Differentiated roles for MreB-actin isologues and autolytic enzymes in Bacillus subtilis morphogenesis (original) (raw)
Abstract
Cell morphogenesis in most bacteria is governed by spatiotemporal growth regulation of the peptidoglycan cell wall layer. Much is known about peptidoglycan synthesis but regulation of its turnover by hydrolytic enzymes is much less well understood. Bacillus subtilis has a multitude of such enzymes. Two of the best characterized are CwlO and LytE: cells lacking both enzymes have a lethal block in cell elongation. Here we show that activity of CwlO is regulated by an ABC transporter, FtsEX, which is required for cell elongation, unlike cell division as in Escherichia coli. Actin-like MreB proteins are thought to play a key role in orchestrating cell wall morphogenesis. B. subtilis has three MreB isologues with partially differentiated functions. We now show that the three MreB isologues have differential roles in regulation of the CwlO and LytE systems and that autolysins control different aspects of cell morphogenesis. The results add major autolytic activities to the growing list of functions controlled by MreB isologues in bacteria and provide new insights into the different specialized functions of essential cell wall autolysins.
Introduction
Most bacteria have an external wall that determines cell shape and is crucial for preventing the cell from bursting due to its high internal turgor pressure. The cell wall is also the target for our best antibiotics (particularly β-lactams), and fragments of the wall are recognized by the innate immune system. In virtually all bacteria the cell wall comprises a single huge sac-like molecule of peptidoglycan (PG, also called murein), which is a network of glycan strands cross-linked by peptide bridges. Gram-positive bacteria, such as Bacillus subtilis, have a multi-layered cell wall that also contains an additional major class of polymers called teichoic acids, which are anionic in nature and are covalently bound either to the PG (wall teichoic acids; WTA) or to membrane phospholipids (lipoteichoic acids; LTA) (Bhavsar and Brown, 2006; Carballido-Lopez and Formstone, 2007; den Blaauwen et al., 2008).
Rod-shaped bacteria enlarge the wall in at least two distinct ways. During growth, the cell elongates along its longitudinal axis, in a process that involves attachment of new glycan strands and cross-linking of peptide side-chains into the pre-existing structure. Elongation alternates with division, in which a plate of new wall material, the division septum, is formed, followed by separation of the daughter cells and maturation of new hemispherical cell poles (den Blaauwen et al., 2008; Haeusser and Levin, 2008; Bramkamp and van Baarle, 2009). Insertion of new PG into the wall requires the action of synthases called penicillin-binding proteins (PBPs) (Matsuhashi et al., 1990; Goffin and Ghuysen, 1998; Scheffers et al., 2004; Scheffers and Pinho, 2005). These enzymes catalyse glycan strand elongation (glycosyl transferase) and/or peptide cross-linking (peptidyl transferase) reactions. PG growth requires the action of hydrolytic enzymes (autolysins) that cleave bonds in the existing PG sacculus to enable surface expansion. These reactions need to be carefully co-ordinated, and PBPs are thought to act in multienzyme complexes, together with autolysins and (in Gram-positives) the enzymes for WTA and possibly LTA synthesis (Holtje, 1996a,b; Kawai et al., 2011; Typas et al., 2012). However, little is known about the precise composition and organization of these putative complexes.
Most bacteria have distantly related homologues of the two major eukaryotic cytoskeletal proteins: actin and tubulin, called MreB and FtsZ respectively. These proteins are key players in organization of the putative complexes involved in elongation (MreB) and division (FtsZ). The MreB proteins were originally thought to form elongated helical structures that spatially controlled the insertion of new wall material (Jones et al., 2001; Daniel and Errington, 2003; Carballido-Lopez and Errington, 2003a; Figge et al., 2004; Graumann, 2004; Carballido-Lopez et al., 2006; Takacs et al., 2010; White et al., 2010). Recently, new highly sensitive and higher resolution imaging methods have suggested that MreB proteins form smaller patches or short arcs that move over the cell surface driven by peptidoglycan synthesis (Dominguez-Escobar et al., 2011; Garner et al., 2011; Reimold et al., 2013). Precisely how MreB proteins regulate PG synthesis to achieve cylindrical cell wall elongation in rod-shaped bacteria remains to be resolved.
To complicate matters, B. subtilis and many other rod-shaped bacteria have multiple MreB isoforms. B. subtilis has three: MreB (in an operon with highly conserved MreC and MreD proteins), Mbl (‘MreB-like’) and MreBH (‘MreB homologue’) (Abhayawardhane and Stewart, 1995; Carballido-Lopez and Errington, 2003a; Defeu Soufo and Graumann, 2004; Carballido-Lopez et al., 2006). Mutations in each of the three genes affect cell shape in different ways. Deletion of all three genes is lethal but lethality can be suppressed, generating mutant cells that have a more or less spherical morphology (Schirner and Errington, 2009; Kawai et al., 2009b). All three proteins associate with each other and with multiple protein partners that have various functions in cell morphogenesis (Jones et al., 2001; Carballido-Lopez et al., 2006; Graumann, 2007; Claessen et al., 2008; Kawai et al., 2009a; 2011; Soufo and Graumann, 2010). It is thought that functional specialization of the three MreB proteins is due at least in part to differences in their spectrum of interacting partners (Carballido-Lopez et al., 2006; Kawai et al., 2011). Although the molecular details of these specializations remain poorly defined.
In previous work we reported that MreBH interacts with the autolysin LytE, and that this interaction is required to distribute LytE to the cylindrical part of the cell as well as at the division septum (Carballido-Lopez et al., 2006). LytE is one of about 35 putative B. subtilis autolysins, which can be grouped into 11 families (Smith et al., 2000). This multiplicity of genes supports the view that autolytic activity is an important cellular function but analysis of gene function is likely to be complicated by functional overlap or redundancy. Interestingly, Bisicchia et al. (2007) discovered that the endopeptidases LytE and CwlO have an overlapping essential role in cell elongation. The genes encoding these enzymes are among a small number of genes that are positively regulated by an essential two-component regulator of cell wall homeostasis, WalKR. The lytE and cwlO synthetic lethality seems to be caused by a lack of d/l-endopeptidase activity in the lateral cell wall, which in turn blocks cell elongation and provokes cell lysis (Hashimoto et al., 2012). While cwlO and lytE probably contribute to WalKR essentiality, it remains to be determined if they constitute the sole cause (Bisicchia et al., 2007).
Autolytic enzymes need to be tightly regulated, although little is presently known about the specific mechanisms involved. Recently, two groups described a novel mechanism of regulation in which ABC-transporter-like complexes regulate the activities of specific endopeptidases (Sham et al., 2011; Yang et al., 2011). The ABC transporter corresponds to a previously described cell division factor called FtsEX. It seems that the ATPase activity of the nucleotide-binding domain protein (FtsE) provokes a conformational change in the transmembrane component (FtsX) (Yang et al., 2012), which in turn activates the PG hydrolytic activity of the cognate autolysins: in Streptococcus pneumoniae this is a direct interaction with the putative PcsB autolysin (Sham et al., 2011); whereas in Escherichia coli activation works through an intermediate periplasmic protein called EnvC, and there are two regulated autolysins, AmiA and AmiB (Yang et al., 2011). In both cases activation of the CW hydrolase(s) at the septum is needed to enable the separation of progeny cells after division, explaining at least in part the deleterious effects of inactivation of the FtsEX complex in both microorganisms.
Previous work showed that the B. subtilis ftsEX genes are not essential for growth and pointed towards a role in regulation of the initiation of sporulation (Garti-Levi et al., 2008). Here we show that FtsEX regulates the activity of one of the major autolysins required for cell elongation in B. subtilis, CwlO, and that the mechanism of regulation is similar to that described for FtsEX proteins in other systems. We also demonstrate that FtsEX/CwlO function is controlled by the Mbl homologue. Differential regulation of LytE vs CwlO explains at least in part the different functional specializations of the MreB isologues. Furthermore, the different phenotypic effects arising from deletions in the lytE or cwlO genes suggest that these endopeptidases have differentiated roles in cell elongation and provide new insights into the control of cell morphogenesis. Another article in this issue (Meisner et al., 2013) describes an independent investigation that yielded results similar and complementary to those described here.
Results
ftsE/X deletions affect cell elongation rather than division in B. subtilis
Garti-Levi et al. (2008) previously showed that ftsEX mutants of B. subtilis are impaired in the initiation of sporulation. They also noted that, unlike the equivalent mutants of E. coli, B. subtilis ftsEX mutants are not significantly affected in cell division. Instead, the cells are slightly shorter and wider. We constructed various ftsE and ftsX mutants and examined their cell phenotype. As reported previously the mutants were indeed wider (cell diameter increased about 23%) and shorter (length reduced about 12%) (Table 1). Under some growth conditions, the normal cylindrical morphology was perturbed, with many cells having a twisted or undulating curved appearance (Fig. 1A). All of these morphological abnormalities were rescued by addition of 20 mM Mg2+ to the medium (Fig. 1A, right panels); a phenotype often observed in mutants with defective peptidoglycan synthesis in the lateral CW (Popham and Setlow, 1995; Murray et al., 1998; Formstone and Errington, 2005; Leaver and Errington, 2005; Carballido-Lopez et al., 2006). In addition to these specific morphological effects, the overall growth rate of ftsEX mutant cultures was reduced, especially in low Mg2+ medium (Fig. S1A). These results suggest that the main role of ftsEX lies in some aspect of cell envelope elongation during vegetative growth.
Table 1.
Cell length and width measurements
| Strain | Genotype | Average cell length (μm)a | % wt | Average cell width (μm)a | % wt |
|---|---|---|---|---|---|
| 168 | Wt | 3.6 ± 0.77 | – | 0.95 ± 0.081 | – |
| 4501 | Δ_ftsX_ | 3.2 ± 0.68 | −12 | 1.17 ± 0.085 | +23 |
| 4503 | Δ_ftsE_ | 3.2 ± 0.63 | −10 | 1.18 ± 0.084 | +24 |
| PDC463 | Δ_cwlO_ | 3.2 ± 0.69 | −11 | 1.18 ± 0.088 | +24 |
| PDC464 | Δ_lytE_ | 4.4 ± 0.97 | +22 | 0.86 ± 0.076 | −9 |
Fig. 1.
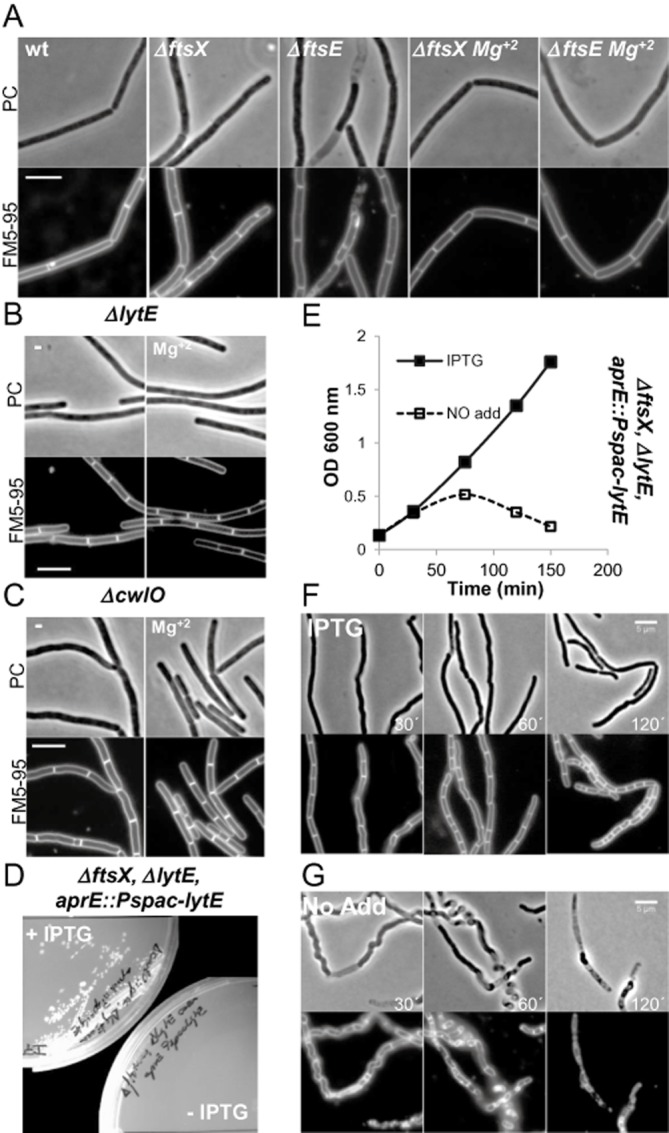
FtsEX mutants are similar to Δ_cwlO_ and synthetic lethal with Δ_lytE_.
A. Cell morphologies of typical fields of wild-type B. subtilis strain 168, Δ_ftsX_ ( 4501 ) and Δ_ftsE_ ( 4503 ) mutant strains growing in a NA plates or in NA plates with supplement of 20 mM Mg2+, as indicated. Scale bar represents 5 μm.
B and C. Cell morphologies of typical fields of strains PDC464 (Δ_lytE_::cat) and PDC463 (Δ_cwlO_ :: spec) cultured on NA plates in the presence or absence of Mg2+ as indicated. Scale bar represents 5 μm.
D. Growth of strain PDC492 (Δ_ftsX_ :: neo Δ_lytE_ :: cat aprE :: Pspac - LytE) on NA plates with or without 0.5 mM IPTG.
E. Growth of strain PDC492 on LB liquid medium in the presence or absence of IPTG. Growth curves (IPTG 0.5 mM, closed symbols; no addition, open symbols).
F and G. Effect of LytE depletion on cell morphology. Phase-contrast micrographs and the corresponding membrane staining images were taken at the indicated times during the growth curves in (E). (F) 0.5 mM IPTG added; (G) no IPTG addition. Scale bar represents 5 μm.
Bacterial two-hybrid analysis
To explore whether the lateral wall localization and elongation phenotype was reflected in interactions of FtsE and FtsX with components of the cell wall elongation system, full-length copies of ftsE and ftsX were cloned into bacterial two-hybrid vectors (Karimova et al., 1998) and screened for interactions against a collection of other proteins. Figure 2A shows that FtsE and FtsX interact with each other but unlike many other cell elongation and division proteins (Wang et al., 1997; van den Ent et al., 2006; Pichoff et al., 2012), no self-interactions were evident. The positive interactions detected for FtsE and FtsX showed that the constructs are at least partially active.
Fig. 2.
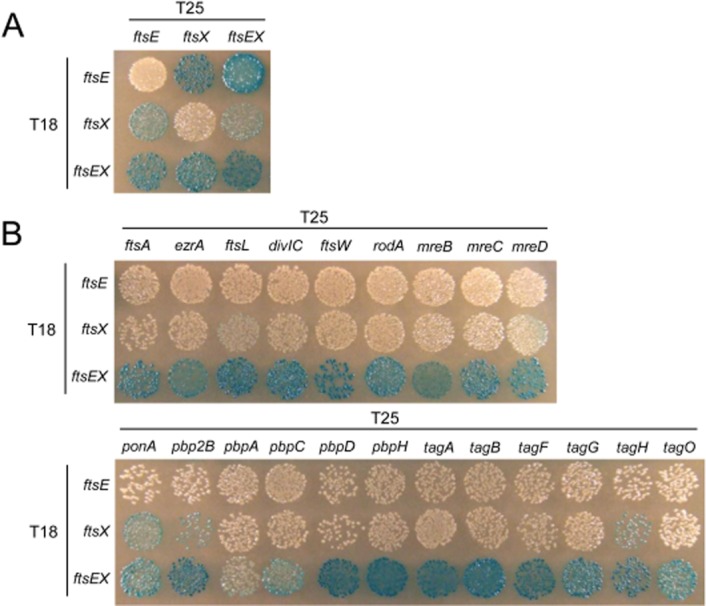
Bacterial two-hybrid analysis of FtsEX protein interactions.
A. Bacterial two-hybrid analysis of interaction between FtsE and FtsX.
B. Bacterial two-hybrid analysis of interaction with FtsE and FtsX.
Escherichia coli strain BTH101 was co-transformed with two-hybrid vector plasmids (pUT18 and pKT25) expressing C-terminal fusions of the cyaA T18 domain to ftsE, ftsX and ftsEX, and N-terminal fusions of cyaA T25 domain to various genes, as indicated. Transformants were spotted onto nutrient agar plates containing X-Gal and incubated at 30°C for 40 h. Blue colouration indicates a positive interaction.
The two-hybrid constructs were then tested for interaction with various other cell wall-associated proteins (Fig. 2B). FtsE showed no strong interactions other than with FtsX. FtsX interacted significantly with three proteins tested: Pbp1a (PonA), Pbp2B and TagH. Interestingly, TagH is the ATP-binding component of the ABC transporter thought to be responsible for export of wall teichoic acid (Lazarevic and Karamata, 1995). Strikingly, when FtsE and FtsX were coexpressed on the same plasmid, interactions with many proteins involved in cell wall synthesis, cell elongation and cell division became apparent (Fig. 3B), suggesting that FtsE and FtsX need to come together before interacting efficiently with partner proteins. The interacting proteins belong to several general classes including enzymes involved in peptidoglycan (PBP) or teichoic acid (Tag) synthesis, as well as proteins implicated in elongation but with other, sometimes unknown, functions (e.g. MreCD). Several other division or cell elongation proteins, and control non-wall-associated proteins were tested, showing no evident interaction with FtsEX: FtsZ, SepF, DivIVA, PbpE, PbpX, MinC, Noc, Soj and Spo0J (not shown). The relative promiscuity of interactions for proteins involved in cell wall-associated functions has been observed previously (Mohammadi et al., 2007; Claessen et al., 2008). More work is needed to determine the extent to which these interactions occur with the native proteins in B. subtilis cells.
Fig. 3.
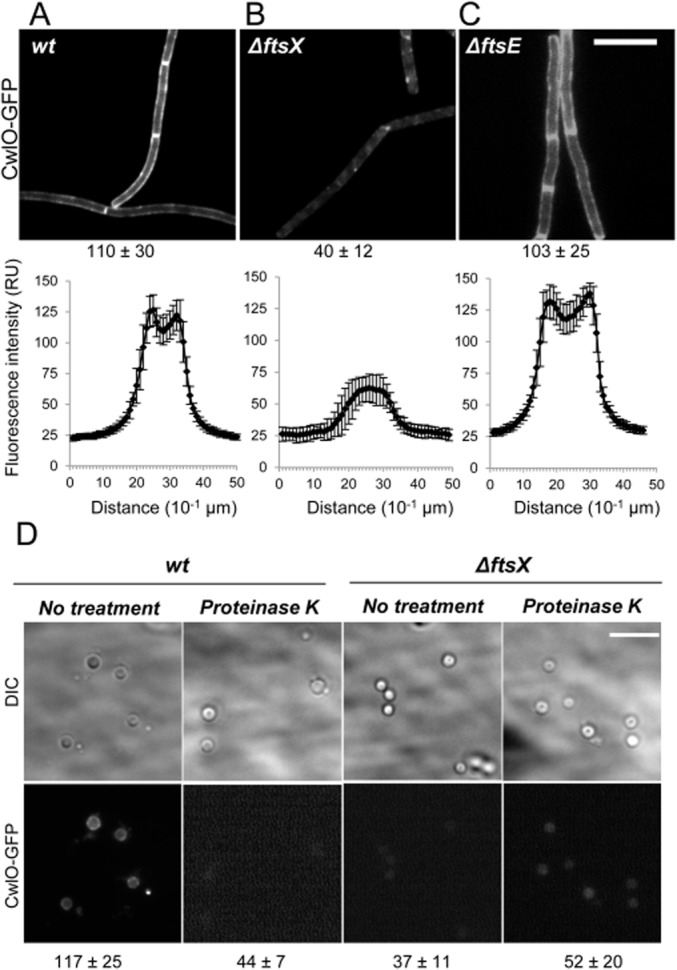
CwlO localizes at the cell membrane in an FtsX-dependent manner.
A–C. Epifluorescence microscopy of strains expressing the fluorescent fusion amyE :: Pxyl - cwlO - gfpsf. The different panels correspond to (A) strain PDC528 (Bs168CA wprA :: hyg, epr :: tet amyE :: Pxyl - cwlO - gfpsf) and isogenic strains (B) PDC560 (Δ_ftsX_), (C) PDC594 (Δ_ftsE_), as indicated. Scale bar represents 5 μm. Fluorescent images were taken with the same acquisition settings and exposure times, with the maximum averaged value of quantified fluorescence intensity over the lateral wall of the cells (see Experimental procedures) indicated below. Lower panels: Profiles of fluorescence intensity corresponding to strains in (A)–(C) respectively. Averaged fluorescence intensity quantified in segments of equal size across the cell's longitudinal axis. The _y_-axis represents fluorescence intensity (relative units, RU), while the _x_-axis represents distance (10−1 μm). Error bars represent standard deviation of fluorescent intensity measurements.
D. Cells of strains PDC528 (wt, left panels) and PDC560 (Δ_ftsX_ :: neo, right panels) were grown in CH media in the presence of 0.5% xylose and protoplasted (see Experimental procedures). Images show the relative fluorescence intensity from CwlO–GFPsf protoplasts treated with proteinase K or not, as indicated. Maximum averaged value of quantified fluorescence intensities across the cells are shown below together with calculated standard deviations. Scale bar: 5 μm.
cwlO and ftsE/X null mutants have similar cell elongation phenotypes and both are synthetic lethal with lytE
CwlO is the closest homologue of the FtsEX-regulated autolysins in both E. coli and S. pneumoniae (Table S1). lytE mutants have a different phenotype from that of ftsEX, having a reduced, rather than increased cell diameter (Carballido-Lopez et al., 2006; Fig. 1B; Table 1). The cell morphology of cwlO mutants has not been described in detail previously. Interestingly, under our growth conditions, cwlO cells had a similar phenotype to that of ftsEX, with short, wide and slightly undulating cells, and again this phenotype was improved by addition of 20 mM Mg2+ to the growth media (Fig. 1C; Table 1).
Bisicchia et al. (2007) previously showed that cwlO mutations have a synthetic lethal cell elongation phenotype when combined with lytE. We confirmed this result (Fig. S2A–D). To further investigate the possible connection of FtsEX with the autolytic enzymes during cell elongation we tested the effects of combining an ftsX deletion with null mutations in cwlO or lytE. A double-deletion mutant of ftsX and cwlO was readily constructed and did not differ in growth or morphology to either of the two single mutants (Fig. S1B). In contrast, attempts to combine lytE and ftsEX mutations were unsuccessful. We therefore generated a conditional mutant for lytE and introduced an ftsX deletion in the presence of inducer (IPTG dependent, PDC492). These cells grew in the presence but not absence of inducer (Fig. 1E). The LytE-depleted cell culture revealed that the cell chains became highly twisted and underwent extensive cell lysis (Fig. 1F and G). These phenotypic effects were similar to those of a cwlO deletion mutant in which lytE was depleted (Bisicchia et al., 2007), consistent with the notion that FtsEX is required for CwlO activity.
FtsX but not FtsE is required for CwlO localization at the lateral cell wall
To test whether FtsEX determines the localization of CwlO in B. subtilis we expressed a CwlO–GFPsf fusion in wt and ftsE or ftsX deletion strains. This took advantage of a superfolding variant of GFP (GFPsf) previously shown to be fluorescent after Sec-mediated transport (Dinh and Bernhardt, 2011). This protein was at least partially functional, because as the only copy of cwlO in cells, it was able to support growth in a lytE deletion strain. CwlO and LytE are both susceptible to degradation by extracellular proteases, WprA and Epr (Yamamoto et al., 2003; Yamaguchi et al., 2004; Hashimoto et al., 2012). Accordingly, we found that the GFP signal for CwlO was considerably enhanced by visualization in a wprA epr mutant background (Fig. S3B). Similarly to the results obtained by Hashimoto et al. (2012) (based on immunofluorescence), CwlO–GFPsf was associated with the cell periphery, along the lateral cell wall, as well as at septa and cell poles (Fig. 3A). Interestingly, in a Δ_ftsX_ background, the GFP fluorescence intensity was low and appeared mainly distributed throughout the cytoplasm, rather than at the cell periphery (Fig. 3B). In contrast, localization in a Δ_ftsE_ mutant strain was associated with the cell periphery. It should be noted that Δ_ftsE_ mutant cells were wider than those of the wild type (Fig. 3C). Fluorescence intensity measurements across typical cells (≥ 50) were plotted (Fig. 3A–C, lower panels) and these supported the peripheral localization in wild-type and Δ_ftsE_ mutant cells, and the lack of CwlO recruitment to the cell envelope in the Δ_ftsX_ mutant.
We then examined the localization of CwlO–GFPsf after stripping the cell wall to produce protoplasts. As for intact cells, the fluorescence signal was associated with the membrane in wt cells but not in the Δ_ftsX_ mutant (Fig. 3D). Treatment of protoplasts with proteinase K eliminated the surface-associated CwlO–GFPsf fluorescence signal in wild-type protoplasts, whereas in the Δ_ftsX_ mutant the weak cytoplasmic signal was unaffected (Fig. 3D, right panels). These experiments support the view that CwlO is exported independently of FtsEX, presumably via its classical _sec_-dependent signal peptide, and then retained on the outside surface of the cytoplasmic membrane, by a mechanism requiring FtsX.
CwlO interacts with FtsX at the cell membrane
To investigate the role of FtsEX in CwlO function, we determined the subcellular localization of CwlO (see Supporting information) in the presence or absence of FtsEX. Cultures of wt, Δ_ftsX_ and Δ_ftsE_ strains were grown at 37°C. At mid-exponential phase, the cultures supernatants' (S) were collected and proteins precipitated by cold-acetone treatment; the culture's pellets were converted to protoplasts by incubation with lysozyme. The protoplasts were collected by centrifugation, leaving a supernatant that constituted the cell wall (W) fraction. Cell membranes (M) and cytoplasmic fractions (C) were obtained from the protoplast pellets (see Experimental procedures). The untreated half of the culture pellet constituted the total (T) fraction. In wt and Δ_ftsE_ cells, CwlO, was detected in both the membrane and cell wall fractions (Fig. 4A). However, in Δ_ftsX_ mutant cells, the CwlO signal was absent from the membrane fraction and reduced in the wall fraction (Fig. 4A). As controls, we examined the distribution of a well-characterized cytosolic protein Soj (Fig. 4A), and an integral membrane protein Pbp2B (Fig. 4A), which were indeed detected predominantly in the cytoplasmic or membrane fractions respectively. When we analysed the presence of CwlO in the supernatant fraction (Fig. 4B), the protein was detected in all three backgrounds in large amounts.
Fig. 4.
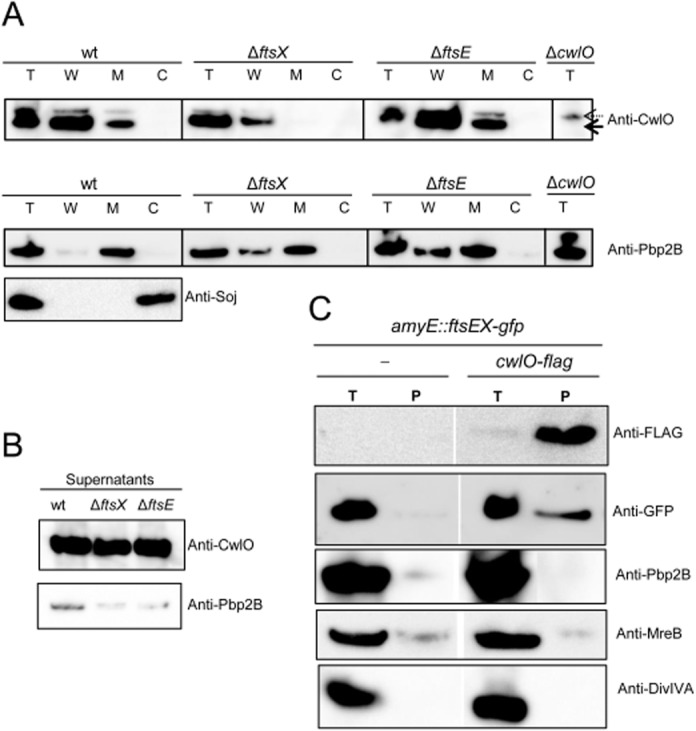
CwlO and FtsX form part of a protein complex in the cell membrane.
A. Reduction in CwlO membrane fraction levels in the absence of FtsX. Fractionation of wt, Δ_ftsE_ and Δ_ftsX_ mutants. Total fraction from Δ_cwlO_ mutant strain was analysed to discard unspecific bands. Solid black arrow indicates CwlO band after Western blotting. Dashed arrow indicates unspecific band. Lower panels correspond to cell fractionation controls using polyclonal antibodies against Pbp2B and Soj membrane and cytosolic proteins respectively.
B. CwlO is detected predominantly in the supernatant fraction.
C. Pull-down of CwlO-FLAG complexes in membranes of exponentially growing cells treated with formaldehyde (described in Experimental procedures). Bands on the Western blots were detected with anti-FLAG, anti-GFP, anti-Pbp2B, anti-MreB and anti-DivIVA antibodies, as indicated. Left lanes correspond to the control strain PDC528 were CwlO remains untagged, while expresses an FtsX–GFP fusion. Right lanes correspond to the strain PDC612 that coexpresses CwlO–FLAG and FtsX–GFP fusion proteins. T, total-cell extract prior cross-linking; P, heated pull-down fraction; the experiment was performed three times with similar results.
To further investigate the interaction between FtsEX and CwlO we used chemical cross-linking followed by pull-down of FLAG-tagged CwlO, then tested for the presence of FtsX protein (Fig. 4C). As expected CwlO-FLAG was only detected in samples containing the cwlO-flag construct. In these samples a single band corresponding to the expected molecular weight (53 KDa) was detected in cross-linked samples after heating (Fig. 4C). In non-heated samples a very faint band was sometimes detected with similar mobility, but no other bands corresponding to high-molecular-weight complexes were detected in the Western blots. Silver staining did reveal a prominent complex with a mass > 100 KDa (Fig. S4). Importantly, anti-GFP anti-serum detected the FtsX–GFP fusion protein in the pull-down samples, but only in the presence of CwlO-FLAG (Fig. 4C). Three other tested cytosolic or membrane-associated proteins, Pbp2B, MreB and DivIVA, were not detected in the pull-down samples. These results suggest that CwlO associates, directly or indirectly, with the membrane component of the ABC transporter, FtsX.
Differential roles of MreB isoforms in control of autolytic activity
We previously reported that LytE interacts with MreBH and that its localization in the lateral cell wall is at least partly dependent on this interaction (Carballido-Lopez et al., 2006). We wondered whether one or other of the MreB isologues control CwlO localization or activity. We recently showed that strains containing only one of the three homologues can be obtained, provided that the remaining protein is overproduced. Availability of these strains provided a means of testing whether the roles of the MreB homologues are differentiated in respect of the control of CwlO or LytE activities. We therefore introduced conditional mutations of the ftsEX or lytE genes into strains expressing single mreB isologues.
The most striking results were obtained for the Mbl-only strain. When a conditional allele of lytE was introduced, the Mbl-only strain showed virtually normal growth, independent of lytE expression (Fig. 5H and K). This result shows that a strain with just Mbl can presumably support the activity of the FtsEX/CwlO system, because this is essential when LytE is not present. In contrast, when ftsEX was depleted in this strain (Fig. 5B and E) growth was abolished and extensive cell lysis occurred. This suggests that Mbl is not able to support efficiently the activity of LytE during cell elongation, and is therefore specific for FtsEX/CwlO.
Fig. 5.
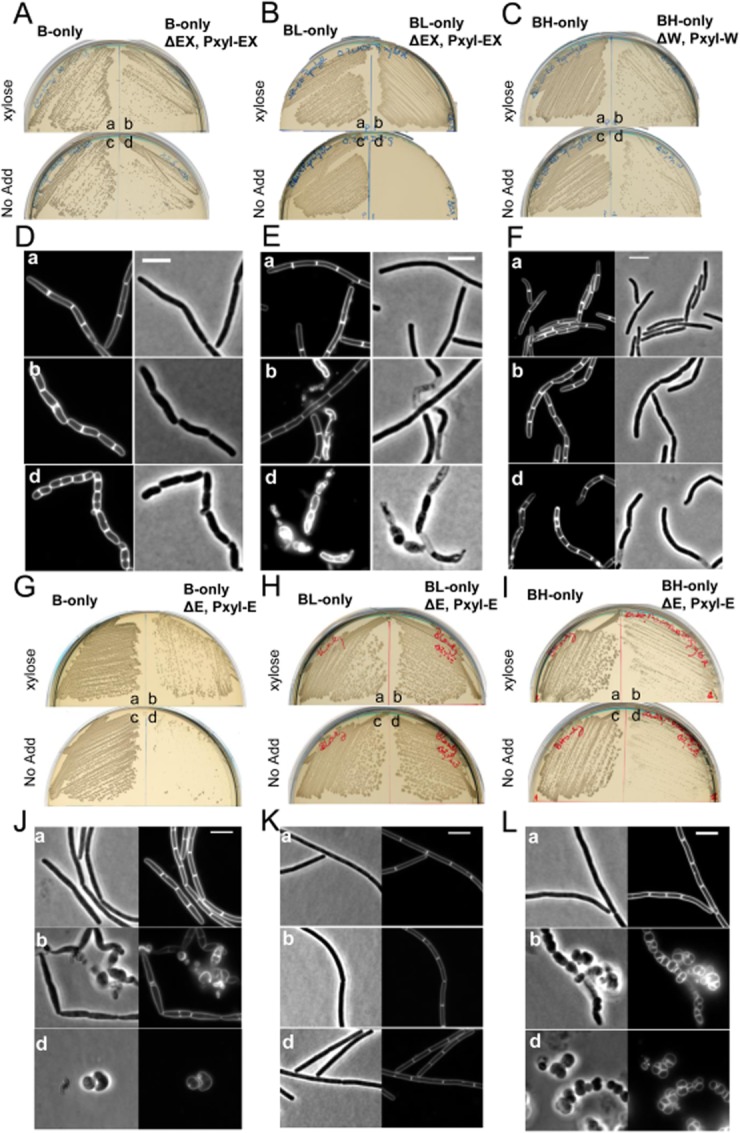
CwlO activity depends on the Mbl actin homologue.
A–C. Growth of strains B-only, BL-only and BH-only (a/c), respectively, and its derivatives PDC664, PDC643 and PDC659 (b/d) on NA 20 mM Mg2+, supplemented with appropriate concentration of IPTG in each case (Kawai et al., 2009a), in the presence (a–b) or absence of xylose (c–d), as indicated.
D–F. Cell morphologies of typical fields of strains in (A)–(C) respectively. Phase-contrast and membrane-stained fluorescent images (FM5-95) of parental strains (a) and its derivatives in the presence (b) or absence (d) of xylose for 3 h at 37°C.
G–I. Growth of strains B-only, BL-only and BH-only (a/c), respectively, and its derivatives (Δ_lytE_ :: spec, aprE :: Pxyl - lytE) PDC688, PDC678 and PDC697 (b/d) on NA 20 mM Mg2+, supplemented with appropriate concentration of IPTG in each case (Kawai et al., 2009a), in the presence (a–b) or absence of xylose (c–d).
J–L. Cell morphologies of typical fields of strains in (G)–(I) respectively. Phase-contrast and membrane-stained fluorescent images (FM5-95) of parental strains (a) and its derivatives in the presence (b) or absence (d) of xylose for 3 h at 37°C.
Strikingly, the results for the MreB- and MreBH-only strains were, in both cases, reciprocal to those obtained for the Mbl-only strain: thus, cell growth and morphology was much more affected in the absence of lytE (Fig. 5G, I, J and L) than ftsEX (Fig. 5A, C, D and F).
We also examined the effects of combining lytE, cwlO or ftsEX mutations with single deletions of each mreB homologue. In general, the results were consistent with those described above (Fig. S5), although they were less clear cut, as expected, because each of these strains still contains two mreB isologues. Nevertheless, it was clear that the effects of an mbl mutation were greatly exacerbated by lytE mutation but not significantly by cwlO or ftsEX mutation, whereas mreB or mreBH mutants tended to show the opposite effect. All of the data support the idea that Mbl is crucial for functioning of the FtsEX/CwlO system, whereas MreB and MreBH are more important for LytE action.
Discussion
FtsEX regulates the CwlO autolysin required for cell elongation in B. subtilis
Previous work on the FtsEX systems of E. coli and S. pneumoniae have demonstrated an unexpected role for the FtsEX ABC-transporter-like protein complex in regulation of specific cell wall autolytic enzymes (Sham et al., 2011; Yang et al., 2011). Our work establishes that FtsEX of B. subtilis has an analogous role, albeit that this system seems to be involved mainly or exclusively with the cell elongation system, rather than cell division. Various phenotypic and functional properties of the FtsEX system, reported here and previously (Garti-Levi et al., 2008) are consistent with a role in elongation: the mutants are shorter and wider than wild-type cells; the phenotype is rescued by high Mg2+ concentrations; the proteins localize in the lateral wall; and the cognate autolysin, CwlO, also seems to be involved in cell elongation.
The similarity of phenotypes of ftsEX and cwlO mutants is also consistent with FtsEX regulating CwlO activity, as is the synthetic lethality of ftsEX with lytE, just as for cwlO and lytE (Bisicchia et al., 2007; Hashimoto et al., 2012). Biochemical experiments also support the notion that FtsEX regulates CwlO: first, we found that CwlO is sequestered to the external surface of the cytoplasmic membrane by a mechanism requiring FtsX; second, protein interactions were detected by both cross-linking and pull-down and cell fractionation experiments. The results lead to a model in which FtsEX regulates CwlO activity by a direct protein–protein interaction (Fig. 6A), similar to those described previously for other organisms (Sham et al., 2011; Yang et al., 2011).
Fig. 6.
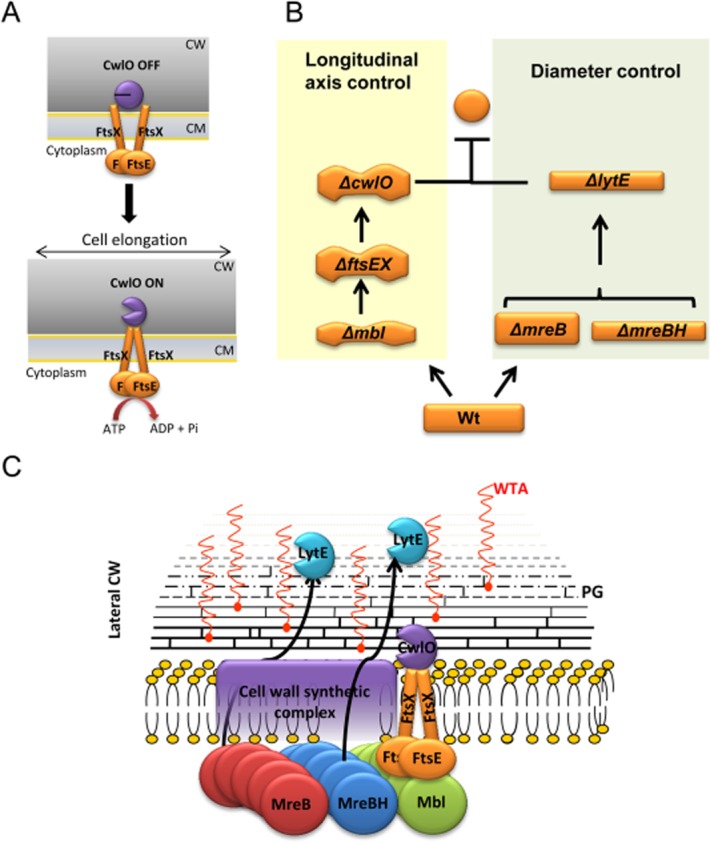
Model for actin cytoskeleton function in co-ordination of CW hydrolytic activities CwlO and LytE during cell elongation.
A. Schematic representation of the FtsEX ATPase cycle during activation of the CwlO CW hydrolase activity in B. subtilis. FtsEX is shown in the model as a tetramer, although other stoichiometries are possible on the basis of the two-hybrid data.
B. Two distinct pathways for CW hydrolytic activity at the lateral cell wall in B. subtilis.
C. Co-ordination by the actin cytoskeleton to ensure the balance between cell wall synthesis and hydrolysis during cell elongation. See text for further details.
Specialized functional roles for MreB isologues in regulation of cell wall autolytic activities
Bacterial actin homologues of the MreB family are thought to orchestrate the synthesis and assembly of the cell wall in most rod-shaped bacteria. The proteins are present in all major lineages of bacteria, as are the genes for cell wall synthesis, suggesting that both the wall and its regulation by MreB were present in bacteria very early in evolution (Trachtenberg, 1998; Carballido-Lopez and Errington, 2003b; Margolin, 2009). Many bacteria have multiple MreB isologues and B. subtilis has three (Jones et al., 2001; Carballido-Lopez et al., 2006). The isologues are thought to have overlapping but differentiated functions, but molecular details of how these functional specializations are achieved remain poorly understood.
Initial attempts to examine the possible involvement of MreB proteins in the FtsEX system were inconclusive because single mutants for any one isologue have two remaining isologues with overlapping and/or complementary functions. However, by taking advantage of recently developed strains containing single MreB isologues (Kawai et al., 2009a) more incisive results were obtained. In particular, it was evident that a strain containing only Mbl (but not MreB- or MreBH-only) grew normally in the presence of the FtsEX/CwlO system. Depletion of its expression resulted in massive cell lysis. In contrast, depletion of lytE showed no phenotypic effect in the Mbl-only strain. This strongly suggests that Mbl can support functioning of the FtsEX/CwlO system but that it cannot support LytE function. The MreB- and MreBH-only strains did not have such clear-cut phenotypes but, in both cases, the results were complementary to the ones obtained with the Mbl-only strain. They tolerated loss of FtsEX or CwlO well, but were extremely sick in the absence of LytE. These results suggest that the MreB homologues are highly differentiated in respect of the control they exert over the FtsEX/CwlO and LytE autolytic systems, with Mbl specialized for regulation of the former and MreB and MreBH for the latter (Fig. 6B).
These results additionally show that the MreB proteins are pivotally involved in the regulation of autolytic activity, in addition to their previously demonstrated roles in control of peptidoglycan and wall teichoic acid synthesis (Formstone et al., 2008; Yamamoto et al., 2008; Kawai et al., 2009b; 2011). They also provide the clearest example so far of how control of different specific enzymes is the likely explanation for the presence of multiple MreB isologues in many bacteria.
Differentiated roles for the CwlO and LytE autolysins in different aspects of cell morphogenesis
It is now well recognized that most bacteria have multiple autolytic enzymes and the presumption is that they are involved in different aspects of cell wall synthesis or remodelling. However, in only a few cases are the specific roles for the autolysins understood. Recent work in B. subtilis and E. coli has shown that the autolysins involved in cell elongation of both organisms are partially redundant, with two proteins of B. subtilis, LytE and CwlO (Hashimoto et al., 2012), and three of E. coli, Spr, YdhO and YebA (Singh et al., 2012) having overlapping functions. Lethal phenotypes are revealed only when both or all of the redundant autolysin genes are deleted (Bisicchia et al., 2007; Singh et al., 2012). Our results provide important new insights into the specialized roles of LytE and CwlO. LytE function is complicated by the fact that it is involved not only in elongation but also in cell separation (Yamamoto et al., 2003; Fukushima et al., 2006). We previously reported that lytE mutants, in addition to forming slightly longer chains of cells, are also slightly thinner than wild-type cells. The basis for this thinning is not clear, although it is interesting that this phenotype is shared by other cell elongation mutants, including ponA (encoding a major peptidoglycan synthase; Murray et al., 1998; Kawai et al., 2009b) and mreBH (which has some role in regulation of LytE localization; Carballido-Lopez et al., 2006). In this article we report that cwlO mutants are different (but similar to ftsEX mutants) in being wider than the wild type as well as being slightly bent or undulated (Fig. 6B). These distinct phenotypes suggest that the proteins have differentiated roles in cell morphogenesis during growth. Irrespective of how they work, one or other activity must be present or, as shown previously and herein, cells stop elongating altogether (Bisicchia et al., 2007; Fig. 1D–G).
The model shown in Fig. 6C illustrates one important difference between the likely roles of CwlO and LytE. Because CwlO is activated by the membrane protein complex FtsEX, its activity is probably restricted to the inner part of the thick (∼ 50 nm) Gram-positive cell wall. Furthermore, requirement for Mbl function suggests that CwlO activity might also be tightly co-ordinated with that of the various peptidoglycan and teichoic acid synthases. Thus, CwlO might contribute intimately to the insertion of newly synthesized wall material, along with the many other proteins associated with the MreB system. In contrast, LytE is probably regulated by some other mechanism. Traditionally, it has been assumed that in Gram-positive bacteria, the multi-layered wall material matures as it migrates outwards, progressively underlain by newly inserted material. The outer layer(s) is thought to be stretched and load bearing, requiring autolytic activity to enable cell growth (Holtje, 1996b; Holtje, 1998; Hayhurst et al., 2008). Although LytE insertion into the wall is probably again co-ordinated with that of wall synthesis by interaction with MreBH and possibly MreB, it has a wall-binding domain that could facilitate its migration outwards during wall maturation. Also, LytE synthesis is regulated in response to various stresses (Bisicchia et al., 2007; Schirner and Errington, 2009; Salzberg et al., 2013). Thus, we envisage the LytE function more as a stress response factor that is synthesized and or recruited when wall expansion is compromised.
We suggest that when CwlO is the major elongation autolysin (lytE mutant), PG synthesis is well co-ordinated with turnover and the normal pattern of growth is achieved, albeit that the cells are narrower than normal. In contrast, when LytE is the only major autolysin, regulated growth and turnover is impaired but LytE activity enables growth albeit in a relatively disordered manner, leading to loss of control over cell width and ability to maintain a consistent longitudinal axis of growth. The differentiation of these two cell phenotypes is reminiscent of the different phenotypes generated by mutations affecting the major MreB isologues, MreB and Mbl (Fig. 6B). As reported some years ago, mbl mutants have a distinctive highly twisted phenotype, whereas mreB mutants tend to maintain control over longitudinal growth, albeit becoming much wider than the wild type (Abhayawardhane and Stewart, 1995; Jones et al., 2001). It now appears that differences in the control of autolytic activity might contribute significantly to these distinct phenotypes. mreB mutants retain Mbl function and therefore FtsEX/CwlO activity, and thereby are still able to regulate PG synthesis and turnover in a highly co-ordinated way, whereas mbl mutants rely on the LytE system which is less tightly co-ordinated with the synthetic machinery. Our results therefore shed new light on the differentiated roles of MreB isologues in cell morphogenesis and about the specialized roles of two major autolytic proteins in the regulation of cell shape.
Experimental procedures
Complete details of all the experimental procedures used are provided in Supporting information.
Bacterial strains and plasmids, and primers
The bacterial strains, plasmids and oligonucleotides sequences used in this study are listed in Tables S2–S4 respectively. All B. subtilis strains used in the reported experiments are derivatives of Bs168CA. The construction of plasmids is described in Supporting information.
Growth conditions and media
Nutrient agar (NA, Oxoid) was used for routine selection and maintenance of both B. subtilis and E. coli strains. For B. subtilis, cells were grown in Luria–Bertani (LB), CH or SMM defined minimal medium (Anagnostopoulos & Spizizen) containing 0.5% xylose or 1 mM IPTG when required, unless stated otherwise. For E. coli, cells were grown in LB medium. Supplements and antibiotics were added as required: 20 μg ml−1 tryptophan, 100 μg ml−1 ampicillin, 5 μg ml−1 chloramphenicol, 5 μg ml−1 kanamycin, 50 μg ml−1 spectinomycin, 0.75 μg ml−1 erythromycin and 10 μg ml−1 tetracycline.
Microscopic imaging
For fluorescence microscopy, cells were grown to mid-exponential phase at 30°C or 37°C and mounted on microscope slides covered with a thin film of 1.2% agarose. See figure legends for specific growth conditions employed for each experiment. Fluorescence microscopy was carried out using Zeiss Axiovert 200M, Nikon Eclipse Ti-U, spinning disk confocal microscope. The images were acquired with Metamorph 6 (Molecular Devices) and FRAP-AI 7 (MAG Biosystems) software, and analysed using ImageJ v.1.44o (National Institutes of Health). When required, cells were incubated in the presence of the membrane dye FM5-95 (90 μg ml−1, Molecular Probes) prior to microscopic examination.
Sample preparation for microscopy
For sample preparation, overnight pre-cultures of B. subtilis were grown in CH medium supplemented with 20 mM MgSO4 (CH-Mg) and appropriate antibiotic selection, from freshly isolated colonies on plates. Day cultures were performed by diluting pre-culture to an OD600 of 0.02 in CH-Mg and grown at 30°C. Expression of fluorescent CwlO–GFPsf fusion was induced by addition of 0.3% xylose. Samples for microscopic observation were taken at mid-exponential phase and immobilized on 1.2% agarose-coated microscope slides.
Protoplast preparation for microscopy
Cells of strains PDC528 (wt, CwlO–GFPsf) and PDC560 (Δ_ftsX::neo_, CwlO–GFPsf) were grown in CH media in the presence of 0.5% xylose. Cells were harvested and resuspended in CH-MSM media in the presence of 0.5% xylose. Cells were protoplasted by incubation with 0.5 mg ml−1 lysozyme during 30 min at 30°C. After CW digestion, the protoplasts suspensions were split in two. One half was treated with proteinase K (10 μg ml−1) for 30 min.
Two-hybrid analysis
To screen for interactions of FtsX and FtsE with various proteins involved in cell wall synthesis or cell division, the ftsX, ftsE and ftsEX coding sequences were amplified by PCR and cloned into two bacterial two-hybrid vectors (pUT18 and pKT25), resulting in a C-terminal fusion of the cyaA (adenylate cyclase) T18 domain, or an N-terminal fusion of the cyaA T25 domain respectively (Karimova et al., 1998). In addition, the coding sequences of ftsA, ezrA, ftsL, divIC, ftsW, rodA, mreB, mreC, mreD, ponA, pbp2B, pbpA, pbpC, pbpD, pbpH, tagA, tagB, tagF, tagG, tagH, tagO were amplified by PCR and cloned into pKT25 generating N-terminal fusions of the T25 domain. Finally, to test the putative interaction between the FtsX extracytoplasmic loop1 and the CW hydrolase CwlO, plasmid pairs encoding the FtsX extracytoplasmic loop1 and either the full-length CwlO or the N-terminal coil-coiled domain coding sequences were co-transformed into BTH101 (cya-99). Co-transformants were spotted onto nutrient agar or minimal media plates, as indicated, containing ampicillin (100 μg ml−1), kanamycin (25 μg ml−1) and 0.004% X-gal. Pictures were taken after 40–72 h of growth at 30°C. Under these conditions, control transformations with empty vectors remained white for up to 72 h of incubation.
Cell fractionation and immunoblotting
Bacillus subtilis cell cultures were grown in LB at 37°C. When cells reached mid-exponential phase, cultures (100 ml) were collected by centrifugation (8000 g for 10 min at 25°C). Culture supernatants' protein content (S) was recovered by cold-acetone precipitation. Pellets were resuspended in 4 ml 1× SMM buffer (0.5 M sucrose, 20 mM MgCl2, 20 mM maleic acid, pH 7); 250 μl 10 mg lysozyme ml−1 (Sigma) and 50 μl complete protease inhibitor (EDTA-free, Roche) were added to cell suspensions and incubated at 37°C for 1 h with gentle shaking. Then cultures were split into two (2 × 2 ml). First half constituted the total fraction (T). The second half was used to obtain the cell wall (CW), membrane (M) and cytoplasmic (C) fractions, as described in detail within Supporting information. Ten micrograms of total protein from each extract were separated by SDS-PAGE on a 4–12% gradient gel (Novex, Life technologies). Separated proteins were analysed by Western blotting as described in detail in Supporting information.
Formaldehyde cross-linking and pull-down of CwlO complexes
Cross-linking and pull-down experiments were performed with some modifications as described by Sham et al. (2011). Briefly, cultures (400 ml) of strains PDC612 (Bs168CA Δ_wprA::hyg_ Δ_epr::tet ΩcwlO-FLAG amyE::Pxyl-ftsEX-gfp_) and PDC613 (Bs168CA Δ_wprA::hyg_ Δ_epr::tet amyE::Pxyl-ftsEX-gfp_ parent negative control) were grown exponentially to OD600 of 0.5. Cells were collected by centrifugation (8000 g for 10 min at 25°C). Cell pellets were washed with 18 ml 1× PBS at 25°C, and cells were collected again by centrifugation. Residual supernatants were removed. Washed pellets were suspended in 19 ml 1× PBS, to which 1200 μl 37% of formaldehyde solution (Sigma) were added. Mixtures were incubated at 37°C for 1 h. Cross-linking reactions were quenched by the addition of 4 ml 1.0 M glycine followed by incubation for 10 min at 25°C. Cells were collected by centrifugation, washed with 20 ml 1× PBS at 25°C and centrifuged again. Pull-down of CwlO-FLAG complexes was performed using an anti-FLAG M2 affinity gel, as described previously (Sham et al., 2011). Identification of protein content in the different fractions was carried out by Western blotting with appropriate anti-sera. A full protocol description can be found within Supporting information.
Acknowledgments
This work was supported by grant BB/G015902/1 from the UK Biotechnology and Biological Sciences Research Council to J.E. and R.A.D. We thank the various group members for helpful discussions, and particularly to Waldemar Vollmer for critical reading of the manuscript. We also thank Yoshikazu Kawai, Robyn Emmins, Heath Murray and Kevin Devine for the gift of strains. We especially thank K. Devine and David Noone for the generous gift of CwlO antibody. We thank David Z. Rudner, Thomas G. Bernhardt and co-workers for communicating their independent discovery of FtsEX function prior to publication, and an anonymous reviewer for very helpful criticisms.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web-site.
References
- Abhayawardhane Y, Stewart GC. Bacillus subtilis possesses a second determinant with extensive sequence similarity to the Escherichia coli mreB morphogene. J Bacteriol. 1995;177:765–773. doi: 10.1128/jb.177.3.765-773.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar AP, Brown ED. Cell wall assembly in Bacillus subtilis: how spirals and spaces challenge paradigms. Mol Microbiol. 2006;60:1077–1090. doi: 10.1111/j.1365-2958.2006.05169.x. [DOI] [PubMed] [Google Scholar]
- Bisicchia P, Noone D, Lioliou E, Howell A, Quigley S, Jensen T, et al. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol Microbiol. 2007;65:180–200. doi: 10.1111/j.1365-2958.2007.05782.x. [DOI] [PubMed] [Google Scholar]
- den Blaauwen T, de Pedro MA, Nguyen-Disteche M, Ayala JA. Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev. 2008;32:321–344. doi: 10.1111/j.1574-6976.2007.00090.x. [DOI] [PubMed] [Google Scholar]
- Bramkamp M, van Baarle S. Division site selection in rod-shaped bacteria. Curr Opin Microbiol. 2009;12:683–688. doi: 10.1016/j.mib.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Carballido-Lopez R, Errington J. The bacterial cytoskeleton: in vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev Cell. 2003a;4:19–28. doi: 10.1016/s1534-5807(02)00403-3. [DOI] [PubMed] [Google Scholar]
- Carballido-Lopez R, Errington J. A dynamic bacterial cytoskeleton. Trends Cell Biol. 2003b;13:577–583. doi: 10.1016/j.tcb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Carballido-Lopez R, Formstone A. Shape determination in Bacillus subtilis. Curr Opin Microbiol. 2007;10:611–616. doi: 10.1016/j.mib.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Carballido-Lopez R, Formstone A, Li Y, Ehrlich SD, Noirot P, Errington J. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell. 2006;11:399–409. doi: 10.1016/j.devcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Claessen D, Emmins R, Hamoen LW, Daniel RA, Errington J, Edwards DH. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol Microbiol. 2008;68:1029–1046. doi: 10.1111/j.1365-2958.2008.06210.x. [DOI] [PubMed] [Google Scholar]
- Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- Defeu Soufo HJ, Graumann PL. Dynamic movement of actin-like proteins within bacterial cells. EMBO Rep. 2004;5:789–794. doi: 10.1038/sj.embor.7400209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh T, Bernhardt TG. Using superfolder green fluorescent protein for periplasmic protein localization studies. J Bacteriol. 2011;193:4984–4987. doi: 10.1128/JB.00315-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, Carballido-Lopez R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Leaver M, Bendezu F, Errington J, de Boer P, Lowe J. Dimeric structure of the cell shape protein MreC and its functional implications. Mol Microbiol. 2006;62:1631–1642. doi: 10.1111/j.1365-2958.2006.05485.x. [DOI] [PubMed] [Google Scholar]
- Figge RM, Divakaruni AV, Gober JW. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol. 2004;51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- Formstone A, Errington J. A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol Microbiol. 2005;55:1646–1657. doi: 10.1111/j.1365-2958.2005.04506.x. [DOI] [PubMed] [Google Scholar]
- Formstone A, Carballido-Lopez R, Noirot P, Errington J, Scheffers DJ. Localization and interactions of teichoic acid synthetic enzymes in Bacillus subtilis. J Bacteriol. 2008;190:1812–1821. doi: 10.1128/JB.01394-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima T, Afkham A, Kurosawa S, Tanabe T, Yamamoto H, Sekiguchi J. A new D,L-endopeptidase gene product, YojL (renamed CwlS), plays a role in cell separation with LytE and LytF in Bacillus subtilis. J Bacteriol. 2006;188:5541–5550. doi: 10.1128/JB.00188-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garti-Levi S, Hazan R, Kain J, Fujita M, Ben-Yehuda S. The FtsEX ABC transporter directs cellular differentiation in Bacillus subtilis. Mol Microbiol. 2008;69:1018–1028. doi: 10.1111/j.1365-2958.2008.06340.x. [DOI] [PubMed] [Google Scholar]
- Goffin C, Ghuysen JM. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann PL. Cytoskeletal elements in bacteria. Curr Opin Microbiol. 2004;7:565–571. doi: 10.1016/j.mib.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Graumann PL. Cytoskeletal elements in bacteria. Annu Rev Microbiol. 2007;61:589–618. doi: 10.1146/annurev.micro.61.080706.093236. [DOI] [PubMed] [Google Scholar]
- Haeusser DP, Levin PA. The great divide: coordinating cell cycle events during bacterial growth and division. Curr Opin Microbiol. 2008;11:94–99. doi: 10.1016/j.mib.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Ooiwa S, Sekiguchi J. Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of D,L-endopeptidase activity at the lateral cell wall. J Bacteriol. 2012;194:796–803. doi: 10.1128/JB.05569-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayhurst EJ, Kailas L, Hobbs JK, Foster SJ. Cell wall peptidoglycan architecture in Bacillus subtilis. Proc Natl Acad Sci USA. 2008;105:14603–14608. doi: 10.1073/pnas.0804138105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtje JV. A hypothetical holoenzyme involved in the replication of the murein sacculus of Escherichia coli. Microbiology. 1996a;142(Part 8):1911–1918. doi: 10.1099/13500872-142-8-1911. [DOI] [PubMed] [Google Scholar]
- Holtje JV. Molecular interplay of murein synthases and murein hydrolases in Escherichia coli. Microb Drug Resist. 1996b;2:99–103. doi: 10.1089/mdr.1996.2.99. [DOI] [PubMed] [Google Scholar]
- Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y, Asai K, Errington J. Partial functional redundancy of MreB isoforms, MreB, Mbl and MreBH, in cell morphogenesis of Bacillus subtilis. Mol Microbiol. 2009a;73:719–731. doi: 10.1111/j.1365-2958.2009.06805.x. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Daniel RA, Errington J. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol Microbiol. 2009b;71:1131–1144. doi: 10.1111/j.1365-2958.2009.06601.x. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Marles-Wright J, Cleverley RM, Emmins R, Ishikawa S, Kuwano M, et al. A widespread family of bacterial cell wall assembly proteins. EMBO J. 2011;30:4931–4941. doi: 10.1038/emboj.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V, Karamata D. The tagGH operon of Bacillus subtilis 168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids. Mol Microbiol. 1995;16:345–355. doi: 10.1111/j.1365-2958.1995.tb02306.x. [DOI] [PubMed] [Google Scholar]
- Leaver M, Errington J. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol Microbiol. 2005;57:1196–1209. doi: 10.1111/j.1365-2958.2005.04736.x. [DOI] [PubMed] [Google Scholar]
- Margolin W. Sculpting the bacterial cell. Curr Biol. 2009;19:R812–R822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuhashi M, Wachi M, Ishino F. Machinery for cell growth and division: penicillin-binding proteins and other proteins. Res Microbiol. 1990;141:89–103. doi: 10.1016/0923-2508(90)90101-u. [DOI] [PubMed] [Google Scholar]
- Meisner J, Montero Llopis P, Sham LT, Garner E, Bernhardt TG, Rudner DZ. FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol Microbiol. 2013;89:1069–1083. doi: 10.1111/mmi.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi T, Karczmarek A, Crouvoisier M, Bouhss A, Mengin-Lecreulx D, den Blaauwen T. The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol Microbiol. 2007;65:1106–1121. doi: 10.1111/j.1365-2958.2007.05851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray T, Popham DL, Setlow P. Bacillus subtilis cells lacking penicillin-binding protein 1 require increased levels of divalent cations for growth. J Bacteriol. 1998;180:4555–4563. doi: 10.1128/jb.180.17.4555-4563.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Shen B, Sullivan B, Lutkenhaus J. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA's self-interaction competes with its ability to recruit downstream division proteins. Mol Microbiol. 2012;83:151–167. doi: 10.1111/j.1365-2958.2011.07923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham DL, Setlow P. Cloning, nucleotide sequence, and mutagenesis of the Bacillus subtilis ponA operon, which codes for penicillin-binding protein (PBP) 1 and a PBP-related factor. J Bacteriol. 1995;177:326–335. doi: 10.1128/jb.177.2.326-335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold C, Defeu Soufo HJ, Dempwolff F, Graumann PL. Motion of variable-length MreB filaments at the bacterial cell membrane influences cell morphology. Mol Biol Cell. 2013 doi: 10.1091/mbc.E12-10-0728. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg LI, Powell L, Hokamp K, Botella E, Noone D, Devine KM. The WalRK (YycFG) and sigma(I) RsgI regulators cooperate to control CwlO and LytE expression in exponentially growing and stressed Bacillus subtilis cells. Mol Microbiol. 2013;87:180–195. doi: 10.1111/mmi.12092. [DOI] [PubMed] [Google Scholar]
- Scheffers DJ, Pinho MG. Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev. 2005;69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers DJ, Jones LJ, Errington J. Several distinct localization patterns for penicillin-binding proteins in Bacillus subtilis. Mol Microbiol. 2004;51:749–764. doi: 10.1046/j.1365-2958.2003.03854.x. [DOI] [PubMed] [Google Scholar]
- Schirner K, Errington J. The cell wall regulator {sigma}I specifically suppresses the lethal phenotype of mbl mutants in Bacillus subtilis. J Bacteriol. 2009;191:1404–1413. doi: 10.1128/JB.01497-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham LT, Barendt SM, Kopecky KE, Winkler ME. Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. Proc Natl Acad Sci USA. 2011;108:E1061–E1069. doi: 10.1073/pnas.1108323108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, SaiSree L, Amrutha RN, Reddy M. Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol Microbiol. 2012;86:1036–1051. doi: 10.1111/mmi.12058. [DOI] [PubMed] [Google Scholar]
- Smith TJ, Blackman SA, Foster SJ. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology. 2000;146(Part 2):249–262. doi: 10.1099/00221287-146-2-249. [DOI] [PubMed] [Google Scholar]
- Soufo HJ, Graumann PL. Bacillus subtilis MreB paralogues have different filament architectures and lead to shape remodelling of a heterologous cell system. Mol Microbiol. 2010;78:1145–1158. doi: 10.1111/j.1365-2958.2010.07395.x. [DOI] [PubMed] [Google Scholar]
- Takacs CN, Poggio S, Charbon G, Pucheault M, Vollmer W, Jacobs-Wagner C. MreB drives de novo rod morphogenesis in Caulobacter crescentus via remodeling of the cell wall. J Bacteriol. 2010;192:1671–1684. doi: 10.1128/JB.01311-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtenberg S. Mollicutes-wall-less bacteria with internal cytoskeletons. J Struct Biol. 1998;124:244–256. doi: 10.1006/jsbi.1998.4063. [DOI] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Huang J, Mukherjee A, Cao C, Lutkenhaus J. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J Bacteriol. 1997;179:5551–5559. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CL, Kitich A, Gober JW. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol Microbiol. 2010;76:616–633. doi: 10.1111/j.1365-2958.2010.07108.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Furuhata K, Fukushima T, Yamamoto H, Sekiguchi J. Characterization of a new Bacillus subtilis peptidoglycan hydrolase gene, yvcE (named cwlO), and the enzymatic properties of its encoded protein. J Biosci Bioeng. 2004;98:174–181. doi: 10.1016/S1389-1723(04)00262-2. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Kurosawa S, Sekiguchi J. Localization of the vegetative cell wall hydrolases LytC, LytE, and LytF on the Bacillus subtilis cell surface and stability of these enzymes to cell wall-bound or extracellular proteases. J Bacteriol. 2003;185:6666–6677. doi: 10.1128/JB.185.22.6666-6677.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Miyake Y, Hisaoka M, Kurosawa S, Sekiguchi J. The major and minor wall teichoic acids prevent the sidewall localization of vegetative DL-endopeptidase LytF in Bacillus subtilis. Mol Microbiol. 2008;70:297–310. doi: 10.1111/j.1365-2958.2008.06397.x. [DOI] [PubMed] [Google Scholar]
- Yang DC, Peters NT, Parzych KR, Uehara T, Markovski M, Bernhardt TG. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc Natl Acad Sci USA. 2011;108:E1052–E1060. doi: 10.1073/pnas.1107780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DC, Tan K, Joachimiak A, Bernhardt TG. A conformational switch controls cell wall-remodelling enzymes required for bacterial cell division. Mol Microbiol. 2012;85:768–781. doi: 10.1111/j.1365-2958.2012.08138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.