CDS Annotation in Full-Length cDNA Sequence (original) (raw)
Abstract
The identification of coding sequences (CDS) is an important step in the functional annotation of genes. CDS prediction for mammalian genes from genomic sequence is complicated by the vast abundance of intergenic sequence in the genome, and provides little information about how different parts of potential CDS regions are expressed. In contrast, mammalian gene CDS prediction from cDNA sequence offers obvious advantages, yet encounters a different set of complexities when performed on high-throughput cDNA (HTC) sequences, such as the set of 60,770 cDNAs isolated from full-length enriched libraries of the FANTOM2 project. We developed a CDS annotation strategy that uses a variety of different CDS prediction programs to annotate the CDS regions of FANTOM2 cDNAs. These include rsCDS, which uses sequence similarity to known proteins; ProCrest; Longest-ORF and Truncated-ORF, which are ab initio based predictors; and finally, DECODER and NCBI CDS predictor, which use a combination of both principles. Aided by graphical displays of these CDS prediction results in the context of other sequence similarity results for each cDNA, FANTOM2 CDS inspection by curators and follow-up quality control procedures resulted in high quality CDS predictions for a total of 14,345 FANTOM2 clones.
During the past decade, large-scale DNA sequencing efforts have produced a wealth of information about genomes. An important step in the analysis of genome information is deciphering the complete coding potential or protein coding sequence (CDS)region of each gene. CDS is a sequence of nucleotides that corresponds with the sequence of amino acids in a protein. A typical CDS starts with ATG and ends with a stop codon. CDS can be a subset of an open reading frame (ORF). In eukaryotes, prediction of CDS regions in genomic sequence is complicated by a low percentage of the genome devoted to CDS and by interruptions of CDS regions by introns. It is not possible at present to predict from genomic sequence the correct distribution of CDS regions that appear in the proteins expressed from a genome. To obtain information about the portion of the mammalian genome that is translated into protein, the mature messengers of the genome's coding potential (full-length mRNAs)must be sampled.
The focus of the RIKEN Mouse Gene Encyclopedia Project is isolation and sequencing of novel full-length cDNAs from the mouse. Recently, the RIKEN Genome Exploration Research Group released the sequences and annotations of 60,770 cDNAs, the result of an international annotation effort termed FANTOM2-MATRICS (Mouse Annotation Teleconference for RIKEN cDNA sequences; The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I & II Team 2002). The primary goals of MATRICS were to provide human-evaluated computational gene assignments (i.e., most informative gene names)and verified descriptive annotations including CDS predictions, for the RIKEN cDNAs, and to identify potential problem clones in the set.
The RIKEN sequences are high-throughput cDNAs (HTC); thus, some of these sequences contain base-call errors. For this reason, accurate CDS prediction from these sequences required algorithms that incorporate sequence error correction. CDS prediction methods for the FANTOM2 clone set improved significantly compared with those used during the annotation of FANTOM1 clones, the first set of 21,076 sequences released as part of the RIKEN Encyclopedia Project (The RIKEN Genome Exploration Research Group Phase II Team and The FANTOM Consortium 2001). For FANTOM1 CDS prediction, we used DECODER (Fukunishi and Hayashizaki 2001), a CDS prediction algorithm developed to compensate for sequencing errors, and thus useful for translating high-throughput cDNA (HTC)sequences. We found that DECODER was effective, but did not predict accurate coding regions in some cases. It was clear that we could increase the accuracy of CDS prediction for the FANTOM2 set by including additional CDS prediction programs, and then allowing annotators to select the most appropriate coding region for each clone. Therefore, to predict CDS region in the FANTOM2 cDNA sequences more accurately, we have developed five additional CDS prediction programs: ProCrest, rsCDS, NCBI CDS predictor, Longest-ORF, and Truncated-ORF. CDS regions for FANTOM2 clones predicted from these programs and DECODER were displayed at the Web-based cDNA annotation system (CAS)developed for MATRICS (http://fantom2.gsc.riken.go.jp). This system allowed users to view CDS prediction results from these different methods within the graphical context of sequence similarity alignments for each clone, and allowed annotators to make high-confidence selection based on multiple evidence. We would mention that the integrated CDS predictions are unique and have not been applied in any other systematic large-scale annotation. Here we report the advantages of the CDS annotation for FANTOM2 cDNAs by using these systems.
RESULTS
Development of CDS Prediction Programs and Curation Strategy
For FANTOM2-MATRICS, we developed five different computational methods to predict the CDS regions of cDNA sequences: rsCDS, the NCBI CDS predictor, ProCrest, Longest-ORF, and Truncated-ORF (see Table 1). In summary, rsCDS and the NCBI CDS predictor incorporate the results of translated amino acid sequence similarity to other proteins for CDS prediction. These two programs differ primarily in their degree of permissiveness for frameshifting, with rsCDS allowing many frameshifts during translation, whereas NCBI CDS predictor allows only a single frameshift per translation. ProCrest predicts CDS regions with using codon usage for each amino acids and tRNA anticodon usage independently. Truncated-ORF predicts partial CDS regions in sequences truncated at their 5′ and/or 3′ ends, and Longest-ORF simply finds the longest ORF (open reading frame)with an ATG start codon. The predicted CDS from these programs together with DECODER were displayed in the Web-based FANTOM2 interface developed for MATRICS. The FANTOM2 interface provides integrated graphical summaries of sequence similarity search results, CDS predictions, comparisons to several motif and protein structure databases, and alignments against the mouse genome assembly. This interface helped MATRICS curators annotate the coding region in the context of biological function. To facilitate CDS annotation, we devised standardized qualifiers to allow annotators to define the CDS region (CDS start and end)and status of a clone (5′ and/or 3′ truncated, immature, UTR).
Table 1.
Features of CDS Prediction Programs
| rsCDS | NCBI CDS | ProCrest | DECODER | Longest-ORF | Truncated-ORF | |
|---|---|---|---|---|---|---|
| Base method (ab initio/sequence similarity) | Homology | Both | Ab initio | Both | Ab initio | Ab initio |
| Detection of frameshift | Yes (any) | Yes (one) | Yes (one) | Yes (any) | No | No |
| Detection of stop codons in frame | Yes | No | No | No | No | No |
| Prediction of partial CDS | Yes | Yes | Yes | Yes | No | Yes |
| Detection of immature | Yes | No | No | No | No | No |
| Usage of sequence quality (phred score) | No | No | Yes | Yes | No | No |
Prior to MATRICS annotation, we developed standardized criteria for annotators to select the best CDS from among the predictions available for each sequence by the different computational methods used. The CDS for each sequence was selected from predictions or manually coded by curators based on consideration of the following factors: homology with known proteins, sequence quality, EST hits, splicing patterns, cluster analysis, repeat sequences, and other supporting evidence displayed in the FANTOM annotation interface. Our annotation policy was to consider as potential CDS regions, only ORFs of at least 100 amino acids in length, unless supported by sequence similarity to known proteins or motifs, including signal peptide and transmembrane domains, or by splicing evidence. If sequences not meeting these criteria were derived from UTR regions or determined from matches to UTRs of known genes or 3′-EST clusters with poly(A)tails, then the sequences were annotated as such, and no CDS region was indicated. Otherwise, the CDS regions of sequences not meeting these criteria were annotated as “Unknown.”
Annotation and Translation of CDS in FANTOM2 Sequences
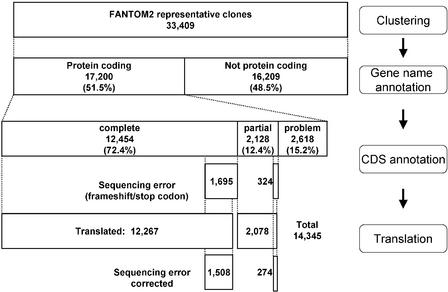
After clustering the FANTOM2 cDNAs, and mapping them onto the mouse genome, 60,770 clones were revealed to represent 33,409 transcriptional units (TUs; The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I & II Team 2002). From each of these TUs, a representative RIKEN clone was chosen based on cDNA sequence and CDS length. Of the 33,409 FANTOM2 representative cDNAs, 17,200 clones were revealed to be protein-coding genes by annotation efforts (see Fig. 1). A total of 12,454 (72.4%) and 2128 (12.4%)clones were annotated as having complete and partial CDS regions, respectively. The remaining 2618 (15.2%) clones of protein-coding genes were annotated as problematic sequences: immature, UTR, artifact, or unknown. Among the 14,582 cDNA clones annotated as complete or partial CDS, 2019 (13.8%)clones include frameshift and/or stop codon errors within the CDS, caused by low sequence quality. The CDS regions for 1782 of these 2019 clones were translated by correction of frameshift and/or stop codon errors by CDS prediction algorithms.
Figure 1.
Flowchart for gene name and CDS annotation steps during MATRICS. The 60,770 FANTOM2 clones were clustered into 33,409 transcriptional units (The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I & II Team 2002). The gene name and CDS of representative clones from each TU were annotated by MATRICS curators. Of 17,200 protein-coding clones, 12,454 and 2128 were annotated as having a complete and partial CDS, respectively. The remaining 2618 clones have problems such as “immature,” “UTR,” or “unknown” (see Methods). Another 187 complete CDS and 50 partial CDS clones could not be translated because of low sequence quality.
The numbers of translated CDS regions predicted by each program for FANTOM2 representative clones are shown in Table 2. rsCDS, DECODER, and ProCrest increased the rate of translations, by correcting sequencing errors, for 1338, 906, and 315 representative clones, respectively. The comparatively higher correction rate of rsCDS results from this program's use of homologous proteins as reference sequences for translation. For clones that represent known genes, there is a high consistency rate between rsCDS prediction and human CDS curation (see Supplementary Table 1, available at www.genome.org). The Longest-ORF was particularly useful for predicting CDS regions where no frameshift or stop codon errors were present (96.7%, 10,409/10,759). Truncated-ORF was designed to specialize in prediction of truncated CDS regions, thus, it predicted partial CDS efficiently (72.8%, 1498/2059). Overall, DECODER shows the highest consistency (79.6%) with human CDS curation. The CDS regions for 101 clones (0.7%)were not predicted by any programs, but were manually determined. As a result of CDS prediction by six different algorithms and subsequent human curation, 14,345 amino acid sequences were generated (12,267 complete and 2078 partial CDS)as FANTOM2 representative protein sequences.
Table 2.
Annotated and Translated CDS by CDS Programs
| Annotated | Translated | rsCDS | NCBI CDS | ProCrest | DECODER | Longest-ORF | Truncated-ORF | Manual | |
|---|---|---|---|---|---|---|---|---|---|
| In-frame (complete) | 10,759 | 10,759 | 5,287 | 8,343 | 9,477 | 9,517 | 10,409 | 0 | 43 |
| In-frame (partial) | 1,804 | 1,804 | 858 | 959 | 750 | 1,000 | 0 | 1,498 | 6 |
| FS/stop (complete) | 1,695 | 1,508 | 1,094 | 3 | 314 | 847 | 0 | 0 | 41 |
| FS/stop (partial) | 324 | 274 | 244 | 0 | 1 | 59 | 0 | 0 | 11 |
| Total (clones) | 14,582 | 14,345 | 7,483 | 9,305 | 10,542 | 11,423 | 10,409 | 1,498 | 101 |
| Sensitivity (%) | — | — | 52.2 | 64.9 | 73.5 | 79.6 | 72.6 | 10.4 | 0.7 |
Comparison of CDS Programs With Human Curation
We extended the comparison of these CDS prediction programs, by calculating the number of times each program predicted CDS regions identical to or different from the curated human CDS (see Fig. 2). It is evident that ProCrest, DECODER, and Longest-ORF (and NCBI CDS to a lesser but still significant extent)predicted the same CDS region that was selected by human curation in the majority of cases (7040, 49.1%), and the most common occurrence was that in which these four programs plus rsCDS predicted the same human-selected CDS (4539, 31.6%). Thus, almost half of the time, predictions from at least three different programs were in agreement with the selected CDS regions by curators (with the exception of Truncated-ORF, for reasons discussed above). Interestingly, the next most frequent case of correlation between predicted CDS and human curation was that in which only the rsCDS algorithm results matched the human selected CDS (5.5%, 791). This indicates the distinctive CDS prediction properties of the rsCDS algorithm. In fact, the combination of predictions by rsCDS and DECODER account for most of the human curated CDS regions (92.1%; see Suppl. Table 2). In addition to rsCDS, there are instances in which the other programs were alone in predicting a CDS identical to the manually annotated CDS, although much less frequently (DECODER, 279; Longest-ORF, 88; NCBI CDS predictor, 74; ProCrest, 67; and Truncated-ORF, 27). Those instances indicate that all the programs are required for CDS prediction. Because the six different programs together worked efficiently to compensate for weak points in any one of the programs alone, we were able to make a large number of FANTOM2 protein sequence predictions.
Figure 2.

Correlation between CDS prediction programs. The frequencies with which different CDS program combinations predicted identically to (colored boxes) or differently from (open boxes) human curated CDS regions are shown. The number of instances for the top three most frequently observed patterns is indicated in bold. The instances of patterns characterized by only a single CDS program predicting identically to human curation are shown in boxed numbers.
Examples of Curated CDS in FANTOM2
In addition to prediction programs, curators used many different types of data to evaluate CDS regions, such as sequence quality, genome mapping, splicing information, EST mapping, predicted transmembrane regions, and protein motifs. The graphical display of this information facilitated detection of potential coding regions by curatorial analyses. Below we demonstrate some advantages of the system for this purpose with examples.
NMD and Splicing Pattern
Faulty mRNAs with truncated coding information (so-called nonsense mRNAs)are recognized and specifically degraded by a process termed nonsense-mediated mRNA decay (NMD) or mRNA surveillance (Hentze and Kulozik 1999). The signal that triggers NMD is a nonsense codon followed by a splicing junction. This rule for termination codon position has broad implications given that, in principle, any intron junction located >50–55 nt downstream of a termination codon could mediate a reduction in mRNA abundance (Nagy and Maquat 1998). The results of FANTOM2 cDNA mapping to the mouse genome assembly allowed us to examine the concurrence of annotated CDS regions with the NMD rule. We used the NMD rule as a method of quality control of the annotated CDS regions. For example, the CDS for clone 4930415J21 was annotated to be located from positions 93 to 1311 by a curator early in MATRICS (Fig. 3). Three programs indicate that the CDS terminates at 1311; however, we changed that to a presumed CDS stop position at 1731, reasoning as follows: firstly, the 3′-most splice junction was located at 1570, that is, 261 bases downstream from 1311; secondly, the phred (sequence quality)score around 1311 was very low.
Figure 3.
Annotation interface for clone 4930415J21 as an example of CDS correction by the NMD rule for termination codon placement. The image shows sequence similarity results for this FANTOM2 clone with protein, predicted CDS, ESTs, genome mapping, and sequence quality. In this case, CDS candidates by rsCDS, ProCrest, DECODER, NCBI CDS predictor, and Longest-ORF are shown from top to bottom. The CDS of this sequence runs from 93 (colored diamond) to 1731 (colored triangle). The TAG stop codon at 1311 (open triangle) is assumed to be false because of low sequence quality. The 3′-most splice junction is located at 1570 (open diamond). The annotated results for gene name and CDS status are shown at the bottom.
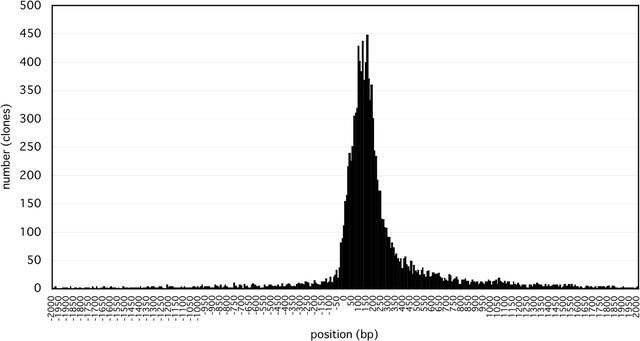
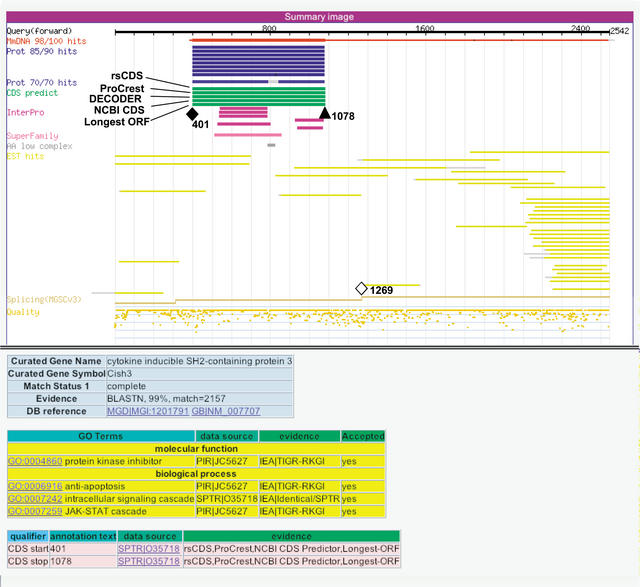
After correcting misannotations that violated the NMD rule, we recalculated the distances between stop codon positions and the splice junction sites nearest to the 3′ ends for all CDS regions selected by human curation. Figure 4 shows a histogram of the positions of stop codons from the last splice junctions for 10,789 clones that were spliced and annotated as having a complete or 5′-truncated CDS. The most frequent fraction was found within 400 bases downstream from the last splice junction. Interestingly, a sizeable number of clones (803, 6.9%)may be a target for NMD because they have a stop codon more than 55 bases upstream from the splice junction. For example, the last splice junction of clone B930030L03 is located at base position 1269; however, its CDS was predicted to be located from positions 401 to 1078 (see Fig. 5). No plausible ORF could be found between the stop codon and the splice junction. It was reported that mRNA degradation by NMD was dependent on translation events in the cytoplasm (Thermann et al. 1998). Clones that appear to violate the NMD rule may represent nuclear mRNA products that escaped from NMD-mediated degradation. Alternatively, a novel mechanism for mRNA stability may apply to these clones.
Figure 4.
The distances between stop codons and last splice junctions. A histogram of the positions of stop codons from last splice junctions for 10,789 clones that were spliced and annotated as having a complete or 5′-truncated CDS is shown. The average number of bases between the positions of the last splice junction and the stop codon for these sequences is 208 bases. For these sequences, it is most common to find the stop codon within 400 bases downstream from the last splice junction.
Figure 5.
Annotation interface for clone B930030L03, a potential target for NMD-mediated instability. The CDS region of this clone was annotated from positions 401 (colored diamond) to 1078 (colored triangle), based on similarity with cytokine inducible SH-2-containing protein 3. The CDS for this clone violates the NMD rule for termination codons; the stop codon position (1078, colored triangle) is located 191 bases upstream from the last splice junction (1269, open diamond); thus, such transcripts in the cell may be targets for degradation by the NMD system.
Selenocysteine-Containing Proteins
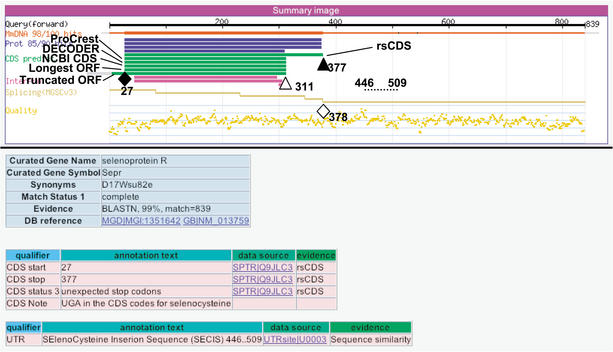
The selenoprotein family is a group of proteins that contain the rare amino acid selenocysteine (Sec). The selenocysteine amino acid becomes incorporated during translation by a process of nonsense suppression of the OPAL (UGA)stop codon (Berry et al. 1991). A sequence in the 3′-UTR of selenoprotein mRNAs that forms an RNA hairpin structure, called the selenoprotein insertion sequence (SECIS), is required for read-through of UGA Sec codons. By sequence similarity to selenoproteins and by searching for the SECIS UTR motif, 15 selenocysteine-containing proteins were found in FANTOM2 sequences. Figure 6 shows an example of RIKEN clone 1110012E09 annotated as Selenoprotein R. The SECIS motif located between base positions 446 and 509 promotes decoding of the OPAL codon at position 311 to selenocysteine. Although rsCDS correctly predicted the CDS region between positions 27 and 377 from sequence similarity to protein data, other programs mispredicted the CDS for this clone. The position of the 3′-most splice junction at 378 also supports that the CDS ends at 377, not at 311 in accordance with the NMD mechanism.
Figure 6.
Annotation interface for clone 1110012E09, Selenoprotein R cDNA. The SECIS motif located between base positions 446 and 509 (dotted line) promotes decoding of the OPAL codon at position 311 (open triangle) to selenocysteine. The CDS region was annotated from positions 27 (colored diamond) to 377 (colored triangle). The last splice junction (378, open diamond) colocates with the stop codon.
Small Proteins
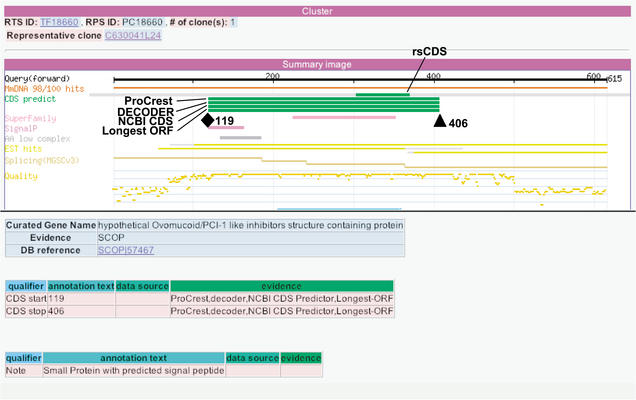
During MATRICS, ORFs <100 amino acids were recorded as CDS regions only when supported by evidence. The CAS graphic interface was useful for annotation of the CDS region of a small protein. The CDS regions of 295 representative clones were curated as proteins <100 amino acids. Of these, 234 clones were annotated by similarity to known proteins, 30 by motifs or domains, and 31 by signal peptide, transmembrane regions, or splicing evidence. Figure 7 shows the RIKEN clone C630041L24 as an example of a small protein with a predicted motif. This sequence has no similarity to known proteins; however, the curated CDS extends from nucleotide positions 119 to 406, based on the predicted signal peptide and an ovomucoid/PCI-1 like inhibitor structure in the region.
Figure 7.
Annotation interface for clone C630041L24, an example of a small protein containing a signal peptide. The CDS region from positions 119 (colored diamond) to 406 (colored triangle) encodes a 96-amino-acid protein. The predicted signal peptide at the N terminus of the CDS indicates that this is a soluble/secreted protein (Grimmond et al. 2003).
DISCUSSION
Recently, two large-scale sequencing projects released significant biological resources for the laboratory mouse, an important animal model for biomedical research. One is the sequences of 60,770 full-length sequenced cDNA clones by the RIKEN Genomic Sciences Center, and the other is a draft sequence of the C57BL/6J genome by the Mouse Genome Sequencing Consortium (Mouse Genome Sequencing Consortium 2002; The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I & II Team 2002).
Because coding sequences occupy just a small fraction of the mammalian genome, it is not an easy task to distinguish all true coding sequences from intergenic regions, and even more challenging to partition coding sequences accurately into units of transcription. For these reasons, genome annotators rely upon transcript-derived sequence to refine gene models and improve coding sequence annotation.
Detecting all coding regions in a genome is of great value, but this provides incomplete information about how the genome's coding potential is realized as expressed products. We now recognize the central importance of transcript variation from the same genomic regions in generating functional diversity from genomes, as the number of genes identified in one sequenced genome after another falls short of original, outdated expectations. This trend seems most prominent in mammalian genomes. Full-length cDNAs, such as those produced by the FANTOM2 project, that encode protein, provide a unique perspective of a genome's coding potential, as they convey the combination of coding sequences that express the genome's functional diversity.
Compared with predicting coding regions in the mammalian genome, it may seem trivial to find the CDS regions in the substantially reduced complexity and concentrated coding potential of cDNAs. However, there are many factors that complicate CDS prediction in cDNA sequences, particularly in the context of a large-scale effort.
First, predicting translation start sites in eukaryote mRNAs is considerably more difficult than it is in prokaryote mRNAs (where the ribosome binding site, the Shine-Dalgarno sequence, is highly conserved), because there is no comparably well-conserved indicator of the translation initiation start site. The Kozak consensus sequence (GCC[A or G]CCATGG; underlined ATG is start codon)is predictive for the majority, but is not universally conserved in eukaryotic mRNAs.
Second, it is difficult to distinguish mRNAs that do not code for protein, so-called noncoding RNAs (ncRNAs), from mRNAs that encode very small proteins. This distinction is complicated further by the presence of truncated sequences derived from untranslated regions of a transcript, or potentially coding transcripts that contain unprocessed introns. There is increasing evidence that the mammalian genome contains a large number of genes transcribed into ncRNAs. In the FANTOM2 data set, there are 16,209 TUs annotated as “unknown EST” or “unclassifiable.” These are candidates for ncRNAs. Detailed analysis for ncRNAs is described in Numata et al. (2003)
Third, sequence quality can significantly affect CDS prediction. The RIKEN mouse encyclopedia project provides a large number of cDNA sequences submitted to a division of the public sequence databases called high-throughput cDNA (HTC). Most of the RIKEN HTCs are high quality sequence; however, some cDNAs have sequence problems, such as frameshifts and stop codon errors caused by low sequence quality, and other cDNA clones were produced from incompletely processed transcripts or have truncated inserts caused by cloning errors.
Fukunishi and Hayashizaki (2001)developed the DECODER program to compensate for cDNA sequencing errors during conceptual translation, and it was used in the FANTOM1 meeting (The RIKEN Genome Exploration Research Group Phase II Team and The FANTOM Consortium 2001). For FANTOM2 annotations, curators choose CDS regions manually from the CDS predictions of six programs. When we compared predictions with CDS regions authorized by human curation, DECODER shows the highest sensitivity (79.6%), however, not perfectly. The rest of the CDS was chosen from other prediction programs. Because we used programs based on different strategies, each program showed distinct features in each type of CDS. The effectiveness of rsCDS was indicated by the large fraction of CDS (5.5%)that was predicted only by the rsCDS algorithm. rsCDS depends on homology to known protein sequences for its predictions. This strategy helps us to correct frameshift and in-frame stop codon errors. Indeed, rsCDS, together with DECODER, contributed greatly to the corrections of base-call errors in the CDS. Another unique algorithm was Truncated-ORF, which specialized in the prediction of partial CDS. It succeeded in predicting partial CDS efficiently, predicting 72.8% of manually annotated partial CDS. Although lower sequence quality in general leads to error-prone CDS prediction, using those six programs on all sequences can compensate for weak points in the different programs.
Besides the six programs used in CDS prediction, viewers for annotation system were helpful for curating CDS accurately. As we described in the section above for NMD and selenocysteine, the mapping information is very informative for CDS annotation. In the FANTOM system, cDNA mapping data to the mouse genome can be seen with predicted CDS candidates in a graphic interface. The search results for known genes, homologs, and motifs/domains are also shown in the interface and help to annotate CDS regions. The sequence quality viewer is another useful tool, helping to assess conflicts between CDS prediction programs or between prediction programs and homologies caused by possible base-call errors. The sequence quality viewer is also a useful tool because we sometimes find a conflict between CDS prediction programs caused by base-call errors.
We have used a CDS annotation strategy that takes advantage of a variety of CDS prediction programs and a convenient graphical display of these predictions in the context of a wide range of sequence similarity results and succeeded in annotating a large, high-quality set of 14,345 mouse protein sequences. This set of translations from transcript CDS regions provides a valuable view of the proteome and of diverse biological area.
All annotation data for the FANTOM2 sequences are available at the RIKEN Web site (http://fantom2.gsc.riken.go.jp/).
METHODS
Sequence Set
The sequences of the FANTOM2 clone set (60,770), which consists of 39,694 new cDNA clones (FANTOM2 new set)and the 21,076 cDNA clones of the FANTOM1 set (The RIKEN Genome Exploration Research Group Phase II Team and The FANTOM Consortium 2001), were used as the data set (The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I & II Team 2002). As FANTOM2 clones contained redundant sequences transcribed from each transcriptional unit, 33,409 representative clones were selected to represent each TU for detailed analysis (The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I & II Team 2002).
CDS Prediction Programs
ProCrest
A new method, called ProCrest (Protein Coding Region Estimator), was developed for estimating the coding region of a transcript. It is based on a new statistical analysis of the protein-coding potential of known mRNA sequences. ProCrest predicts CDS regions using codon usage for each amino acid and tRNA anticodon usage independently.
rsCDS
rsCDS (relational search for CoDing Sequence)is a predictor for CDS status and CDS regions that is based on the homology searches. It was developed to identify coding regions of known genes or those that are homologous to known genes very accurately even if there are frameshift errors. As rsCDS is based on the assumption that subject sequences in the database contain no errors, incorrect predictions can still arise from errors in the database.
The basic algorithm of rsCDS is as follows. First, a given cDNA sequence is subjected to a BLAST search against a protein database. Then each of the multiple hits is aligned by FASTY. According to these alignments, one of the following sequence statuses is assigned from “Complete CDS,” “5′-truncated,” “3′-truncated,” “Alternative N-terminal,” “Alternative C-terminal,” “5′ Immature,” “3′ Immature,” “Coding potential with predicted start codon,” “Coding potential with predicted stop codon,” “Reverse,” “?,” or “No BLAST match.” In addition, frameshift errors and base-call errors that turn amino acids into stop codons are predicted. If the output status is “Complete CDS,” “Truncated,” or “Alternative Terminal,” a coding region is also predicted based on the alignment. If the status is “Complete CDS,” the positions corresponding to the N and C terminus of the protein hit are assigned as the start and end of the CDS, respectively. If the status is “Truncated,” the region from the truncated 5′ end to the position corresponding to C terminus of the protein hit, or that from the position corresponding to the N terminus of the protein hit to the truncated 3′ end is assigned. If the status is “Alternative Terminal,” either the 5′ or the 3′ end of the CDS is determined according to the N or C terminus of the protein hit, and the other end is determined by one of the appropriate triplets TAA, TAG, TGA, or ATG (see Suppl. figures in detail).
NCBI CDS Predictor
In the NCBI CDS predictor, two sets of results for each cDNA sequence are used to assess whether it contains a complete CDS. The cDNA sequence is compared with all proteins in GenBank, and all ORFs in the cDNA sequence are identified. If an alignment with a protein that begins with methionine coincides with a start codon in an ORF, that region is selected as the CDS. Otherwise, if a protein alignment terminates at the stop codon of a set of ORFs, then the most likely ORF is used. In both of these cases, the ORF is required to be at least half the protein's length (the low threshold is chosen because eliminating short but functional alternate splice forms is undesirable). If there are no protein matches coinciding with any of the termini of an ORF, then the most likely ORF is used. The NCBI CDS predictor considers a cDNA to be chimeric if two proteins from different LocusLink sets align to the same cDNA at >96% identity over nonoverlapping intervals. We consider a cDNA to be frameshifted if, upon BLASTX alignment with two proteins from different organisms, a gap of 1 or 2 nt is recorded in the same position on the cDNA. Lastly, the predictor considers a cDNA to be incompletely processed if an ORF that coincides with a protein start extends over <30% of the length of the protein.
DECODER
DECODER is an amino acid translation program for full-length cDNA sequences with frameshift errors. It introduces artificial frameshifts into the given sequence and calculates the likelihood that the generated protein is an actual protein, taking into account the sequencing accuracy (phred score)at the position of the frameshift, Kozak sequence, codon usage, and the position of the potential start codon. Then the most likely protein and the corresponding CDS region are determined. For further details, see Fukunishi and Hayashizaki (2001).
Longest-ORF and Truncated-ORF
The Longest-ORF simply finds the longest ORF, whereas the Truncated-ORF algorithm assumes that the 5′ and/or 3′ end of a given cDNA sequence and its coding region are truncated. First it determines which of the potential stop codons is located nearest to the 5′ end for each of three reading frames. The one located farthest from the 5′ end is assumed to be the authentic one, yielding a 5′-truncated CDS. Similarly, the algorithm determines which potential start codon is located nearest to the 3′ end for each of three reading frames, and the one located farthest from the 3′ end is assumed to be the correct one, yielding a 3′-truncated CDS.
Viewers Used for the CDS Curation
CAS
CAS (cDNA annotation system)was developed for functional annotation for RIKEN mouse full-length cDNA sequences. A graphic interface shows information for each cDNA sequence, for example, DNA and protein sequence similarities, matches to several motif and protein structure databases, alignment against the mouse genome, sequence quality, and predicted CDS. For further details, see Kasukawa et al. (2003). CAS is available at the RIKEN Web site (http://fantom2.gsc.riken.go.jp/).
ITOP
ITOP (Inspecting Transcript Object in Phred/phrap)provides sequence quality information for cDNA sequences in three views: (1)ESECONSED, providing a view of quality scores with each nucleotide; (2)MOSAIC, providing a position of each read on the contig and the sequencer name; (3)a graph view, showing quality scores using a line graph. For details, see Kasukawa et al. (2003). The ITOP system is available at http://fantom2.gsc.riken.go.jp/ITOP/ as well as from the FANTOM viewer.
CDS Curation Strategy in MATRICS
In the FANTOM2 MATRICS, CDS and gene names were assigned at the same time. To control the quality of the human curation, we provided curators guidelines for CDS determination. First, curators should follow the basic steps: (1)Choose CDS at least 100 amino acids (303 bases)long, except when the region is supported by a particular reason, such as protein or domain homology. (2)Choose the most likely CDS region from predicted CDS regions. (3)Check one of the boxes in status 1 (5′-UTR, 3′-UTR, noncoding RNA, artifact, unknown) when reliable CDS cannot be determined.
We prepared the following CDS qualifiers for describing CDS regions and suspected cloning problems.
CDS Start
Base position of the CDS start codon, ATG in most cases. For 5′-truncated clones, this describes the start position of the coding frame.
CDS Stop
Base position of the CDS stop. This describes the last position of the coding frame for 3′-truncated clones.
CDS Status 1
Status of the location of the sequence in the corresponding gene: (1)5′-UTR for clones containing only 5′-untranslated region; (2)3′-UTR, for clones containing only 3′-untranslated region; (3)noncoding RNA, for a noncoding transcript that seems to have biological function; (4)artifact, for not derived from mouse mRNA; (5)unknown.
CDS Status 2
Status for completeness and orientation of the CDS: (1)5′-truncated; (2)3′-truncated; (3)reverse.
CDS Status 3
Problems in the CDS region: (1)frameshift, for an insertion or deletion of one or more bases caused by base-calling error; (2) immature, for introns in the CDS region; (3)unexpected stop codon, for stop codon that appears in the middle of coding regions, caused by base-calling error.
CDS Note
Curatorial comments about CDS or clone problems.
Clone Classification Based on Gene Name and CDS Data
Clones were classified based on their curated CDS and gene name. Gene name categorization was followed as described in The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I & II Team (2002). CDS categories were determined based on CDS status. First, clones were classified as follows: “complete,” “5′-truncated,” “3′-truncated,” “5′-3′-truncated,” “immature,” “5′-UTR,” “3′-UTR,” “non-coding,” “unknown,” and “artifact.” Then, those were grouped into three CDS categories: (1)”complete” into “Complete CDS”; (2)“5′-truncated,” “3′-truncated,” and “5′-3′-truncated” into “Partial CDS”; (3)“immature,” “5′-UTR,” “3′-UTR,” “noncoding,” “unknown,” and “artifact” into “Problem.”
Acknowledgments
We thank Lukas Wagner for useful discussion. This study has been supported by the Research Grant for the RIKEN Genome Exploration Research Project from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government to Y.H.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1060303.
Footnotes
[Supplemental material is available online at www.genome.org.\]
References
- Berry, M.J., Banu, L., Chen, Y.Y., Mandel, S.J., Kieffer, J.D., Harney, J.W., and Larsen, P.R. 1991. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature 353**:** 273-276. [DOI] [PubMed] [Google Scholar]
- The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I & II Team. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420**:** 563-573. [DOI] [PubMed] [Google Scholar]
- Fukunishi, Y. and Hayashizaki, Y. 2001. Amino acid translation program for full-length cDNA sequences with frameshift errors. Physiol. Genom. 5**:** 81-87. [DOI] [PubMed] [Google Scholar]
- Grimmond, S.M., Miranda, K.C., Yuan, Z., Davis, M.J., Hume, D.A., Yagi, K., Tominaga, N., Bono, H., Hayashizaki, Y., Okazaki, Y., et al. 2003. The mouse secretome. Functional classification of the proteins secreted into the extracellular environment. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Hentze, M.W. and Kulozik, A.E. 1999. A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96**:** 307-310. [DOI] [PubMed] [Google Scholar]
- Kasukawa, T., Furuno, M., Nikaido, I., Bono, H., Hume, D.A., Bult, C., Hill, D.P., Baldarelli, R., Gough, J., Kanapin, A., et al. 2003. Development and evaluation of an automated annotation pipeline and cDNA annotation system. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420**:** 520-562. [DOI] [PubMed] [Google Scholar]
- Nagy, E. and Maquat, L.E. 1998. A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem. Sci. 23**:** 198-199. [DOI] [PubMed] [Google Scholar]
- Numata, K., Kanai, A., Saito, R., Kondo, S., Adachi, J., Wilming, L.G., Hume, D.A., RIKEN GER Group and GSL Members, Hayashizaki, Y., and Tomita, M. 2003. Identification of putative noncoding RNAs among the RIKEN mouse full-length cDNA collection. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- The RIKEN Genome Exploration Research Group Phase II Team and The FANTOM Consortium. 2001. Functional annotation of a full-length mouse cDNA collection. Nature 409**:** 685-690. [DOI] [PubMed] [Google Scholar]
- Thermann, R., Neu-Yilik, G., Deters, A., Frede, U., Wehr, K., Hagemeier, C., Hentze, M.W., and Kulozik, A.E. 1998. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 17**:** 3484-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://fantom2.gsc.riken.go.jp/; FANTOM2.
- http://fantom2.gsc.riken.go.jp/db/; FANTOM2 cDNA annotation database.