Two Creutzfeldt–Jakob disease agents reproduce prion protein-independent identities in cell cultures (original) (raw)
Abstract
Human Creutzfeldt–Jakob disease (CJD) and similar neurodegenerative diseases such as sheep scrapie are caused by a variety of related infectious agents. They are associated with abnormal host prion protein (PrP), which is assessed by limited proteolysis to yield resistant PrP bands (PrP-res). Although PrP-res has been posited as the infectious agent, purified PrP-res itself is not infectious. To establish the independence of CJD agent characteristics from those of PrP-res, two different mouse-passaged CJD strains were propagated in neuronal cell lines whose PrP-res patterns differ markedly from each other and from those found in infected brain. In mouse brain, the fast CJD strain, FU, elicits many PrP-res deposits, whereas the slow SY strain elicits few. Both strains evoked PrP-res in cultured murine cells, although SY induced PrP-res only transiently. PrP-res patterns in FU- and SY-infected GT1 cells were identical, and were significantly different from those in brain and in N2a cells. Nevertheless, all FU-infected cell lines reproduced their original fast disease in mice, even after extensive subculture, whereas SY-infected cells produced only slow disease. These data indicate PrP-res neither encodes nor alters agent-specific characteristics. PrP-res was also a poor predictor of infectivity because SY cells that had lost PrP-res were ≈10-fold more infectious than PrP-res-positive cultures. Furthermore, FU titers increased 650-fold, whereas PrP-res remained constant. Passaged FU-infected cells had titers comparable to brain, and >30% of cells displayed abundant cytoplasmic PrP-res aggregates that may trap agent. The continuous substantial replication of CJD in monotypic cells will further the discrimination of agent-specific molecules from pathological host responses to infection.
Keywords: transmissible encephalopathies, agent strains, in vitro propagation, high infectivity, in situ detection
One of the most effective ways to investigate the molecular nature of viruses and establish their life cycle has been to propagate them in simplified tissue culture systems. This goal is particularly important in the case of transmissible spongiform encephalopathies, where the nature of the infectious agent remains enigmatic. Although the protease-resistant host membrane prion protein (PrP-res) has been posited to encode infectivity and agent-specific characteristics, no purified, recombinant, transgenic, or amplified form of PrP-res has been capable of reproducibly transmitting infection (1). Until now, it has been difficult to achieve infectivity levels in cultured cells as high as those found in Creutzfeldt–Jakob disease (CJD)-infected degenerating brain. Initial reports in CJD and scrapie showed low infectivity in brain cells cultured from infected animals (2, 3). Furthermore, many of these cultures lost infectivity after extensive passage, and malignant transformation was often seen in CJD cultures (4). The frequent loss of agent could result from rapid cellular growth outstripping agent replication, because the effective doubling time for CJD and scrapie agents can be very prolonged (5). Moreover, if only a few of the original brain-derived cells were infectious, overgrowth by uninfected or resistant cells could compromise agent propagation. Higher scrapie titers have been reported in PC12 cells but these were nondividing cultures (6).
Continuous culture of N2a and other neuroblastoma cell lines after in vitro infection with a scrapie agent (Chandler/RML strain) yielded persistently infected cells, although infectious titers were still low. These cells also often lost their infectivity unpredictably, and had to be reinfected anew (3, 7). Levels of infection were generally ≈1 LD50 per 100 cells or less, and some cultures that were infectious for mice failed to show abnormal PrP (PrP-res) in Western blots of whole-cell lysates. PrP-res is typically, but not always associated with high levels of agent replication (5, 8). There was also no consistent assay of PrP-res in individual cells in the various laboratories (9). Nevertheless, while not practical for the high-yield production of infectious agent, these cultures were valuable in elucidating PrP metabolism, and removal of PrP after treatment with selected compounds (e.g., 10–12).
More recently, murine hypothalamic GT1 and neuroblastoma N2a cell lines have yielded more reproducible long-term infection with the Chandler/RML scrapie agent, yet these lines have been resistant to infection by other scrapie strains (13). Thus, these studies underscore agent-specific requirements that are unrelated to PrP polymorphisms or levels of PrP expression. Because tissue culture models have concentrated on sheep-derived scrapie agents propagated in mice, particularly the Chandler/RML strain, our interest in human agents led us to determine the infectivity of CJD strains in two GT1 sublines and in N2a cells overexpressing murine PrP (14). We used two distinct mouse-passaged CJD agents that while indistinguishable by their brain PrP-res-banding patterns on Western blots, display vastly different titers, incubation times, and pathologic brain changes in mice (15, 16). Here, we show that both the slow SY agent, and the fast virulent FU CJD agent, can infect GT1 cells. FU can also replicate in more rapidly dividing N2a cells. FU-infected GT1 lines showed increasing infectivity with passage by bioassay, and attained titers close to those found in brain (≈1LD50 per cell). These two strains continued to produce their slow and fast phenotypes in mice despite extensive passage in vitro. However, in these simplified cultures, SY and FU remained indistinguishable by their PrP-res banding or glycosylation patterns. Instead, the PrP-res-banding profiles were determined by cell type. These findings are problematic for the prion hypothesis where abnormal PrP folding or glycosylation, and hence PrP-res band patterns, are postulated to encode each agent strain (17–19).
Materials and Methods
Cell Cultures and in Vitro Infection. Murine GT1–1 and GT1–7 sublines from the immortalized hypothalamic GT1 cells (14) were trypsinized and were split 1:4 or 1:5 weekly. N2a58 neuroblastoma cells overexpressing WT murine PrP (14) were trypsinized and were split 1:10 every 5 days. Cell lines were fed high-glucose DMEM with 10% heat-inactivated FBS with 1% penicillin-streptomycin (GIBCO). Cells at 50% confluence in six-well plates were exposed for 24 h to 1% or 2% normal or infected brain homogenates in medium, were washed, grown to 90% confluence, and were then transferred to a 25-cm2 flask at passage in vitro 1 (p1). Infected GT1 sublines were not subcloned. N2a58 cells were diluted at p1 and were seeded at 0.7 cells per well in 96-well plates. Single-cell clones were screened for PrP-res (detected in 21% of the clones, n = 48), and a clone with the strongest signal (FU N2a58H1) was selected for further expansion in parallel with a clone from mock N2a cells (N2a58#1). Whole-cell lysates were treated with for 30 min with proteinase K (PK) at 25–30 μg/ml to obtain maximal amounts of PrP-res for Western blotting (5). Protein was determined by Bio-Rad DC protein assay, and quantitation of PrP bands was performed on films in the linear range (20). For deglycosylation, SDS-denatured proteins were digested with 10 units of PNGase F (Sigma) overnight at 37°C. If necessary, proteins of >12 kDa were quantitatively precipitated with six volumes of EtOH and centrifugation at 19,000 × g for 45 min at 4°C. PrP on blots was evaluated with four PrP antibodies: (i) a rabbit antibody raised against the N terminus (amino acids 1–50) of PrP (IBL, Gunma, Japan), (ii) a rabbit polyclonal antibody recognizing PrP-res amyloid (21), and (iii) two goat polyclonal antibodies recognizing the carboxyl end of PrP amyloid (amino acids 91–200; C-20 and M-20, Santa Cruz Biotechnology). Biotinylated lectins (Vector Laboratories) were used to verify removal of sugar residues (20).
Infectivity Bioassays. Washed cells were homogenized through a 26-gauge needle, and were briefly bath-sonicated and freeze-thawed twice at –70°C. PrP-overexpressing TGA20 mice (5–10 per experimental group, gift of C. Weissmann, Scripps Research Institute, Palm Beach, FL; ref. 22) were inoculated intracerebrally with 30 μl of homogenates. There are ≈3 × 105 brain cells in 30 μl of a 1% brain homogenate, and FU CD-1 brain contains ≈2 × 109 LD50 per g (15), i.e., ≈1 LD50 per brain cell. Two end point titrations of FU in TGA20 mice yielded the same terminal titers (≈3 × 109 LD50 per gm of brain). In contrast, there are only ≈105 LD50 per gm of SY brain (15). Pictures of ill mice were taken with digital Nikon DS500 and Sony DCR movie cameras.
In Situ Detection of Pathologic PrP. Because cells detached and became morphologically disrupted by using GdnHCl and PK treatments, we modified our methods for PrP-res detection in paraffin sections (21). Cells in flasks were fixed in situ with 4% fresh paraformaldehyde in PBS for 10 min, were scraped, pelleted at 4,000 × g for 10 min, and were then fixed for an additional 12–16 h before paraffin embedding. Slides were autoclaved in citrate buffer (21), digested with 0.002–0.004% trypsin for 7–15 min, quenched, exposed to antibodies, and then developed with Vector red. FU- and mock-infected control cells were mounted on the same slide.
Results
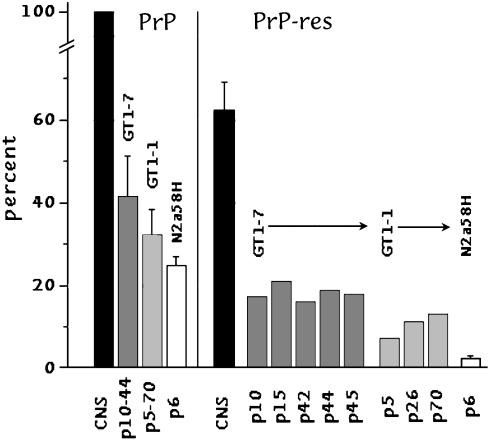
To address the question of whether PrP-res itself encodes intrinsic agent properties, we examined different cell lines infected with two distinct CJD agents. The results obtained in these highly simplified cell cultures can help clarify this profound question. We first determined the amount of total PrP, and of PrP-res, relative to that found in FU-infected mouse brain by using whole-cell lysates to avoid inaccuracies from subcellular fractionation. Fig. 1 Left shows the amount of total PrP in brain as a 100% standard per mg of cellular protein. The expression of PrP was less in the sublines than in brain, and, as can be seen from the SEMs, varied <2-fold when sampled every 10–15 passages. Mock controls showed the same amounts of total PrP (data not shown). PrP-res in each FU-infected cell line was also less than in brain (Right), and varied little after its first appearance. Notably, PrP-res in different passages of GT1–7 and GT1–1 all showed <2-fold changes in PrP-res with progressive subculture. Representative data for PrP-res at different passages are shown in Fig. 2_A_. N2a58H cells with the lowest expression of PrP had the lowest amount of PrP-res, and brain PrP-res was 62% of total PrP, in good accord with previous determinations (15). Mock controls, treated with normal brain in parallel, showed no PrP-res (Fig. 2 B, lanes 11 and 12, and C, lanes 4–6).
Fig. 1.
Quantitation of total PrP (Left) and PrP-res (Right) in different passages as compared with FU-infected brain (where total PrP is taken as the 100% standard). Insignificant differences in total PrP expression in each cell line are apparent by the small SEMs. Assay of individual cell passages (p) after infection also show little variation in PrP-res (<2-fold) in GT1 cells.
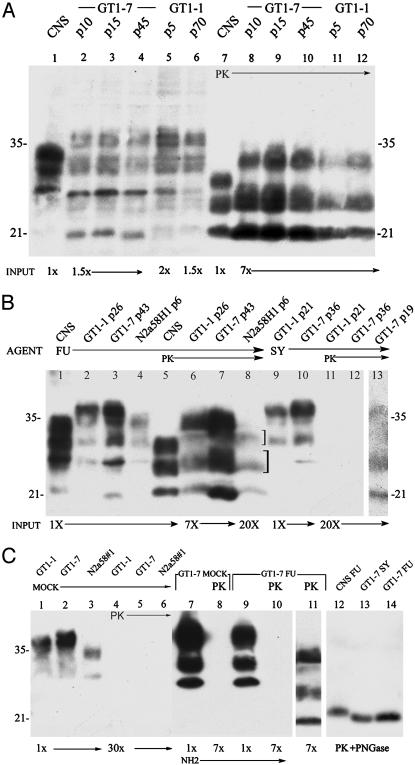
Fig. 2.
Western blots of whole-cell lysates from brain (CNS) and cell lines at different passages. The cell line, CJD agent strain, passage number, and relative protein load applied to each lane is indicated. (A) Total PrP (lanes 1–6) and PrP-res (lanes 7–12; digested with PK) show insignificant changes in PrP-res with extended passages of FU. (B) Relevant passages with relatively low PrP and PrP-res in N2a58H1 cells, and loss of PrP-res in SY GT1–1 cells after p19. (C) Mock controls show no PrP-res (lanes 4–6), and complete removal of N-terminal PrP sequences (lane 10), but not PrP core amyloid sequences (lane 11) after PK digestion. PK followed by PNGase removed glycosyl residues: SY and FU in GT1–7 cells show the same _M_r band (lanes 13,14) that is ≈1.5 kDa lower than in the CNS (lane 12). PrP antibody was M20 in A, and C20 in B. Both give the same pattern. The NH2-specific antibody is shown in lanes 7–10. Glycoform ratios of the three major PrP-res bands (±10%, higher to lower _M_r) were 1:1.1:0.5 (brain), 0.5:2.1:2 (GT1–7), 1:2.9:3 (GT1–1), and 1:1.3:1.1 (N2a58H1) by using optimal PK digestions and loads. The different glycoform ratios specified cell types but not strain characteristics, as in mammalian tissues (1).
Total cellular PrP, as well as PrP-res, had markedly different banding profiles by Western blotting in cell lines as compared with brain. In FU- and SY-infected mouse brain, PrP-res shows identical band mobilities and glycoform ratios (15), and hence does not discriminate these two very different agents. Fig. 2 shows total PrP from brain (Fig. 2 A and B, lane 1) and representative passages of GT1 and N2a sublines (Fig. 2 A lanes 2–6, and B, lanes 2–4). There are striking differences in intensity and _M_r of bands. There are also obvious differences between the _M_r of PrP-res bands as well as their glycoform ratios in brain and cell lines (Fig. 2 A, lanes 7–12, and B, lanes 5–8 and legend). Both FU-infected GT1 sublines show the same PrP-res pattern, although FU-infected GT1–7 have 2-fold more Prp-res than GT1–1 cells (Fig. 2 A, lanes 8–12). GT1–7 cells challenged with slow SY showed only a weak PrP-res signal and this signal appeared only at later passages 13–19 (Fig. 2_B_, lane 13). This PrP-res signal was lost after p19 (lane 12). Moreover, the _M_r of PrP-res bands from FU and SY in GT1–7 cells were the same in three independent analyses, again indicating the PrP-res pattern is cell-type- rather than agent-specific.
To assess whether the more resistant amyloid core was retained and all of the susceptible N-terminal PrP was completely digested, blots were probed with antibodies to both the amyloid and NH2 regions of PrP. The two amyloid core antibodies showed the same bands, whereas after PK, even at 7× gel loads, no N-terminal PrP was detectable (Fig. 2_C_, lanes 7–11). The cell-type-specific PrP-res band patterns did not change with passage (Fig. 2 A). We also tested whether the different sizes of PrP-res bands in cells were due to differential glycosylation. This posttranslational modification has been proposed to encode strain-specific properties (18, 19), despite the fact that PrP deglycosylation alters neither the infectious titer nor the strain characteristics of a CJD agent (23). Fig. 2_C_, lanes 12–14, shows the complete deglycosylation of representative PrP-res samples. The higher _M_r bands in GT1 cells as compared with brain were due to increased glycosylation. There was only a single low-_M_r band after deglycosylation with PNGase. Lectin staining also verified the deglycosylation of PrP-res, but not of many other PK-resistant proteins (Fig. 6, which is published as supporting information on the PNAS web site). Despite agent strain differences, deglycosylated PrP-res was the same in FU- and SY-infected GT1–7 cells, as in brain. However, GT1 cells showed a 1- to 2-kDa lower _M_r than infected brain, presumably caused by sugar residues on PrP during PK digestions. Hence, it might be predicted, according to the prion hypothesis, that the agent strain passaged in GT1 cells should give rise to a variant agent strain when inoculated into mice.
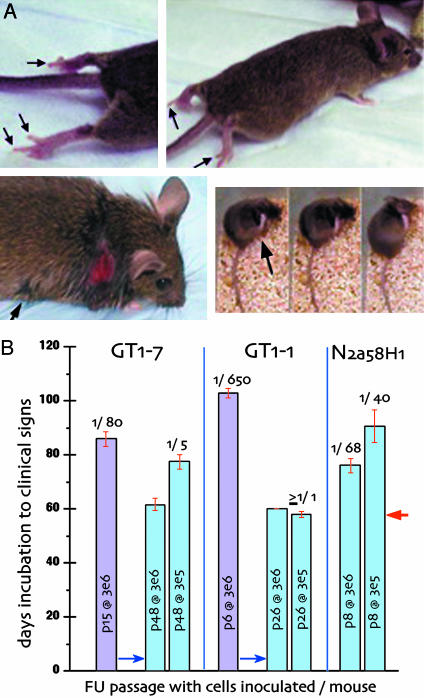
To determine infectivity titers and strain-specific characteristics, we inoculated high-PrP TGA20 indicator mice that succumb more rapidly to scrapie than do WT mice (22). Because end point titers of our CJD agents were the same in TGA20 as in WT CD-1 mice, TGA20 infectious doses were equivalent. As in scrapie, TGA20 mice inoculated with FU and SY brain homogenates also yielded a shorter incubation time than CD-1 mice, yet the large >200 day differences in incubation time between the FU and SY strains were maintained in TGA20 mice. With TGA20 mice, the incubation time to clinical signs was 60 days by using the maximal dose of FU brain (30 μl of a 1% homogenate). In the linear range of brain dilution (10–2 to 10–6) the infectious dose could be determined by using the curve fit y = 30067_e_ + 9·e_^(–0.12906_x) where y is the log infectious doses, and x is the incubation in days (R = 0.9988). This finding is equivalent to ≈17 days per log change (8), and was used to determine FU-infectious doses on a per cell basis for cultures at early and later passages. Infection with FU brain also provoked an unusual clinical syndrome with hind leg paresis in TGA20 mice, often before coat ruffling or hunching (Fig. 3_A_). This FU clinical sign is unique, and not seen in RML scrapie-infected TGA20 mice. Additionally, maximal doses of SY brain gave a very prolonged incubation time in TGA20 mice, with the same stereotypic scratching syndrome (Fig. 3_A_) as found in WT mice (15). SY also failed to induce the hind leg paresis of FU. Incubation time to SY disease was 318.2 ± 4.72 (SEM) and 315.3 ± 7.9 days in two independent experiments (n = 15) with maximal brain doses. Because SY has only 1/10,000th of FU brain infectivity, this incubation time bioassay was less precise, with ≈40 days per log dilution of brain homogenate.
Fig. 3.
TGA20 strain assays. (A) FU hind leg paresis (Upper). Note extension of hind digits and supine paw (dragged). (Lower) SY infection with stereotypic scratching leading to neck wound. Patches of scratched rough hair also elsewhere (e.g., arrow, Left). (Right) Three consecutive movie frames (<2 sec per frame) demonstrating rapid leg movement on neck (arrow) with rest of body and tail immobile during scratching. Mice are killed before the skin breaks, but in this case the wound rapidly developed overnight. (_B_) Infectivity of FU tissue cultures (GT1–7, GT1–1, and N2a58H1). Each bar shows the passage _in vitro_ (p) and the number of cells inoculated per mouse. Blue bars show determinations at two tenfold dilutions. The LD50 per cell is shown at the top of each bar. The minimal incubation time (red arrow) indicates of ≥3 × 105 LD50 per sample; and further cell dilutions may show significantly >1 LD50 per GT1–1 cell. Titers of FU were the same in TGA20 and WT CD-1 mice (see Materials and Methods).
All FU- and SY-infected tissue cultures yielded the behavior only of the parental FU or SY agent strain, despite the different patterns of PrP-res they evoked in culture. Fig. 3_B_ shows FU incubation times with their corresponding infectious doses per cell. The different cell passages assayed, and number of cells inoculated per mouse, are indicated in each bar, and the minimal time for clinical signs was 60 days (arrow) in some cultures. A 10-fold cell dilution was compared to verify infectious doses (blue bars). Although FU-infected GT1–7 cells (Left) displayed comparable amounts of PrP-res at p15 and p48, GT1–7 showed ≈15-fold more infectivity at p48 (1 LD50 per five cells). GT1–1 cells showed an even more dramatic change in infectivity. There was an ≈650-fold increase in infectivity between p6 and p26 (Center), yet PrP-res increased <2-fold at these passages (Fig. 1). Low PrP-res N2a58H1 cells also had equivalent infectivity as high-PrP-res GT1–7 cells at p15, and contained ≥10 times more infectivity than p6 GT1–1 cells expressing higher levels of PrP-res. Thus, whereas PrP-res did not accurately predict the infectious titer, it was a reasonable indicator of continued FU, but not SY replication. Most importantly, the slower growing GT1–1 cells had the same high titer as brain. Passaged GT1–1 cells infected with FU had at least 1 LD50 (or one infectious dose) per cell, which is equivalent to the assayed infectivity per cell in end stage FU brain. Preservation of high titers in FU-infected GT1 cells was also confirmed in later cell passages by using WT CD-1 mice. Similar homogenates from FU-infected GT1–1 cells at p70 as well as from FU GT1–7 cells at p95 also showed the same high titers as found in TGA20 mice.
SY inoculations showed persistent infection, despite the absence of detectable PrP-res in later passages. The SY-infected GT1–7 subline was infectious both at p15, when PrP-res was detectable, and also at p36 when it was not. Moreover, the incubation time was shorter in late-passage PrP-res-negative cells than in PrP-res-positive earlier subcultures (338 versus 370 days, or an ≈1 log increase in the negative cells). Both SY passages produced only the SY scratching syndrome, with no hind leg paresis. Thus, SY in culture also faithfully reproduced its strain characteristics after extended cell passages. Mice inoculated with GT1–1 control cells are still alive at >550 days, as would be expected for uninfected cells.
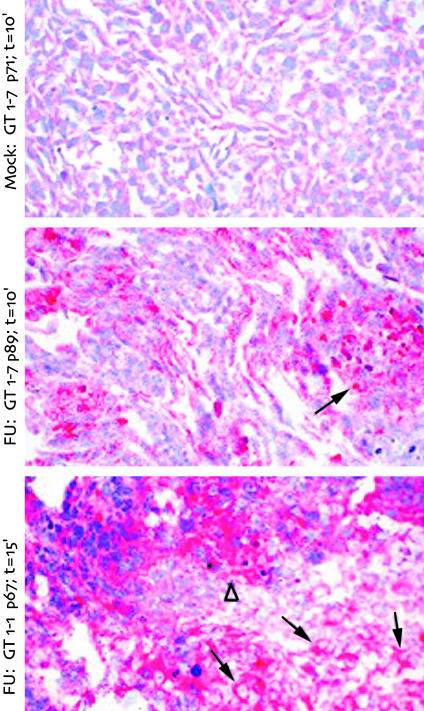
To visualize PrP-res in individual cells, and thereby estimate the percentage of infected cells, we evaluated cell pellet sections treated with trypsin. Limited trypsin digestion left only the amyloid core of pathologic PrP intact. Fig. 4 shows representative mock- and FU-infected cells probed with an antibody to the carboxyl portion of PrP-res. The N-terminal antibody, as in Western blots of Fig. 2_C_, showed no signal, indicating complete removal of nonamyloid portions of PrP (data not shown). Mock cells displayed no abnormal PrP-res aggregates (Top). In contrast, FU-infected GT1–7 and GT1–1 cells displayed abundant PrP-res aggregates in >30% of cells. In GT1–7 cells, PrP-res formed compact aggregates within the cytoplasm (arrow, Middle). Approximately 30% of the cells showed these PrP-res aggregates. This percent is probably an underestimate of infected cells because sections will not sample all such aggregates. FU-infected GT1–1 cells exhibited a strong, but more diffuse staining throughout the cytoplasm (arrows, Bottom), although a few cells showed the more compact PrP-res aggregates (▵). A high proportion of FU GT1–1 cells were PrP-res-positive (≥50%), and because cells were fixed in situ before scraping, the closely associated positive cells may reflect cell to cell spread of agent. There were a few more pyknotic nuclei in infected GT1 sublines as compared with mock controls, possibly due to high levels of infection and/or secondary to pathologic PrP-res accumulation, but the vast majority of cells were morphologically normal. Thus, these cells can be advantageous for agent-specific studies, because they are not visibly compromised by the degeneration found in end stage brain. Although not quantitative, these in situ results are consistent with previously determined ratios of ≈100,000 PrP-res molecules to each infectious dose (5).
Fig. 4.
In situ detection of PrP-res in mock- and FU-infected cell lines at the indicated passages (p). Mock cells produced no PrP-res (Top). GT1–7 cultures showed cytoplasmic aggregates of PrP-res (arrow, Middle). In GT1–1 intracellular PrP-res spread more diffusely (arrow) in many adjacent cells, possibly indicating cell-to-cell spread, except in a few cells with more aggregated deposits (▵). Only a few pyknotic dark blue nuclei are seen in infected cultures despite high PrP-res levels. Trypsin digestion times are indicated, and more extensive digestion in the GT1–1 line did not decrease the PrP-res signal.
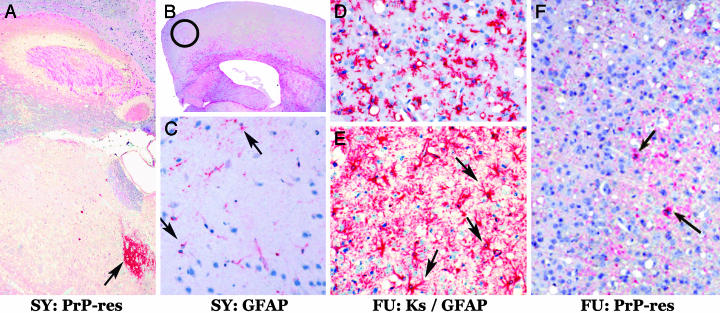
Mice inoculated with cell and brain homogenates produced essentially identical strain-specific lesions in TGA20 and WT CD-1 mice. In CD-1 mice, SY lesions are not found in the cerebrum, but are remarkably confined to the medial thalamus (15, 16). In contrast, FU induces vacuolization throughout many regions of the cerebrum (15, 16). TGA20 mice recapitulated these markedly different strain-specific patterns as shown in Fig. 5. Thalamic, but not cortical lesions, were produced by all SY inocula as representatively shown in Fig. 5 A–C, whereas all FU inocula produced severe cortical vacuolization with astrogliosis and massive accumulation of microglia (Fig. 5 D–F).
Fig. 5.
SY and FU agents breed true after extended in vitro propagation. Representative end stage neuropathology of SY (A–C) and FU (D–F) in TGA20 mice. (A) PrP antibodies show PrP-res only in the thalamus (red, arrow) but not the rest of the cerebrum. (B) Antibody to glial fibrillary acidic protein (GFAP) (21) shows no cortical astrogliosis (red) in SY mice. Red astrocytes, normally found in the white matter, are apparent. Circle denotes the same cortical region from different mice shown in C–F.(C) SY-infected GT1–7 cell homogenates produced no cortical vacuolization and only rare small GFAP-positive cells (arrows); the same result was obtained by using SY brain homogenates. Microglia, assessed by keratan sulfate antibody (Ks) staining (21), were not found with SY inocula (data not shown). (D) FU infection elicited many keratan sulfate-positive (red) microglia after inoculation of FU-infected GT1–7 cell homogenates. Note the many vacuoles. The pathology was the same by using FU GT1–1, FU N2a58H1, or FU brain homogenates. (E) FU infection also elicited intense GFAP staining (red) of many astrocytic fibers as well as hypertrophic (gemistocytic astrocyte) cell bodies (arrows). This massive astrocytosis was apparent by the naked eye in all FU infections regardless of source (cells or brain). (F) Many cortical vacuoles are again seen after FU GT1–1 inocula. All FU-infected TGA20 mice displayed only a few small deposits of PrP-res, which contrasts with the frequent larger deposits in CD-1 mice (15, 16). TGA20 mice also show a higher (red) background of normal PrP because they overexpress this protein by 8-fold versus WT mice (22). The cytological reduction in PrP-res deposits was also confirmed by a 4-fold reduction of PrP-res per mg of brain in TGA20 versus WT mice by Western blotting (data not shown). Nevertheless, vacuolization was comparable in TGA20 and CD-1 mice, and spongiform change does not invariably correspond to PrP-res levels (21).
Discussion
We have achieved robust CJD infections of cell lines, obtaining uniquely high levels of CJD agents. Persistent infection was established in three different cell lines by using the FU CJD agent. The less virulent SY agent successfully infected the GT1–7 subline. Both CJD agents evoked detectable pathologic PrP-res only after five or more tissue culture passages. Whereas PrP-res indicated persistent infection, the amount was not quantitatively related to infectivity. In particular, PrP-res remained constant, and did not reflect the substantial (650-fold) increase in infectivity with prolonged passages of FU. The levels of FU infectivity after extended passage in the GT1–1 subline were of the same order of magnitude as found in FU-diseased brain (1 LD50 per cell), and were 400-fold higher than in the only other CJD-infected culture titered in animals (24). Infectivity here was also ≥100-fold higher than that found in most comparable murine scrapie-infected cultures (3, 7, 24).
As in the brain, SY and FU PrP-res banding and glycosylation patterns were indistinguishable in infected monotypic GT1–7 cells. In sharp contrast, the PrP-res patterns of the each different cell type (GT1, N2a58, and brain) infected with the single FU agent yielded its own unique PrP-res pattern. Thus, the PrP-res band profile depended on the cell type rather than the agent strain. Despite the marked changes in PrP-res, and continuous long-term passage of agent in cells with distinctive PrP-res profiles, the infectious agents produced only the clinical syndrome and incubation time of the parental strain. Clearly, the agent strain bred true, whereas the murine PrP-res pattern did not. This finding, among others reviewed (1), strongly militates against the proposal that PrP secondary and tertiary conformation, as indicated by PrP-res band patterns, somehow encodes the agent strain (17–19). It also seems unlikely that sugar residues encode agent properties because deglycosylation of infectious CJD brain fractions had no effect on agent strain characteristics (23), and different PrP glycoforms here discriminated only the cell type, but not the FU and SY strains.
The foregoing culture results are entirely consistent with agent characteristics being independent of PrP. These results include classic experiments showing the preservation of strain-specific scrapie information after serial propagation in unrelated species (25). Sheep scrapie strains propagated in mice, rats, and hamsters (all with different PrPs), reproduced their identical strain-specific phenotype when reinoculated into sheep. The incubation times, clinical signs, and neuropathology were the same as those seen with the original sheep agent. Because material from mouse had also been serially passaged, the authors were able to rule out residual sheep material in these donor mouse brains. Other strains have also bred true after extended passages in a foreign species (26). Moreover, the epidemic U.K. bovine spongiform encephalopathy agent from cows, humans, domestic cats, nyala, and kudu all reproduce the same bovine spongiform encephalopathy-specific lesion profile and transmission characteristics in congenic mice, i.e., the bovine spongiform encephalopathy agent also maintains host independent characteristics (27). Although we cannot inoculate humans with our passaged CJD agents, we have tested our FU strain after multiple mouse passages by reinoculation into rats. FU has remained remarkably stable, giving the unique neuropathology as found in its first human-to-rat transmission (L.M., unpublished data). Additionally, FU passaged in mice continues to elicit an early innate immune response well before PrP-res is detected (28), and this finding implicates infection by a foreign pathogen rather than host protein. All of these data are difficult to reconcile with any version of the prion hypothesis, or the assumption that a transmissible spongiform encephalopathy strain must lose its individual identity after passage in a different species.
Many additional experiments have demonstrated separation of most pathologic PrP from infectious particles (1, 29), and over the last 20 years no form of PrP itself has ever reproduced infection in normal animals (1). Hence, one cannot exclude a viral agent, particularly because intact viruses are often resistant to nuclease digestion, and intact, but not CJD-specific, endogenous retroviral particles cosediment with CJD infectivity after nuclease treatment (29). Furthermore, most PrP can be dissociated from infectivity, whereas treatments that disrupt cosedimenting viral particles reduce CJD infectivity by >99.5% (29). However, it is the biology of these agents: their evolution (21), spread, cell specificity, latency, virus-like interference capabilities (15, 16, 30), and occasional mutation (31), as well as the principle of parsimony, which continues to indicate a viral causative agent.
What then is the function of pathologic PrP in infection and why does it accumulate? Clearly, some PrP is needed for agent replication because PrP-null mice are resistant to infection (32). Host PrP may serve as a receptor (33) or docking site essential for agent replication or completion of its life cycle (34), but other required cell-specific factors remain unknown. Whereas PrP-res levels do not reliably predict infectivity titers, they can indicate persistent infection. On the other hand, myeloid microglia have high levels of infectivity, but only marginally detectable PrP (8), and the level of cellular PrP needed for infection remains unknown. The infected cell lines here displayed abundant cytoplasmic PrP-res during robust agent replication, implicating agent interaction with PrP-rich intracellular membranes. Thus, pathological PrP may be the end result of a host defense mechanism that ultimately traps infectious particles in an insoluble mass of amyloid junk (33, 34). This conclusion seems plausible because infectious agent is released when PrP amyloid is disaggregated (1, 29). A PrP-trapping mechanism may also protect these agents from clearance and detection during cell-to-cell surface spread (35). Many viruses subvert antibody recognition by assuming a cloak of host membrane proteins.
The propagation of CJD strains in monotypic cell lines can simplify discovery of cellular genes that are up- or down-regulated in comparison with mock controls. The study of purified cell types, such as microglia, has already revealed that many genes involved in innate immunity are activated by CJD infection (8, 28, 36). Similar gene array and RT-PCR studies of neuroectodermal cells in vitro may uncover additional diagnostic markers of CJD infection. These or similar cell lines may be useful for rapid assessment of human CJD tissues as well as for drug screening. More importantly, the absence of profound degenerative changes in infected neuronal lines, as compared with brain, should simplify detection of intrinsic agent components. High-yield CJD cell lines are likely to provide a better substrate for separation and analysis of agent nucleic acids, which we presume encode specific strains.
Supplementary Material
Supporting Figure
Acknowledgments
We thank Sheldon Penman for editing and clarifying the manuscript, and Mark Chernyak for assistance with immunocytochemistry. This work was supported by National Institutes of Health Grants NS 12674 and NS 34569, and Department of Defense Grant DAMD17-03-1-0360.
Abbreviations: CJD, Creutzfeldt–Jakob disease; PrP, prion protein; Prp-res, resistant PrP bands; PK, proteinase K; p_n_, passage in vitro n; GFAP, glial fibrillary acidic protein.
References
- 1.Manuelidis, L. (2003) Viral Immunol. 16**,** 123–139. [DOI] [PubMed] [Google Scholar]
- 2.Manuelidis, E. E., Fritch, W. W., Kim, J. H. & Manuelidis, L. (1987) Proc. Natl. Acad. Sci. USA 84**,** 871–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Race, R. (1991) Curr. Top. Microbiol. Immunol. 172**,** 181–193. [DOI] [PubMed] [Google Scholar]
- 4.Manuelidis, L., Murdoch, G. & Manuelidis, E. (1988) Ciba Found. Symp. 135**,** 117–134. [DOI] [PubMed] [Google Scholar]
- 5.Manuelidis, L. & Fritch, W. (1996) Virology 215**,** 46–59. [DOI] [PubMed] [Google Scholar]
- 6.Rubenstein, R., Carp, R. & Callahan, S. (1984) J. Gen. Virol. 65**,** 2191–2198. [DOI] [PubMed] [Google Scholar]
- 7.Borchelt, D., Scott, M., Taraboulos, A., Stahl, N. & Prusiner, S. (1990) J. Cell Biol. 743–752. [DOI] [PMC free article] [PubMed]
- 8.Baker, C. A., Martin, D. & Manuelidis, L. (2002) J. Virol. 10905–10913. [DOI] [PMC free article] [PubMed]
- 9.Caughey, B., Neary, K., Buller, R., Ernst, D., Perry, L. L., Chesebro, B. & Race, R. E. (1990) J. Virol. 64**,** 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Priola, S., Caughey, B., Raymond, G. & Chesebro, B. (1994) Infect. Agents Dis. 2–3**,** 54–58. [PubMed] [Google Scholar]
- 11.Vey, M., Pilkuhn, S., Wille, H., Nixon, R., DeArmond, S. J., Smart, E. J., Anderson, R. G. W., Taraboulos, A. & Prusiner, S. B. (1996) Proc. Natl. Acad. Sci. USA 93**,** 14945–14949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mange, A., Nishida, N., Milhavet, O., McMahon, H. E. M., Casanova, D. & Lehmann, S. (2000) J. Virol. 74**,** 3135–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bosque, P. J. & Prusiner, S. B. (2000) J. Virol. 74**,** 4377–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishida, N., Harris, D. A., Vilette, D., Laude, H., Frobert, Y., Grassi, J., Casanova, D., Milhavet, O. & Lehmann, S. (2000) J. Virol. 74**,** 320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manuelidis, L. (1998) Proc. Natl. Acad. Sci. USA 95**,** 2520–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manuelidis, L. & Lu, Z. Y. (2000) Neurosci. Lett. 293**,** 163–166. [DOI] [PubMed] [Google Scholar]
- 17.Prusiner, S. B. (1997) Science 278**,** 245–251. [DOI] [PubMed] [Google Scholar]
- 18.Aguzzi, A. & Weissmann, C. (1997) Nature 389**,** 795–798. [DOI] [PubMed] [Google Scholar]
- 19.Collinge, J., Sidle, K., Meads, J., Ironside, J. & Hill, A. (1996) Nature 383**,** 685–667. [DOI] [PubMed] [Google Scholar]
- 20.Sklaviadis, T., Manuelidis, L. & Manuelidis, E. E. (1986) Proc. Natl. Acad. Sci. USA 83**,** 6146–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manuelidis, L., Fritch, W. & Xi, Y. G. (1997) Science 277**,** 94–98. [DOI] [PubMed] [Google Scholar]
- 22.Fischer, M., Rulicke, T., Raeber, A., Sailer, A., Moser, M., Oesch, B., Brandner, S., Aguzzi, A. & Weissmann, C. (1996) EMBO J. 15**,** 1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 23.Manuelidis, L., Sklaviadis, T. & Manuelidis, E. E. (1987) EMBO J. 6**,** 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butler, D., Scott, M., Bockman, J., Borchelt, D., Taraboulos, A., Hsiao, K., Kingsbury, D. & Prusiner, S. (1988) J. Virol. 62**,** 1558–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zlotnik, I & Rennie, J. C. (1965) J. Comp. Pathol. 75**,** 147–157. [DOI] [PubMed] [Google Scholar]
- 26.Kimberlin, R. H., Walker, C. A. & Fraser, H. (1989) J. Gen. Virol. 70**,** 2017–2025. [DOI] [PubMed] [Google Scholar]
- 27.Bruce, M. (2003) Br. Med. Bull 66**,** 99–108. [DOI] [PubMed] [Google Scholar]
- 28.Baker, C., Lu, Z. & Manuelidis, L. (2004) J. Neurovirol. 10**,** 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manuelidis, L., Sklaviadis, T., Akowitz, A. & Fritch, W. (1995) Proc. Natl. Acad. Sci. USA 92**,** 5124–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manuelidis, L. & Lu, Z. Y. (2003) Proc. Natl. Acad. Sci. USA 100**,** 5360–5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruce, M. E. & Dickinson, A. G. (1987) J. Gen. Virol. 68**,** 79–89. [DOI] [PubMed] [Google Scholar]
- 32.Büeler, H., Aguzzi, A., Sailer, A., Greiner, R.-A., Autenried, P., Auget, M. & Weissmann, C. (1993) Cell 73**,** 1339–1347. [DOI] [PubMed] [Google Scholar]
- 33.Manuelidis, L. (1994) Transfusion 34**,** 915–928. [DOI] [PubMed] [Google Scholar]
- 34.Manuelidis, L. (1997) Ann. Inst. Pasteur (Paris) 8**,** 311–326. [Google Scholar]
- 35.Manuelidis, L., Zaitsev, I., Koni, P., Lu, Z.-Y., Flavell, R. & Fritch, W. (2000) J. Virol. 74**,** 8614–8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baker, C. & Manuelidis, L. (2003) Proc. Natl. Acad. Sci. USA 100**,** 675–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figure