choose inefficient mitochondria? Biochim Biophys Acta. 2011;1807:633–642. doi: 10.1016/j.bbabio.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 25.Jensen DE, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 26.Ventii KH, et al. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res. 2008;68:6953–6962. doi: 10.1158/0008-5472.CAN-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eletr ZM, Wilkinson KD. An emerging model for BAP1's role in regulating cell cycle progression. Cell Biochem Biophys. 2011;60:3–11. doi: 10.1007/s12013-011-9184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harbour JW, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiesner T, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43:1018–1021. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bott M, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43:668–672. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Misaghi S, et al. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol Cell Biol. 2009;29:2181–2192. doi: 10.1128/MCB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishikawa H, et al. BRCA1-associated protein 1 interferes with BRCA1/BARD1 RING heterodimer activity. Cancer Res. 2009;69:111–119. doi: 10.1158/0008-5472.CAN-08-3355. [DOI] [PubMed] [Google Scholar]
- 33.Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J Biol Chem. 2009;284:34179–34188. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheuermann JC, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu H, et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol Cell Biol. 2010;30:5071–5085. doi: 10.1128/MCB.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kristie TM, Liang Y, Vogel JL. Control of alpha-herpesvirus IE gene expression by HCF-1 coupled chromatin modification activities. Biochim Biophys Acta. 2010;1799:257–265. doi: 10.1016/j.bbagrm.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knez J, Piluso D, Bilan P, Capone JP. Host cell factor-1 and E2F4 interact via multiple determinants in each protein. Mol Cell Biochem. 2006;288:79–90. doi: 10.1007/s11010-006-9122-x. [DOI] [PubMed] [Google Scholar]
- 38.Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27:107–119. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 39.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yokoyama A, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narayanan A, Ruyechan WT, Kristie TM. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc Natl Acad Sci U S A. 2007;104:10835–10840. doi: 10.1073/pnas.0704351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med. 2009;15:1312–1317. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith ER, et al. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25:9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stokes MP, et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci U S A. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 46.Kaelin W., Jr . Molecular Biology of Clear Cell Renal Carcinoma. In: Figlin RA, Rathmell WK, Rini BI, editors. Renal Cell Carcinoma. New York: Springer; 2012. p. 27. [Google Scholar]
- 47.Xu X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chapman-Fredricks JR, et al. Adult renal cell carcinoma with rhabdoid morphology represents a neoplastic dedifferentiation analogous to sarcomatoid carcinoma. Ann Diagn Pathol. 2011;15:333–337. doi: 10.1016/j.anndiagpath.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 49.Gokden N, et al. Renal cell carcinoma with rhabdoid features. Am J Surg Pathol. 2000;24:1329–1338. doi: 10.1097/00000478-200010000-00002. [DOI] [PubMed] [Google Scholar]
- 50.Abdel-Rahman MH, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011;48:856–859. doi: 10.1136/jmedgenet-2011-100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Testa JR, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–1025. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wysocka J, Reilly PT, Herr W. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol Cell Biol. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo G, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat Genet. 2012;44:17–19. doi: 10.1038/ng.1014. [DOI] [PubMed] [Google Scholar]
- 54.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 56.Peña-Llopis S, et al. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 2011;30:3242–3258. doi: 10.1038/emboj.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 58.Steiger JH. Tests for Comparing Elements of a Correlation Matrix. Psychological Bulletin. 1980;87:245–251. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Data 1
Suppl. Data 2
Suppl. Data 3
Suppl. Data 4
Supp. Note, Figures, and Tables