Shadow Enhancers as a Source of Evolutionary Novelty (original) (raw)
. Author manuscript; available in PMC: 2014 Dec 5.
Published in final edited form as: Science. 2008 Sep 5;321(5894):1314. doi: 10.1126/science.1160631
The dorsal-ventral patterning of the Drosophila embryo is controlled by Dorsal, a sequence-specific transcription factor that is related to mammalian NF-κB. Previous ChIP-chip assays predicted that as many as a third or even half of all Dorsal target genes contain multiple enhancers for the same or similar expression pattern. Here we show that some of these secondary enhancers, or “shadow enhancers”, produce gene expression patterns that overlap those produced by the primary enhancers in transgenic embryos. We suggest that shadow enhancers help ensure the precision of embryonic patterning and discuss their significance in the evolution of genetic novelty.
The dorsal-ventral patterning of the early Drosophila embryo is controlled by a sequence-specific transcription factor, Dorsal, which is related to mammalian NF-κB (1). Dorsal works in concert with the products encoded by two of its target genes, Twist and Snail, to regulate gene expression in the early embryo. ChIP-chip assays identified a few hundred binding clusters for Dorsal, Twist, and Snail scattered throughout the Drosophila genome (2). Over 35 of these clusters function as authentic enhancers when tested in transgenic embryos.
ChIP-chip assays predicted that many of the Dorsal target genes contain two separate enhancers for the same or similar expression pattern. This prediction was experimentally confirmed for vnd and miR-1 (2). vnd contains two enhancers that mediate expression in the presumptive neurogenic ectoderm, while miR-1 contains at least two enhancers for expression in the ventral mesoderm. In both cases, the secondary enhancers map within 5 kb of the transcription start site.
However, some of the potential secondary enhancers identified by the ChIP-chip assays are predicted to map quite far from Dorsal target genes. For example, brinker (brk) is regulated by a known enhancer located in the 5′ flanking region (3). A potential secondary enhancer maps within the major intron of a neighboring gene, Atg5, located ~13 kb downstream of the brk transcription start site. A ~1 kb genomic DNA fragment encompassing the Atg5 intron was tested for enhancer activity in transgenic embryos (Fig. 1A). It directs broad lateral stripes of lacZ reporter gene expression, similar to the endogenous brk expression pattern that is recapitulated by the previously identified 5′ enhancer.
Figure 1.
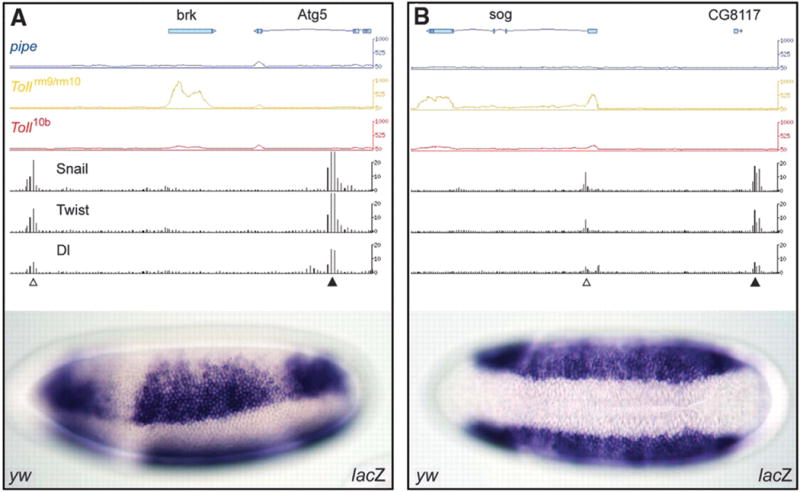
Identification of shadow enhancers. (A) Genome browser showing the Brinker locus (brk) and neighboring gene, Atg5 (http://flybuzz.berkeley.edu/cgibin/gbrowse/fly4_3/). The first three lines from the top, blue, yellow and red, show the levels of steady-state RNAs in the pipe, Tollrm9/rm10 and _Toll_10b mutants, respectively. brk transcripts are absent in pipe and _Toll_10b mutants, but present in Tollrm9/rm10 mutants. Atg5 is inactive in all mutants, suggesting that the intronic enhancer is dedicated to brk regulation. The last three lines show the distribution of Snail, Twist, and Dorsal based on whole-genome ChIP-chip assays. The leftmost cluster (open arrowhead) coincides with the known, primary enhancer in the 5′ flanking region. A second cluster (filled arrowhead) is detected 13 kb downstream of the brk transcription start site within Atg5. A ~1 kb genomic DNA fragment encompassing the 3′ binding cluster (shadow enhancer) was tested in transgenic embryos (filled arrowhead). The 5′ enhancer was tested previously (5). Both fragments function as authentic enhancers to generate lateral stripes of gene expression in the neurogenic ectoderm.
(B) Same as A, except that the sog locus is shown. sog transcripts are predominantly detected in Tollrm9/rm10 mutants where the gene is fully active (see the blue, yellow and red lines, which show the results of the whole-genome tiling arrays). ChIP-chip assays identify two clusters of Dorsal, Twist, and Snail binding sites, within intron 1 and 3′ of the neighboring gene, CG8117. Both genomic DNA fragments function as authentic enhancers to direct lateral stripes of gene expression. Gene prediction models are displayed above each graphical presentation. Each 3′ end is labeled with triangle.
A similar situation is seen for the Dorsal target gene, sog. Bioinformatics methods identified an intronic enhancer that recapitulates the normal sog expression pattern in the presumptive neurogenic ectoderm (3). ChIP-chip assays identified this enhancer, as well as a second cluster of Dorsal, Twist, and Snail binding sites located 20 kb 5′ of the sog transcription start site, downstream of a neighboring gene (Fig. 1B). The newly identified binding cluster generates broad lateral stripes of gene expression in transgenic embryos, similar to those produced by the intronic enhancer. The shadow enhancers identified in this study are almost certainly dedicated to the regulation of the brk and sog transcription units since the associated genes, CG8117 and Atg5, respectively, are not significantly expressed in the early embryo (Fig. 1; supplemental Fig. 1).
ChIP-chip assays suggest that as many as a third or even half of all Dorsal target genes might be regulated by secondary enhancers (Supporting online Tables S1 and S2). We propose the term “shadow enhancer” for remote secondary enhancers mapping far from the target gene and mediating activities overlapping the primary enhancer. Phylogenetic comparisons suggest that the brk and sog shadow enhancers are evolving nearly twice as fast as the primary enhancers mapping within or near the two genes (supplemental Fig. 2). Despite these different rates of divergence, the overall structures of the shadow enhancers are clearly related to their respective primary enhancers (supplemental Fig. 3). Given the conservation of the shadow enhancers in all 12 sequenced drosophilids, it is likely that they are essential for fitness.
Why are Dorsal target genes regulated by shadow enhancers? They might help ensure precise and reproducible patterns of gene expression during embryogenesis. It is possible that shadow enhancers are pervasively used in animal development. For example, the mouse sonic hedgehog gene is regulated in the floorplate of the embryonic neural tube by two separate enhancers with slightly distinct activities (4). Shadow enhancers can explain why deletions of well-defined enhancers sometimes produce no apparent phenotypes (e.g., 5).
The evolution of _cis_-regulatory DNAs is a major mechanism of animal diversity (e.g., 6). However, there is the potential problem that such change could compromise essential genetic activities. Shadow enhancers have the potential to evolve novel binding sites and achieve new regulatory activities without disrupting the core patterning functions of key developmental control genes.
Supplementary Material
Supplemental Data
References
- 1.Stathopoulos A, et al. Cell. 2002;111:687. doi: 10.1016/s0092-8674(02)01087-5. [DOI] [PubMed] [Google Scholar]
- 2.Zeitlinger J, et al. Genes Dev. 2007;15:385. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Markstein M, et al. Proc Natl Acad Sci USA. 2002;99:763. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeong Y, et al. Development. 2006;133:761. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- 5.Xiong N, Kang C, Raulet DH. Immunity. 2002;16:453. doi: 10.1016/s1074-7613(02)00285-6. [DOI] [PubMed] [Google Scholar]
- 6.Jeong S, et al. Cell. 2008;132:783. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 7.This study was funded by the NIH (GM46638) and Moore Foundation.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data