The meddling microbes midst our medicines (original) (raw)
Abstract
It is not surprising that the complex metabolic machinery of the gut microbiome has accidental, or directed, ability to alter our medicines and influence their efficacy. What is not known is the extent to which this has contributed to drug failures or contraindications, or to the derailment of clinical trials. Some studies are unraveling the mechanisms by which the microbiota alter specific drugs, such as digoxin, and contribute to variations in efficacies between patient populations. Microbiome profiling, therefore, may well become an inevitable arm of all clinical trials in the future.
Keywords: digoxin, Eggerthella lenta, microbiota, medicines
A recent New York Times article titled “Do clinical trials work?” discussed the failure of the angiogenesis inhibitor Avastin in a randomized double-blind, placebo-controlled trial involving 600 brain cancer patients.1 The article went on to state that our uncertainty about such drugs “is the norm, not the exception,” attesting the distinctive physiology and pathology of human subjects, and the unique pharmacodynamics of compounds in different individuals. To improve the odds for successful phase III clinical trials, companies now conduct tests to identify and enroll smaller sub-populations with the appropriate “genetic or molecular” attributes, the likely responders. The other arm of the uncertainty, as detailed by a number of studies, is likely the influence of the gut microbiota on various drugs.
The same month, a study by Haiser et al. detailed the mechanism by which the gut bacterium Eggerthella lenta modifies the cardiac drug digoxin, and renders it inactive (Fig. 1).2 Digoxin, a cardiac glycoside derived from the foxglove plant, is used for the treatment of heart conditions including atrial fibrillations and atrial flutter. The narrow therapeutic range of digoxin (0.5–2.0 ng/ml) and related glycosides, and the risk of toxicity, limit their usefulness. In a subset of patients, cardiac glycosides are substantially converted to reduction products, such as dihydrodigoxin and dihydrodigoxigenin (“digoxin reduction products;” DRPs), that have decreased affinity for the target, the α-subunit of sarcolemmal Na+K+-ATPase.
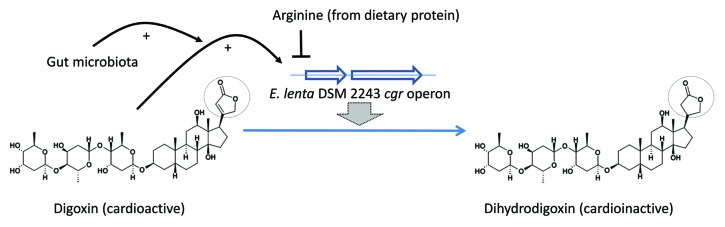
Figure 1. Digoxin is reduced to the cardioinactive dihydrodigoxin by E. lenta strains harboring the cgr operon. Digoxin induces cgr expression, and normal gut microbiota potentiate this induction; dietary protein, and arginine, inhibit cgr expression and digoxin reduction.
As early as 1969, Herman and Repke speculated that the conversion was mediated by gastrointestinal microorganisms, and this was confirmed by Lindenbaum et al. in 1981.3,4 In their study, volunteers orally administered digoxin, compared with subjects receiving the drug intravenously, had greater urinary DRP levels.4 Further, stool cultures from subjects that had increased urinary DRP (DRP excretors) converted digoxin to DRP in vitro, while those from non-excretors did not. Antibiotic administration to the excretors doubled their serum digoxin concentrations, and decreased urinary DRP levels.
Follow-up studies from the same group identified the gram-positive, non-sporulating, gut anaerobe Eggerthella lenta (then known as Eubacterium lentum) as the sole organism responsible for digoxin reduction, and implicated a role for arginine in regulating the growth of the organism and its ability to reduce digoxin.5 Their studies also noted that the presence of the organism was not an absolute predictor for digoxin conversion. Other studies by the same group showed geographic differences in digoxin inactivation, with DRP production being observed in 13–14% of healthy volunteers from South India and Bangladesh, compared with 35–36% of subjects from New York.6,7 Further, greater numbers of urban subjects in India were converters, compared with rural subjects.
Haiser et al. confirmed digoxin reduction by E. lenta DSM2243, and its inhibition by arginine.4 Using RNA-seq, they identified a two-gene operon that was highly upregulated by digoxin, as well as by related compounds with an α,β-unsaturated butyrolactone ring. The two genes, cgrA and cgrB, encode proteins with homology to bacterial cytochromes, and may utilize digoxin as an alternate electron acceptor. Digoxin-induced cgr expression was inhibited by arginine.
While a direct enzymatic role for CgrAB in the conversion of digoxin was not established, three related strains of E. lenta that lacked the cgr locus failed to reduce digoxin. Further, ex vivo digoxin reduction and qPCR assays on stool samples from 20 unrelated healthy subjects showed a positive correlation between high reducers and the presence of the cgr operon. E. lenta DSM2243 mono-association in germ-free mice induced cgr expression, and digoxin administration resulted in decreased serum and urine digoxin compared with animals mono-associated with a _cgr-_negative strain; a high protein diet, likely via increased arginine levels, increased serum digoxin levels in DSM2243-administered animals. Thus, the presence of _cgr_-positive E. lenta in the stool is predictive of increased digoxin inactivation, and the degree of digoxin reduction can be influenced by diet and microbiota.
One can well imagine a frisson of indignation from E. lenta in response to the name given to the two gene operon (cgr for cardiac glycoside reductase); the bacterium likely has a completely different purpose for these genes, matters not of the heart. Haiser et al. showed that digoxin itself failed to support E. lenta growth in vitro, and hypothesized that digoxin reduction was an “off-target” response. In its ability to induce the cgr operons, does digoxin mimic some other secondary metabolite of E. lenta? Could such compounds play a role in the physiology of E. lenta carriers, or can such products be modified for therapeutic purposes? The related actinomycete Streptomyces spa. produces γ-butyrolactones for quorum-sensing, although, unlike digoxin, these compounds have a completely reduced lactone ring (akin to the reduced digoxin compounds that failed to induce cgr expression); however, an unsaturated lactone ring is present in some of the biosynthetic intermediates.
Several review articles discuss microbiota-mediated transformation of various drugs, from acetaminophen to sulfasalazine, and make the case for a “metagenomic basis for therapeutics.”8,9 Thus, our distinct gut populations likely herald an inevitable future of personalized medicine.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Footnotes
References
- 1.Leaf C. Do Clinical trials work? The New York Times (Online Ed.). 2013 July 13; http://www.nytimes.com/2013/07/14/opinion/sunday/do-clinical-trials-work.html?pagewanted=all&_r=0
- 2.Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–8. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herrmann I, Repke K. Transformation of cardenolides by microorganisms in the intestine. In: Schubert K, editor. Proceedings of the 2nd Symposium Über Biochemische Aspekte der Steroidforschung; 1968; Berlin, Germany. Berlin: Akademie-Verlag; 1969. p. 115–119. [Google Scholar]
- 4.Lindenbaum J, Rund DG, Butler VP, Jr., Tse-Eng D, Saha JR. Inactivation of digoxin by the gut flora: reversal by antibiotic therapy. N Engl J Med. 1981;305:789–94. doi: 10.1056/NEJM198110013051403. [DOI] [PubMed] [Google Scholar]
- 5.Saha JR, Butler VP, Jr., Neu HC, Lindenbaum J. Digoxin-inactivating bacteria: identification in human gut flora. Science. 1983;220:325–7. doi: 10.1126/science.6836275. [DOI] [PubMed] [Google Scholar]
- 6.Alam AN, Saha JR, Dobkin JF, Lindenbaum J. Interethnic variation in the metabolic inactivation of digoxin by the gut flora. Gastroenterology. 1988;95:117–23. doi: 10.1016/0016-5085(88)90299-5. [DOI] [PubMed] [Google Scholar]
- 7.Mathan VI, Wiederman J, Dobkin JF, Lindenbaum J. Geographic differences in digoxin inactivation, a metabolic activity of the human anaerobic gut flora. Gut. 1989;30:971–7. doi: 10.1136/gut.30.7.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson ID. Drugs, bugs, and personalized medicine: pharmacometabonomics enters the ring. Proc Natl Acad Sci U S A. 2009;106:14187–8. doi: 10.1073/pnas.0907721106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haiser HJ, Turnbaugh PJ. Is it time for a metagenomic basis of therapeutics? Science. 2012;336:1253–5. doi: 10.1126/science.1224396. [DOI] [PubMed] [Google Scholar]