Transient accumulation of Okazaki fragments as a result of uracil incorporation into nascent DNA (original) (raw)
Abstract
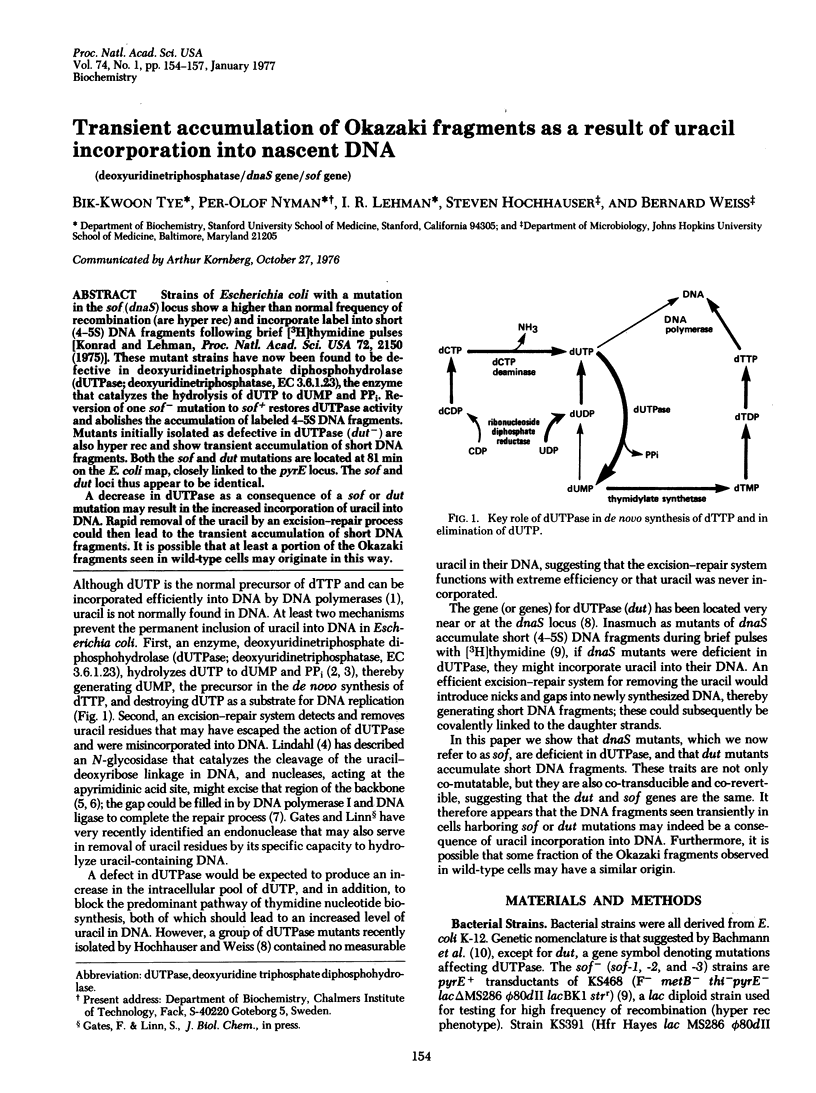
Strains of Escherichia coli with a mutation in the sof (dnaS) locus show a higher than normal frequency of recombination (are hyper rec) and incorporate label into short (4-5S) DNA fragments following brief [3H]thymidine pulses [Konrad and Lehman, Proc. Natl. Acad. Sci. USA 72, 2150 (1975)]. These mutant strains have now been found to be defective in deoxyuridinetriphosphate diphosphohydrolase (dUTPase; deoxyuridinetriphosphatase, EC 3.6.1.23), the enzyme that catalyzes the hydrolysis of dUTP to dUMP and PPi. Reversion of one sof- mutation to sof+ restores dUTPase activity and abolishes the accumulation of labeled 4-5S DNA fragments. Mutants initially isolated as defective in dUTPase (dut-) are also hyper rec and show transient accumulation of short DNA fragments. Both the sof and dut mutations are located at 81 min on the E. coli map, closely linked to the pyrE locus. The sof and dut loci thus appear to be identical. A decrease in dUTPase as a consequence of a sof or dut mutation may result in the increased incorporation of uracil into DNA. Rapid removal of the uracil by an excision-repair process could then lead to the transient accumulation of short DNA fragments. It is possible that at least a portion of the Okazaki fragments seen in wild-type cells may originate in this way.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BERTANI L. E., HAEGGMARK A., REICHARD P. ENZYMATIC SYNTHESIS OF DEOXYRIBONUCLEOTIDES. II. FORMATION AND INTERCONVERSION OF DEOXYURIDINE PHOSPHATES. J Biol Chem. 1963 Oct;238:3407–3413. [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessman M. J., Lehman I. R., Adler J., Zimmerman S. B., Simms E. S., Kornberg A. ENZYMATIC SYNTHESIS OF DEOXYRIBONUCLEIC ACID. III. THE INCORPORATION OF PYRIMIDINE AND PURINE ANALOGUES INTO DEOXYRIBONUCLEIC ACID. Proc Natl Acad Sci U S A. 1958 Jul 15;44(7):633–640. doi: 10.1073/pnas.44.7.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GREENBERG G. R., SOMERVILLE R. L. Deoxyuridylate kinase activity and deoxyuridinetriphosphatase in Escherichia coli. Proc Natl Acad Sci U S A. 1962 Feb;48:247–257. doi: 10.1073/pnas.48.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadi S. M., Goldthwait D. A. Endonuclease II of Escherichia coli. Degradation of partially depurinated deoxyribonucleic acid. Biochemistry. 1971 Dec 21;10(26):4986–4993. doi: 10.1021/bi00802a024. [DOI] [PubMed] [Google Scholar]
- Konrad E. B., Lehman I. R. Novel mutants of Escherichia coli that accumulate very small DNA replicative intermediates. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2150–2154. doi: 10.1073/pnas.72.6.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk M., Kinross J. Conditional lethality of recA and recB derivatives of a strain of Escherichia coli K-12 with a temperature-sensitive deoxyribonucleic acid polymerase I. J Bacteriol. 1972 Mar;109(3):971–978. doi: 10.1128/jb.109.3.971-978.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki R., Arisawa M., Sugino A. Slow joining of newly replicated DNA chains in DNA polymerase I-deficient Escherichia coli mutants. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2954–2957. doi: 10.1073/pnas.68.12.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R., Weiner J. H., Weiner A., Kornberg A. Ten proteins required for conversion of phiX174 single-stranded DNA to duplex form in vitro. Resolution and reconstitution. J Biol Chem. 1975 Aug 10;250(15):5859–5865. [PubMed] [Google Scholar]
- Shapiro R., Klein R. S. The deamination of cytidine and cytosine by acidic buffer solutions. Mutagenic implications. Biochemistry. 1966 Jul;5(7):2358–2362. doi: 10.1021/bi00871a026. [DOI] [PubMed] [Google Scholar]
- Verly W. G., Rassart E. Purification of Escherichia coli endonuclease specific for apurinic sites in DNA. J Biol Chem. 1975 Oct 25;250(20):8214–8219. [PubMed] [Google Scholar]