The RAB5-GEF Function of RIN1 Regulates Multiple Steps During Listeria monocytogenes Infection (original) (raw)
. Author manuscript; available in PMC: 2015 Jan 2.
Published in final edited form as: Traffic. 2014 Sep 4;15(11):1206–1218. doi: 10.1111/tra.12204
Abstract
Listeria monocytogenes is a food-borne pathogenic bacterium that invades intestinal epithelial cells through a phagocytic pathway that relies on activation of host cell RAB5 GTPases. L. monocytogenes must subsequently inhibit RAB5, however, in order to escape lysosome-mediated destruction. Relatively little is known about upstream RAB5 regulators during L. monocytogenes entry and phagosome escape processes in epithelial cells. Here we identify RIN1, a RAS effector and RAB5-directed GEF, as a host cell factor in L. monocytogenes infection. RIN1 is rapidly engaged following L. monocytogenes infection and is required for efficient invasion of intestinal epithelial cells. RIN1-mediated RAB5 activation later facilitates the fusion of phagosomes with lysosomes, promoting clearance of bacteria from the host cell. These results suggest that RIN1 is a host cell regulator that performs counterbalancing functions during early and late stages of L. monocytogenes infection, ultimately favoring pathogen clearance.
Keywords: Listeria monocytogenes, MET, RIN1, RAB5, phagosome
INTRODUCTION
Invasive bacterial pathogens gain entry by manipulating host cell endocytic machinery (1). Bacterial proteins commandeer host proteins, referred to as host cell factors, throughout the infection process. A variety of host cell factors are required for entry, downstream events controlling survival, replication and spread. Depending on the pathogen, host cell entry is accomplished through macropinocytosis (2), clathrin-dependent endocytosis (3, 4) or caveolin-mediated uptake (5). The entry processes are predominantly actin-dependent (6), reflecting the need for cytoskeletal remodeling to accommodate large cargo (7). Although various genera of invasive bacteria employ distinct sets of host cell factors for entry into host cells, there is a striking commonality in their reliance on small GTPases (8, 9).
Listeria monocytogenes is a Gram-positive, food-borne pathogen that invades intestinal epithelial cells, spreads laterally through the gut epithelium and traverses the intestinal barrier, eventually disseminating to distal organs (10). In immune-compromised individuals L. monocytogenes can cause severe listeriosis with symptoms ranging from gastroenteritis to bacterial meningitis, and mortality rates of approximately 30% (11). In pregnant women, L. monocytogenes can lead to spontaneous abortions and neonatal infections (10).
L. monocytogenes uses two surface internalin (Inl) proteins to bind epithelial host cells, and utilizes clathrin-dependent mechanisms to enter these non-professional phagocytes (4). InlA interacts with CDH1 (E-cadherin) (12) while InlB interacts with MET, a receptor tyrosine kinase (13). Either internalin is sufficient for epithelial cell invasion, although both are needed for optimum entry efficiency (14). Engagement with InlB stimulates MET, leading to the recruitment and activation of signal transduction proteins including RAS (15, 16). L. monocytogenes infection triggers activation of the downstream RAS effectors PI3K and RAF (15, 17), but other RAS effectors commonly activated following MET stimulation have not been studied in this context. L. monocytogenes attachment also triggers RAB5 activation through an unknown mechanism, a step required for efficient internalization by receptor-mediated phagocytosis (18, 19).
Host cells mount a bactericidal response that also employs RAB5, in this case to fuse phagosomes with lysosomes and destroy the internalized bacterium before it can replicate (20, 21). To avoid this fate, L. monocytogenes uses a cytolysin to escape the phagosome and enter the cytoplasm (22) where replication takes place. The L. monocytogenes surface protein ActA promotes actin polymerization to cloak bacteria from the host autophagic clearance systems and to propel bacteria through the cytoplasm to facilitate protrusive entry into adjacent cells (23, 24). To allow time for phagosomal escape, the L. monocytogenes GAPDH protein ADP ribosylates RAB5, rendering this GTPase non-responsive to activation by guanine nucleotide exchange factors (GEFs) (21, 25). RAB5 subjugation is essential for L. monocytogenes to escape into the cytosol and replicate (21). Hence, while RAB5 facilitates invasion it subsequently promotes bacterial killing in phagolysosomes. This requires L. monocytogenes to switch from promoting to suppressing RAB5 activity for a successful infection. Little is known, however, about the role of host cell RAB5 regulators during L. monocytogenes invasion and spread.
RIN1 is a RAS effector involved in receptor tyrosine kinase endocytosis and trafficking (26, 27). Through its VPS9 domain, RIN1 functions as a GEF with specificity for RAB5 GTPases, promoting internalization and degradation of activated receptors (28–30). RIN1 also binds and activates ABL non-receptor tyrosine kinases that regulate actin cytoskeleton remodeling (31). An intricate balance between these two RIN1 effectors (RAB5 and ABL) determines the rate and route of receptor internalization. (27, 32). The RAB5-GEF activity of RIN1 is exerted independently of RABGEF1 (a.k.a. Rabex5), another RAB5 GEF regulating endocytic processes in the cell (33).
Because L. monocytogenes uses a growth factor receptor tyrosine kinase to enter host cells (15, 34), and because RAS proteins signal through RIN1 to regulate RAB5 during endocytosis (27, 29), we examined whether RIN1 functions as a host cell factor for L. monocytogenes. We found that RIN1 facilitates L. monocytogenes intestinal epithelial cell entry through its RAB5-GEF function and that loss of RIN1 impaired invasion. RIN1 plays a strikingly different role post-invasion by accelerating RAB5-dependent fusion of L. monocytogenes containing phagosomes with lysosomes.
RESULTS
MET-mediated L. monocytogenes invasion is facilitated by RIN1-mediated activation of RAB5
Engagement of L. monocytogenes InlB with host cell MET stimulates receptor tyrosine kinase activity, leading to activation of RAS proteins and the downstream MAP kinase cascade (15). We tested whether L. monocytogenes binding also activates the RAS effector RIN1 in HeLa cells, a human cervical cancer cell line widely used as a model for epithelial cell invasion. Following growth factor stimulation by RAS, the RIN1 protein becomes phosphorylated by ABL tyrosine kinases (27). We observed the same signaling mark (RIN1-pY36) as early as 2.5 minutes following addition of L. monocytogenes to HeLa cells (Figure 1A, Figure S1A). Phosphorylation was ABL-dependent, as judged by reduced phosphorylation in the presence of the ABL-specific kinase inhibitor imatinib. This result suggests that RIN1 signaling is engaged early in the process of host cell invasion by InlB binding to MET.
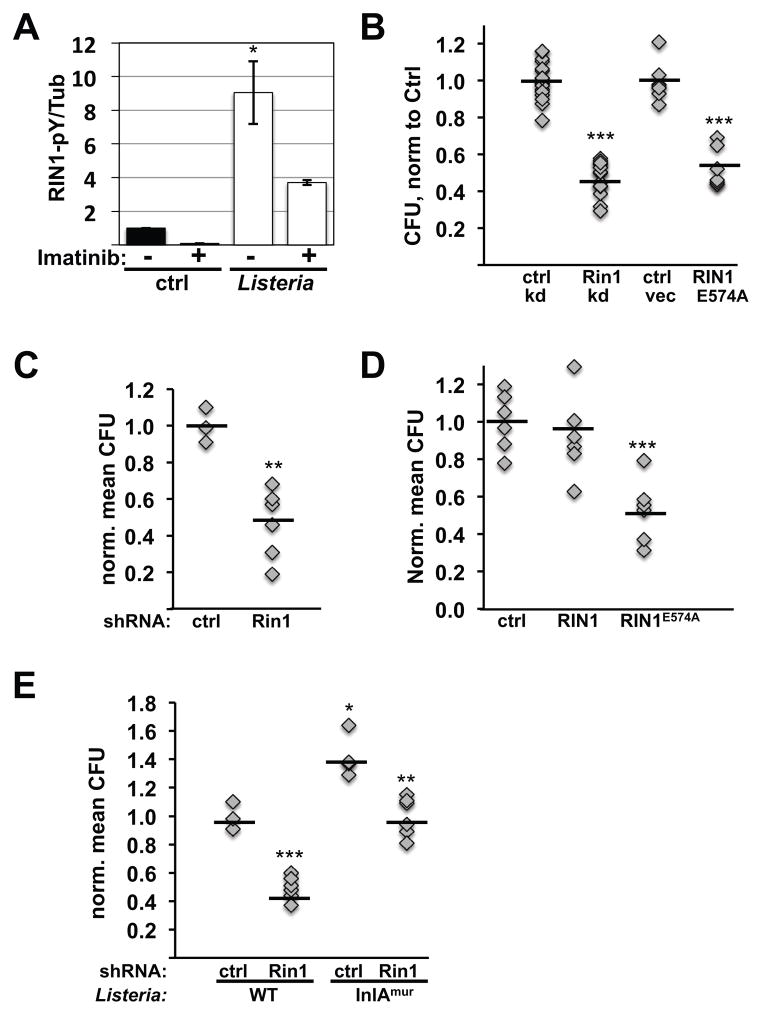
FIGURE 1. RIN1-to-RAB5 signaling is required for efficient L. monocytogenes entry into epithelial cells.
A. HeLa cells were infected with L. monocytogenes (500 MOI; 2.5 min) and lysates probed with anti-RIN1pY36, anti-RIN1 and anti-tubulin (Tub). A short time and high MOI were used to detect the transient phosphorylation that occurs upon infection. RIN1pY36 levels were normalized to total RIN1 and tubulin (Figure S1). The ABL tyrosine kinase inhibitor imatinib was used at 5μM. n=2 experiments, p=0.03. B. Control, Rin1 shRNA (Rin1 kd) or RIN1E574A transduced IEC-18 cells were infected at 50 MOI for 1 hour and extracellular bacteria killed using gentamicin. Cell lysate dilutions were plated to determine colony-forming units (CFUs) (ctrl-kd n=17, Rin1-kd n=16, 4 independent experiments, p=1.8×10−16; ctrl-vec and RIN1E574A n=6, 2 independent experiments, p=2.6×10−6). C. Invasion assay (as in B) using IEC-6 Cdx2 cells transduced with control or Rin1-shRNA. Cell lysate dilutions were plated to obtain CFUs (ctrl n=3, Rin1-kd n=6, p=0.003). D. IEC6-Cdx2 cells transduced with control, RIN1 or RIN1E574A vectors were subjected to invasion assays as in B and CFUs determined (ctrl n=9, RIN1 and RIN1E574A n=10, 2 independent experiments, p=2.3×10−6). E. Control or Rin1-shRNA transduced IEC-6 Cdx2 cells were infected (10 MOI) with wild type or InlAmur L. monocytogenes and extracellular bacteria killed using gentamicin. Cell lysate dilutions were plated to obtain CFUs (ctrl n=4, Rin1-kd n=6; wild type L. monocytogenes p=6.7×10−5, InlAmur p=0.002). InlAmur L. monocytogenes invaded more efficiently than wild type (p=0.009). Note: Rin1=rat protein; RIN1=human protein. (*p ≤ 0.05, **p ≤ 0.005, ***p ≤ 0.0005)
We next sought to determine whether RIN1 was necessary for cell invasion by L. monocytogenes. For infection assays, we initially used IEC-18, a rat intestine epithelial cell line that resembles epithelial cells of the intestinal barrier that L. monocytogenes invades in vivo (35). Rin1 silencing significantly reduced the efficiency at which L. monocytogenes invaded IEC-18 cells (Figure 1B, Figure S1B). A similar effect was seen using IEC-6 Cdx2, a rat intestinal epithelial cell line that stably over-expresses the homeobox domain transcription factor, Cdx2 (36). These cells show enhanced polarization with tight and adherens junctions, making them an especially useful model of an intestinal epithelium. Rin1-silenced IEC-6 Cdx2 cells were also compromised in their ability to be invaded by L. monocytogenes (Figure 1C, Figure S1D). The same effect of RIN1 silencing was seen in HeLa cells (Figure S2A). These findings implicate RIN1 as a host cell factor employed during L. monocytogenes invasion.
Given the established requirement for RAB5 activation during L. monocytogenes invasion (18) we next tested whether the RAB5-GEF function of RIN1 contributes to invasion efficiency. Expression of a GEF domain mutant that prevents RIN1-mediated RAB5 activation, RIN1E574A, significantly decreased L. monocytogenes invasion in all three cell lines tested (Figures 1B, 1D and Figures. S1C, S1E, S2B). In contrast, a mutation that disrupts RIN1 signaling through ABL tyrosine kinases, RIN1QM (37), did not affect L. monocytogenes entry into IEC-18 cells (Figure S2C). To rule out the possibility that other RAB5-GEFs were reduced as a side effect of RIN1 knockdown, we assessed the levels of Rabgef1, a ubiquitous RAB5 GEF also called Rabex5, and Rin2, a RIN1 paralog. Protein levels of Rabgef1 and Rin2 were unaltered in Rin1 silenced IEC-6 Cdx2 cells (Figure S2D). These results suggest that MET-dependent L. monocytogenes entry is facilitated by RIN1-mediated activation of RAB5. Notably, RIN1 over-expression had little effect on L. monocytogenes entry into these cells (Figs. 1D, S2B, S2C), possibly because another host cell factor is rate limiting for invasion.
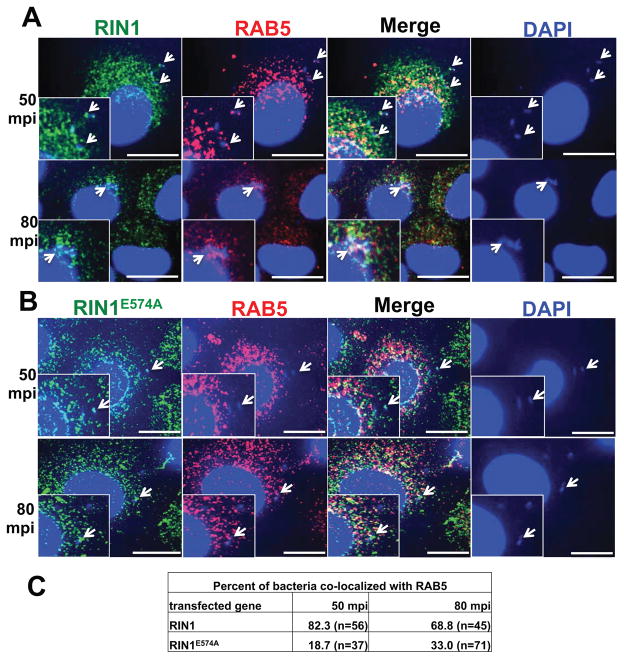
Having shown that RAB5 activation by RIN1 in epithelial cells is required for efficient L. monocytogenes invasion, we reasoned that RIN1 and RAB5 could potentially be recruited to the host cell surface and phagosomes during bacterial entry. We visualized RIN1 and Rab5 at 50 and 80 minutes post infection (mpi). We observed co-localization of both proteins with bacteria during early (50 mpi) as well as later (80 mpi) stages of entry into IEC-6 Cdx2 cells expressing wild type RIN1 (Figure. 2A, C). Recruitment of Rab5 was diminished, however, in IEC-6 Cdx2 cells expressing RIN1E574A (Figure. 2B, C). These observations provide further support for the role of RIN1 as a host cell factor needed for efficient RAB5 activation during L. monocytogenes entry into epithelial cells.
FIGURE 2. RIN1 and RAB5 are recruited to L. monocytogenes during invasion.
A and B. IEC-6 Cdx2 cells transduced with RIN1 (A) or RIN1E574A (B) were plated on collagen, infected with L. monocytogenes at MOI 100 for 50 or 80 minutes at 37°C. The cells were washed thoroughly, fixed and stained for RIN1 (green) and RAB5 (red). Host cell nuclei and L. monocytogenes were visualized by DAPI (blue). Arrows show RAB5 and RIN1 recruitment to L. monocytogenes. Insets show magnified versions of the regions pointed by the arrows. Scale bar: 10 μm. C. Quantification of percentage total bacteria co-localizing with RAB5 at each time point in RIN1 and RIN1E574A IEC-6 Cdx2 cells, representative of two independent experiments.
Dual receptor-mediated entry also relies on host cell RIN1
L. monocytogenes entry into mammalian cells is mediated by two distinct ligands, InlA and InlB, which bind to host cell CDH1 (E-cadherin) and MET, respectively (12, 16). Because of sequence variation between human CDH1 and rodent Cdh1 proteins, L. monocytogenes InlA does not recognize mouse or rat epithelial cells. In order to examine the course of L. monocytogenes invasion involving both host surface receptors, as occurs during the normal course of infection, we utilized a murinized version of the bacterium (L. monocytogenes InlAmur). This previously characterized strain shows enhanced affinity to mouse and rat Cdh1 and consequently more efficient internalization (38). As expected, InlAmur L. monocytogenes invaded IEC-6 Cdx2 cells more efficiently than the wild type strain (Figure 1E). Rin1 silencing still decreased invasion efficiency, however, demonstrating the importance of RIN1 signaling even when both host cell receptors are available and compatible.
RIN1 accelerates the host cell bactericidal response
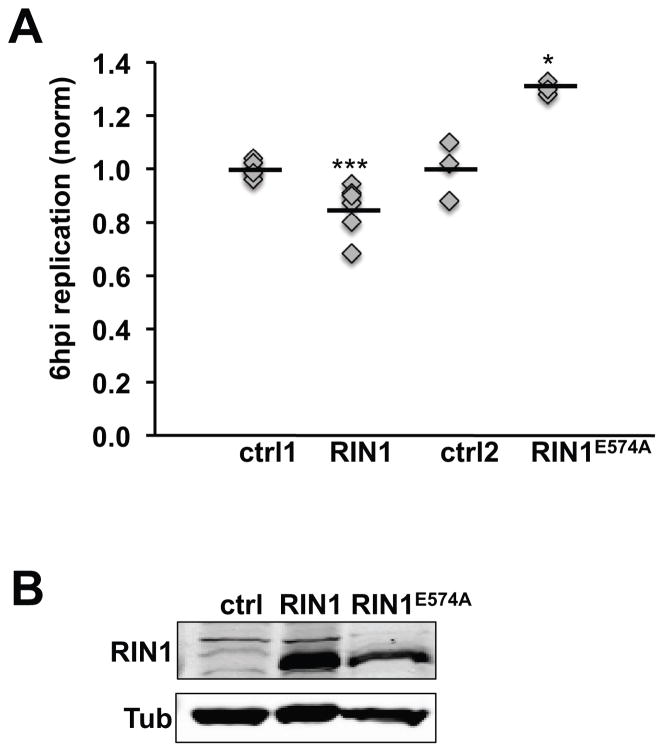
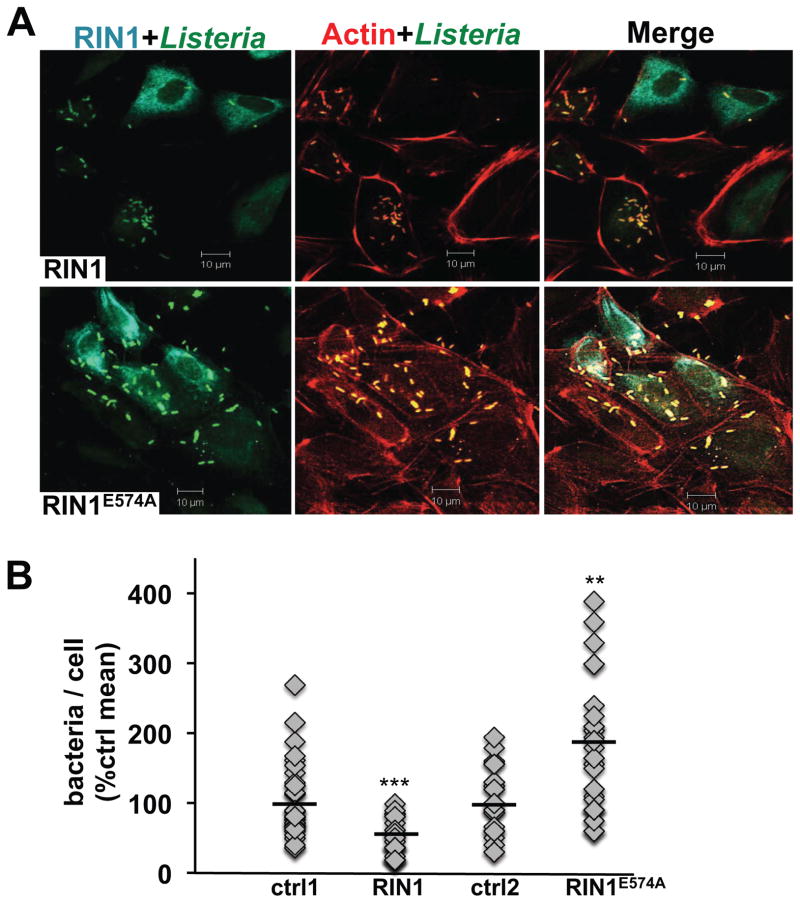
Following L. monocytogenes entry, host cells mount a bactericidal response that includes fusing phagosomes to bacteria-degrading lysosomes through RAB5-mediated trafficking. L. monocytogenes avoids this fate by inhibiting host cell RAB5, and by escaping the phagosome to replicate in the cytoplasm. We postulated that RIN1 might promote the bactericidal response through enhanced RAB5 activity. Indeed, normalizing for differences in cell entry efficiency, RIN1 over-expression led to a small but significant decrease in L. monocytogenes escape to the cytoplasm and replication at 6 hours post-infection in IEC-18 cells (Figure 3A, B). In contrast, expression of the GEF-mutant RIN1E574A led to an increase in cytoplasmic L. monocytogenes, confirming that the effects of RIN1 in this assay are RAB5-dependent. Phagosome escape is a prerequisite for L. monocytogenes actin polymerization and actin-based motility. We used co-localization of L. monocytogenes with actin as a measure of phagosome escape and replication. Although RIN1 over-expression did not appear to alter the rate of actin coating in IEC-18 cells, it did lead to lower totals of cytoplasmic actin-coated bacteria as early as three hours post-infection compared to the control group (Figure 4A, B). Conversely, the RIN1E574A mutant increased the number of actin-coated bacteria, supporting our hypothesis that RAB5 activation by RIN1 contributes to the host cell strategy for preventing L. monocytogenes escape and replication. The same effect was seen at six hours post infection (data not shown).
FIGURE 3. RIN1-to-RAB5 signaling inhibits L. monocytogenes replication.
A. IEC-18 cells transduced with control, RIN1 or RIN1E574A were infected with L. monocytogenes (50 MOI) after which extracellular bacteria were killed with gentamicin. At 1 hour post-infection (hpi) cells were either lysed or incubated in medium with low gentamicin before lysing at 6 hpi. Dilutions of the lysates were plated to obtain CFUs. Intracellular replication was measured as the fold change in CFU (6 hpi vs. 1 hpi) (ctrl1 and RIN1: n=6, p=1×10−30; ctrl2 and RIN1E574A: n=3, p=0.01). Values are representative of two independent experiments. B. Immunoblot evaluation of RIN1 expression. (*p ≤ 0.05, **p ≤ 0.005, ***p ≤ 0.0005)
FIGURE 4. RIN1 inhibits phagosome escape.
A. IEC-18 cells transduced with control, RIN1 or RIN1E574A vector were infected with L. monocytogenes (20 MOI) and extracellular bacteria killed with gentamicin, then incubated in medium with low gentamicin for 2 hours. Fixed cells were stained for RIN1 (cyan), actin (red) and L. monocytogenes (green). Scale bar: 10 μm. B. Quantified total bacteria co-localized with actin per cell (actin coating correlates with cytoplasmic localization). Values are representative of two independent experiments and normalized to percent of the control mean. (ctrl1 n=133, RIN1 n=81, p=4 × 10−6, ctrl2 n=70, RIN1E574A n=62, p=0.0012; *p ≤ 0.05, **p ≤ 0.005, ***p ≤ 0.0005)
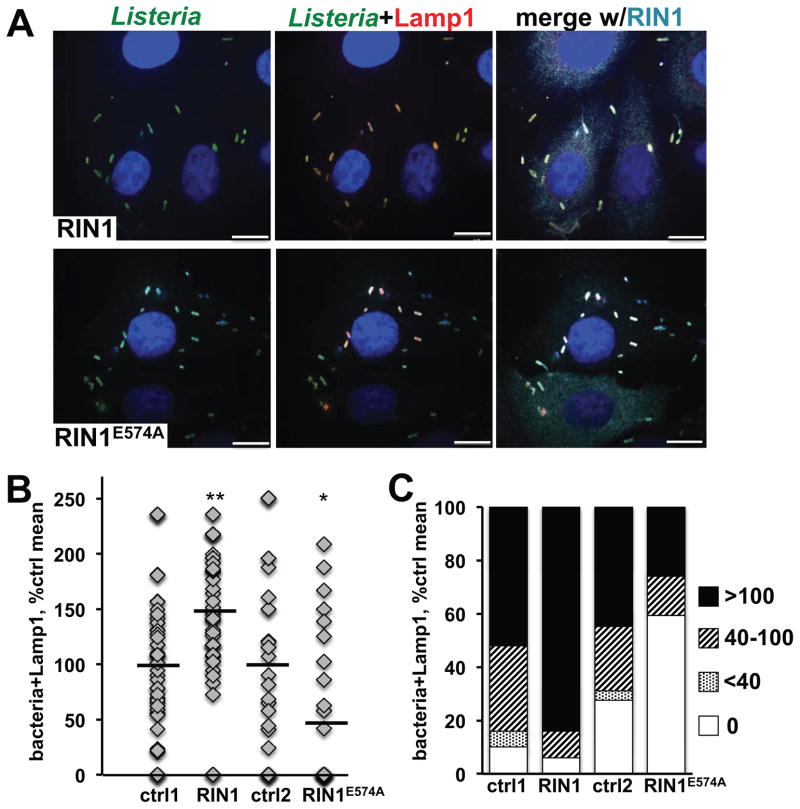
Host cells eliminate _L. monocytogenes_-containing phagosomes by fusing them with lysosomes, a process initiated by phagosome acquision of the lysosomal marker Lamp1. To test whether RIN1 promotes this RAB5-dependent bactericidal pathway, therefore, we quantified the co-localization of bacteria with Lamp1 at 2.5 hpi as an early measure of L. monocytogenes clearance. RIN1 over-expression increased co-localization of L. monocytogenes with Lamp1 in IEC-18 cells, compared to control cells, whereas expression of the GEF-defective mutant RIN1E574A caused a decrease in the fraction of intracellular bacteria co-localized with Lamp1 (Figure 5 A, B and C, S3A). The fraction of Lamp1-negative L. monocytogenes in these images may represent early escapers to the cytoplasm, but this could not be verified without actin staining.
FIGURE 5. RIN1-to-RAB5 signaling promotes lysosome fusion.
A. IEC-18 cells transduced with control, RIN1 or RIN1E574A vectors were infected with L. monocytogenes (20 MOI) and extracellular bacteria killed with gentamicin followed by incubation in medium with low gentamicin for 1.5 hours. Fixed cells were stained for RIN1 (cyan), L. monocytogenes (green) and Lamp1 (red). Nuclei were stained with DAPI (blue). Scale bar: 10 μm. B. and C. Quantification of lysosome-localized bacteria in infected IEC-18 cells. B. Each point is the percentage of total bacteria co-localizing with Lamp1 in a given field. C. Each bar is the percent co-localization in distribution categories. Values are representative of two independent experiments and normalized to percent of control mean (ctrl1 n=147, RIN1 n=124, p=9×10−4, ctrl2 n=161, RIN1E574A=105, p=0.03; *p ≤ 0.05, **p ≤ 0.005, ***p ≤ 0.0005)
To examine whether Lamp1-positive bacteria went on to fuse with lysosomes, we looked at co-localization of GFP-L. monocytogenes with dextran-Texas red-loaded lysosomes (Figure S3B). The co-localization of bacteria with fluorescent dextran confirmed that at least a portion of the internalized L. monocytogenes were undergoing lysosome fusion in these cells. Hence, RIN1 signaling through RAB5 increases the fusion of bacteria-laden phagosomes with host cell lysosomes, counteracting the effect of L. monocytogenes proteins that block RAB5 activation to improve escape efficiency. (25)
Autophagy, an intracellular destruction mechanism distinct from Lamp1-marked lysosome uptake, provides another way for host cells to clear invasive bacteria (39). L. monocytogenes normally evades autophagy during pathogenesis, at least in part by assembling an actin cloak (40, 41). We examined whether autophagy contributed to the RIN1-mediated bactericidal response by examining co-localization of L. monocytogenes with the autophagy marker LC3 in infected IEC-6 Cdx2 cells over-expressing RIN1 (Figure S3C and D). There was little discernable co-localization of L. monocytogenes with LC3, suggesting that autophagy plays an insignificant role in rapid clearance of L. monocytogenes by RIN1 signaling.
Cell-cell spread of L. monocytogenes is impeded by the RIN1-to-RAB5-mediated bactericidal response
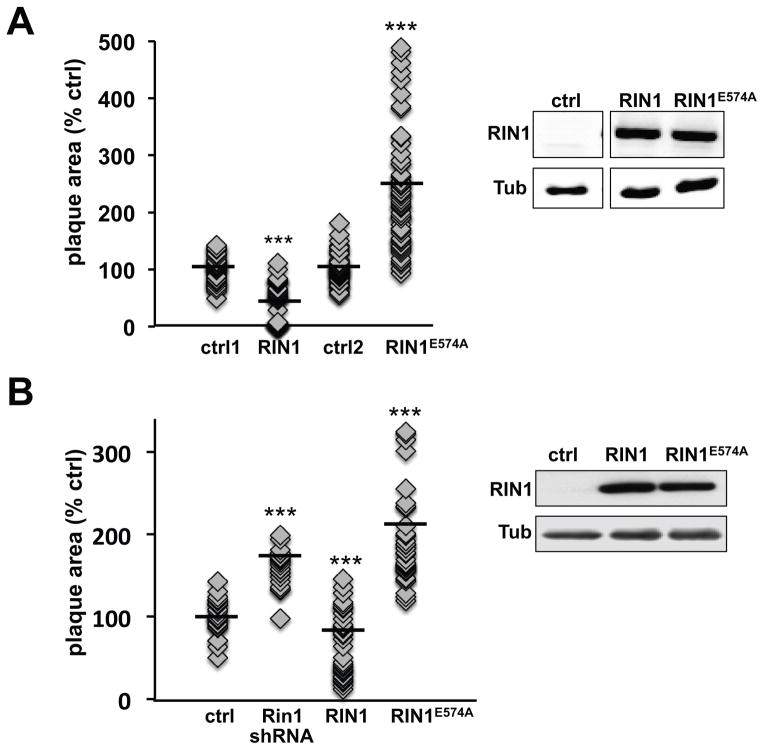
We expected that changes in the rate of phagosome escape would be reflected in the efficiency of cell-cell spread by L. monocytogenes. This was tested by plaque assay following L. monocytogenes infection. RIN1 over-expressing IEC-18 cells produced plaques of smaller average area than control, implicating a defect in L. monocytogenes cell-cell spread (Figure 6A, Figure S4A). In contrast, Rin1 silencing (Figure S4A) or expression of RIN1E574A (Figure 6A, Figure S4A) led to larger plaques compared to control cells, demonstrating that the bactericidal contribution of RIN1 signal transduction results in a cell-spread defect following L. monocytogenes infection. This result was confirmed in IEC-6 Cdx2 cells (Figure 6B). The ABL-binding mutant of RIN1, RIN1QM, had no effect on replication and plaque formation, when compared with wild type RIN1 (Figure S4B).
FIGURE 6. RIN1 inhibits bacteria spread between cells.
A. IEC-18 cells infected with L. monocytogenes (50 MOI) were trypsinized and transferred to a monolayer of IEC-18 cells transduced with control, RIN1 or RIN1E574A vectors in low gentamicin medium and overlaid with DMEM+0.7% agarose. After 48 hours plaque areas were measured using ImageJ (NIH). Each point is a single plaque area normalized to percent of control mean (ctrl1 n=55, RIN1 n=50, p=3×10−19; ctrl2 n=65, RIN1E574A n=75, p= 2×10−19). Values were obtained from two independent experiments. RIN1 expression confirmed by immunoblot (right). B. Plaque assay as described in A but using IEC-6 Cdx2 cells transduced with control, Rin1-shRNA, RIN1 or RIN1E574A vector (ctrl n=29, Rin1-kd n=21, p=7.1×10−13, RIN1 n=36, p=0.0001, RIN1E574A n=37, p=1.9×10−12). Values were obtained from two independent experiments. Expression confirmed by immunoblot (right). (*p ≤ 0.05, **p ≤ 0.005, ***p ≤ 0.0005)
Intercellular attachments, such as tight junctions, play an important role in maintaining polarity and establishing the barrier function of intestinal epithelia. Junction slackening is required for efficient L. monocytogenes spread from primary host cells to adjacent cells. Because tight junction organization and turnover are regulated by RAB5-dependent mechanisms (42), we tested the possibility that the cell-spread defects we observed in RIN1 modified cells were, at least in part, due to altered junctions. A disruption of tight and adherens junction organization was readily apparent following L. monocytogenes infection (Figure S5A). Staining of collagen-attached IEC-6 Cdx2 cells for the tight junction associated protein Tjp1 (ZO-1) and adherens junction protein Cdh1 (E-Cadherin) revealed no organizational remodeling in cells over-expressing RIN1 or RIN1E574A (Figure S5B, C). Examination of IEC-6 Cdx2 cells attached to Matrigel or fibronectin matrix also showed no difference in tight junction organization (data not shown). Total levels of the junction proteins Tjp1 and Cdh1 remained unaltered upon RIN1 over-expression or expression of RIN1E574A (data not shown), suggesting that the L. monocytogenes cell-spread defects we observed were primarily due to alterations in the RAB5-dependent host cell bactericidal response.
DISCUSSION
A common feature of bacterial invasion mechanisms is the commandeering of trafficking GTPases, especially RAB5, by pathogen-encoded proteins (9). In addition, multiple pathogens are known to inhibit RAB5 activity in order to evade lysosome-mediated degradation. Although there has been some progress in characterizing host protein involvement during these stages of infection (43–45), little is known about the role played by host cell regulators of RAB GTPases during infection. A recent report showed that RIN1 over-expression can partially rescue the effects of RAB5 inactivation during macrophage entry by Pseudomonas aeruginosa (44), but this was not examined further.
During L. monocytogenes invasion, InlB-mediated entry is dependent on RAB5 activation (18), but the bacterium must subsequently block RAB5 activation in order to evade clearance by fusion of its phagosome shelter with the lysosome (25). Any defect in this critical switch from RAB5 activation to inactivation can derail the course of infection. In this study we characterized the contributions of RIN1, an epithelial cell RAS effector and RAB5-GEF, during L. monocytogenes invasion, bactericidal escape and cell-cell spread. Importantly, we validate the role of RIN1 using silencing of the endogenous gene and by specifically disabling the RIN1-GEF function.
Our results suggest a model in which MET-dependent invasion of intestinal epithelial cells by L. monocytogenes rapidly engages RIN1, and RIN1-GEF activity is required for efficient entry (Figure 7). At later time points, however, RAB5 activation by RIN1 facilitates L. monocytogenes clearance from the cell by driving the fusion of bacteria-loaded phagosomes with degrading lysosomes. Even a partial escape from this bactericidal response can provide the pathogen an opportunity to replicate and spread to adjacent cells. Somewhat surprisingly, although RAB5 has been implicated in formation and turn over of tight junctions (42), RIN1 alterations had no apparent effect on the stability of cell-junctions. Future studies should evaluate the potential effect of other RAB5 regulators on cell junction stability during L. monocytogenes infection.
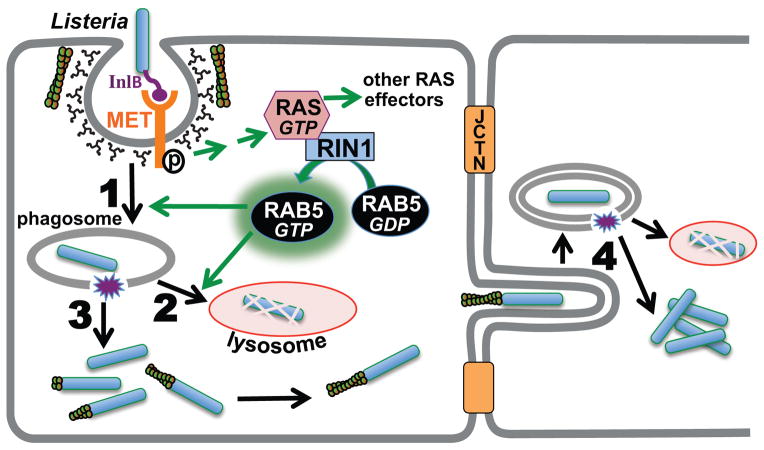
FIGURE 7. RIN1 is a RAB5 activator that controls the course of L. monocytogenes infection.
InlB binding to MET initiates clathrin-mediated invasion while also triggering activation of RAS and its effectors. RIN1 activates RAB5 by guanine nucleotide exchange. RAB5 facilitates L. monocytogenes internalization (1) but also promotes the fusion of phagosome-encased bacteria with lysosomes (2). This step clears bacteria before its escape and replication in the cytoplasm (3) and blocks spread to adjacent cells (4). Disrupting RIN1 function (silencing or GEF-interfering mutant) lowers invasion efficiency, but impedes the bactericidal response.
InlB-dependent invasion by L. monocytogenes is particularly relevant during the establishment of fetoplacental listeriosis and for traversing the blood brain barrier (14, 46). The pronounced and multi-stage contributions of RIN1 during L. monocytogenes infection of an intestinal epithelium model system strongly implicate RIN1 as a host cell factor for in vivo infections by L. monocytogenes. We note that numerous other bacteria utilize RAB5 to enter host cells, or they modulate RAB5 activity while directing pathogen-containing vesicles to cellular compartments favorable for pathogen replication and propagation (9). A few pathogens encode their own GEF and/or GAP activities for modulating small-GTPases (2, 47), but most appear to rely on host cell components. Based on our findings, RIN1 should be considered as a potential host cell factor for this larger group of pathogens.
The increased incidence of listeriosis in immunecompromised individuals and pregnant women underscores the need for early treatments that effectively prevent the occurrence of septicemia, bacterial meningitis, spontaneous abortion or stillbirths (48). This necessitates the identification and validation of host cell regulatory proteins required during pathogen infection. Based on its multiple levels of involvement at vulnerable stages of infection, RIN1 would appear to be a promising candidate for this approach. Finally, a thorough understanding of the host proteins that mediate internalization and trafficking is critically important for developing L. monocytogenes as a tumor vaccine vector (49).
MATERIALS AND METHODS
Expression constructs
All RIN1 expression constructs were made in lentivirus vectors. RIN1 wild type, RIN1E574A and RIN1QM in pM4-blastR vector (50), (37). Lentivirus generation and transduction were performed as previously described (26). For stable Rin1 knockdown, mRin1 shRNA (Sigma Aldrich) in pM4-blastR was used.
Cell culture and reagents
HeLa cells and rat Intestine Epithelial Cells (IEC-18 and IEC-6 Cdx2, from Enrique Rozengurt and James Sinnett-Smith, UCLA Dept. Medicine) (35) were cultured in DMEM (Media Tech) with 10% Fetal Bovine Serum (Hyclone) and 1% Penicillin Streptomycin (Invitrogen). HeLa cells stably expressing M4-blastR constructs were established by lentivirus infection followed by selection with 4 μg/ml blasticidin (Invitrogen). Lentivirus expressing LC3-EGFP construct for autophagy was obtained as a kind gift from Dr. Jeff F. Miller’s laboratory (UCLA). The virus was titered to an MOI=10 and cells were selected for infection using 2 μg/ml puromycin. 10 μg/ml fibronectin (Sigma Aldrich) or 100 μg/ml collagen (Sigma Aldrich) were used to coat the culture dishes and cover slips as indicated. L. monocytogenes strain 10403S was used in most experiments performed. EGDe strain was used to compare murinized L. monocytogenes (from Dr. Sarah D’Orazio, U. Kentucky College of Medicine) with the matched wild type strain. GFP-L. monocytogenes strain 10403S (from Dr. Jeff F. Miller, UCLA) was used to observe co-localization with dextran. L. monocytogenes strains were grown at 30°C.
Gene Silencing
HeLa cells with stable silencing of RIN1 were established by infection with pLKO.1-shRIN1-puroR and selection with 2 μg/ml puromycin (Invivogen) (51). IEC-18 and IEC-6 Cdx2 cells were subject to stable silencing of Rin1 using Rin1 shRNA obtained from PLKO-mRin1 1680 shRNA-puroR vector (Sigma) and cloned into pM4-BlastR with a U6 promoter. The shRNA sequence is: CCGGCTCCTGTTAGAAGCTGAGTATCTCGAGATACTCAGCTTCTAACAGGAGTTTTTG.
Expression levels were evaluated by western blotting.
Gentamicin Protection Assay
L. monocytogenes (10403S or EGDe) invasion was measured by Gentamicin protection assays. 0.75×106 cells were plated on fibronectin coated 6-well plates. Cells were infected with the indicated MOI of L. monocytogenes for 60 minutes at 37°C, 5% CO2. Extracellular bacteria were killed for one hour with 150 μg/ml gentamicin (Sigma Aldrich) at 37°C, 5% CO2. Cells were trypsinized and lysed in 0.1% Triton, in the presence of DNase and appropriate dilutions of the lysate were plated on LB-Agar plates. Agar plates were stored at 30°C for about 20 hours before colony counts were obtained. For replication assays, cells were transferred to 10 μg/ml gentamicin following killing of extracellular bacteria and lysed at 3 hours or 6 hours post-infection in order to obtain colony counts as described above. All statistical analyses were performed using Student’s t-test.
Plaque Assays to Assess Cell-Spread
To measure L. monocytogenes cell spread, wild type IEC-18 or IEC-6 Cdx2 cells were infected with L. monocytogenes at MOI=50 for one hour as described above. Extracellular bacteria were killed using 150 ug/ml gentamicin. Cells were trypsinized and added to a confluent monolayer of wild type or mutant cells at a ratio of 1:10 in the presence of 10 μg/ml gentamicin, followed by a three hour incubation at 37°C, 5% CO2. The cells were washed and an overlay of 0.7% Agarose, 1X DMEM and 10 μg/ml gentamicin was laid on the monolayer. After 48 hours of incubation at 37°C, plaques were visualized and plaque areas measured using ImageJ software (NIH). All statistical analyses were performed using Student’s t-test.
Preparation of Whole Cell Lysates for Immunoprecipitation and Immunoblotting
HeLa, IEC-18 or IEC-6 Cdx2 cells were lysed in NP-40 lysis buffer (150 mM NaCl, 50 mM Tris pH 8.0, 1% NP-40) containing 1 mM PMSF, 10 μg/ml leupeptin, 1 μM pepstatin and 1 mM sodium orthovanadate (Sigma Aldrich). 50 ug of the lysates were prepared in 5X SDS Laemmlli buffer and boiled for 5 minutes, before loading on an 8% SDS PAGE.
Immunoprecipitation, immunoblotting and immunofluorescence
Antibodies used for immunoblotting and their sources were- Tubulin (Sigma Aldrich, #T6074-200UL), human RIN1 (Mouse mAb, clone #C9E11, Colicelli lab, AbPro), mouse Rin1 (mouse mAb, Colicelli lab), pY36 RIN1 (31) (Rabbit pAb, Colicelli Lab, Biosource International), Rabex-5 (Sigma Aldrich, #R5405), RIN2 (Genetex, #GTX117153), sheep-anti-mouse-HRP (Amersham Biosciences, #NA931), goat-anti-rabbit-IRDye 800 (Li-Cor Biosciences, #926-32211) and goat-anti-mouse-IRDye 680 (Li-Cor Biosciences, #926-32220).
For immunoblotting, proteins were transferred to Nitrocellulose membranes overnight. The membranes were blocked with 5% milk in PBST (0.1% Tween-20) followed by incubation with primary and secondary antibodies at room temperature. The membranes were washed with PBST between incubations. Imaging and quantification were done using a Li-Cor Odyssey scanner.
For immunoprecipitation experiments, mRin1 (mouse mAb, clone B9A12, Colicelli Lab) and RIN2 (Rabbit pAb, Colicelli Lab) antibodies were used. Cells were lysed in NP-40 lysis buffer containing protease and phosphatase inhibitors. Lysates were incubated with the antibody and protein A- agarose (Fisher) beads overnight at 4°C. Following incubation, beads were washed in lysis buffer, boiled in SDS sample loading buffer and run on 8% SDS PAGE, followed by immunoblotting.
The antibody sources for immunofluorescence experiments were- RIN1 (Mouse mAb, clone #C9E11, Colicelli lab, AbPro), RIN1 (Sigma, #HPA035491), L. monocytogenes (Abcam, #ab35132), pan-RAB5 (Abcam, #ab18211), ZO-1 (Zymed, #61-7300), Lamp1 (Santa Cruz, #SC-19992), phalloidin-rhodamine 0.2 uM (Invitrogen), 70,000 MW dextran-Texas red, lysine fixable (Invitrogen, #D1864), E-Cadherin (BD Transduction Laboratories, #610181), goat-anti-rabbit Alexa-Fluor 568 (Invitrogen, #A11036) and goat-anti-mouse Alexa Fluor 647 (Invitrogen, #A21245), goat-anti-mouse Alexa Fluor 488 (Invitrogen, #A11001), goat-anti-rat Alexa Fluor 647 (Invitrogen, #A21247). For imaging experiments, 0.75×106 cells were plated on fibronectin or collagen coated cover slips overnight. The cells were infected with L. monocytogenes as indicated, followed by killing of extracellular bacteria with 150 ug/ml gentamicin and incubation for the described times at 10 ug/ml gentamicin. The cells were washed and fixed in 4% paraformaldehyde, permeabilized in 0.1% Triton-X-100 detergent, quenched in 50 mM NH4Cl and blocked in 10% goat serum (Gibco). For RAB5 staining experiments, cells were fixed and permeabilized in methanol for 5 minutes at −20°C and the rest of the procedure was followed as specified above. Cells were then incubated with primary and secondary antibodies at room temperature. Fluorescently labeled phalloidin was added along with the secondary antibody staining. DAPI staining was performed for 5 minutes following secondary antibody incubation. Confocal microscopy (Zeiss Pascal) was used to image the cells.
Microscopy
Fixed and stained cells were examined on a laser scanning confocal microscope (Axiovert 200M, Carl Zeiss LSM 5 Pascal) or spinning disc confocal microscope (Yokagawa CSU-22 spinning disc, Hamamatsu C9100-13 EMCCD camera, Zeiss Aiovert 200M). Cells were imaged with a plan/neofluar 100x oil lens, NA 1.3 (Carl Zeiss) and 8-bit digital images were captured using a cooled charge-coupled device (transmitted light channel for lsm5 camera, Zeiss). LSM 5 Pascal software (release version 4.2 sp1) was used to process the images. All images were quantified in a double-blinded fashion and all statistical analyses were performed using Student’s t-test.
Supplementary Material
Supplemental
Acknowledgments
The authors would like to thank the following for valuable advice and suggestions: Dr. Sarah D’Orazio (U. Kentucky College of Medicine), Enrique Rozengurt and James Sinnett-Smith (UCLA, Dept. Medicine), Jeffrey Malloy (UCLA) and Charlie Ho (UCLA). We also thank the following for technical assistance: Ken Dekitani, Joshua Weinreb and Danit Tarashandegan, who performed the double blind quantification of confocal images and plaques.
References
- 1.Humphries AC, Way M. The non-canonical roles of clathrin and actin in pathogen internalization, egress and spread. Nature reviews Microbiology. 2013;11(8):551–560. doi: 10.1038/nrmicro3072. [DOI] [PubMed] [Google Scholar]
- 2.Patel JC, Galan JE. Differential activation and function of Rho GTPases during Salmonella-host cell interactions. The Journal of cell biology. 2006;175(3):453–463. doi: 10.1083/jcb.200605144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanchard E, Belouzard S, Goueslain L, Wakita T, Dubuisson J, Wychowski C, Rouille Y. Hepatitis C virus entry depends on clathrin-mediated endocytosis. Journal of virology. 2006;80(14):6964–6972. doi: 10.1128/JVI.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pizarro-Cerda J, Kuhbacher A, Cossart P. Entry of Listeria monocytogenes in mammalian epithelial cells: an updated view. Cold Spring Harbor perspectives in medicine. 2012;2(11) doi: 10.1101/cshperspect.a010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norkin LC, Anderson HA, Wolfrom SA, Oppenheim A. Caveolar endocytosis of simian virus 40 is followed by brefeldin A-sensitive transport to the endoplasmic reticulum, where the virus disassembles. Journal of virology. 2002;76(10):5156–5166. doi: 10.1128/JVI.76.10.5156-5166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dramsi S, Cossart P. Intracellular pathogens and the actin cytoskeleton. Annual review of cell and developmental biology. 1998;14:137–166. doi: 10.1146/annurev.cellbio.14.1.137. [DOI] [PubMed] [Google Scholar]
- 7.Bonazzi M, Vasudevan L, Mallet A, Sachse M, Sartori A, Prevost MC, Roberts A, Taner SB, Wilbur JD, Brodsky FM, Cossart P. Clathrin phosphorylation is required for actin recruitment at sites of bacterial adhesion and internalization. J Cell Biol. 2011;195(3):525–536. doi: 10.1083/jcb.201105152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agaisse H. Investigating the involvement of host factors involved in intracellular pathogen infection by RNAi in Drosophila cells. Methods in molecular biology. 2008;415:395–402. doi: 10.1007/978-1-59745-570-1_23. [DOI] [PubMed] [Google Scholar]
- 9.Stein MP, Muller MP, Wandinger-Ness A. Bacterial pathogens commandeer Rab GTPases to establish intracellular niches. Traffic. 2012;13(12):1565–1588. doi: 10.1111/tra.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farber JM, Peterkin PI. Listeria monocytogenes, a food-borne pathogen. Microbiological reviews. 1991;55(3):476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Dennis SB, Hartnett E, Paoli G, Pouillot R, Ruthman T, Wilson M. FDA-iRISK--a comparative risk assessment system for evaluating and ranking food-hazard pairs: case studies on microbial hazards. Journal of food protection. 2013;76(3):376–385. doi: 10.4315/0362-028X.JFP-12-372. [DOI] [PubMed] [Google Scholar]
- 12.Bonazzi M, Lecuit M, Cossart P. Listeria monocytogenes internalin and E-cadherin: from structure to pathogenesis. Cellular microbiology. 2009;11(5):693–702. doi: 10.1111/j.1462-5822.2009.01293.x. [DOI] [PubMed] [Google Scholar]
- 13.Veiga E, Cossart P. Listeria InlB takes a different route to met. Cell. 2007;130(2):218–219. doi: 10.1016/j.cell.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Disson O, Grayo S, Huillet E, Nikitas G, Langa-Vives F, Dussurget O, Ragon M, Le Monnier A, Babinet C, Cossart P, Lecuit M. Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature. 2008;455(7216):1114–1118. doi: 10.1038/nature07303. [DOI] [PubMed] [Google Scholar]
- 15.Li N, Xiang GS, Dokainish H, Ireton K, Elferink LA. The Listeria protein internalin B mimics hepatocyte growth factor-induced receptor trafficking. Traffic. 2005;6(6):459–473. doi: 10.1111/j.1600-0854.2005.00290.x. [DOI] [PubMed] [Google Scholar]
- 16.Bierne H, Cossart P. InlB, a surface protein of Listeria monocytogenes that behaves as an invasin and a growth factor. Journal of cell science. 2002;115(Pt 17):3357–3367. doi: 10.1242/jcs.115.17.3357. [DOI] [PubMed] [Google Scholar]
- 17.Tang P, Rosenshine I, Finlay BB. Listeria monocytogenes, an invasive bacterium, stimulates MAP kinase upon attachment to epithelial cells. Molecular biology of the cell. 1994;5(4):455–464. doi: 10.1091/mbc.5.4.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiwani S, Wang Y, Dowd GC, Gianfelice A, Pichestapong P, Gavicherla B, Vanbennekom N, Ireton K. Identification of components of the host type IA phosphoinositide 3-kinase pathway that promote internalization of Listeria monocytogenes. Infection and immunity. 2012;80(3):1252–1266. doi: 10.1128/IAI.06082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosse T, Ehinger J, Czuchra A, Benesch S, Steffen A, Wu X, Schloen K, Niemann HH, Scita G, Stradal TE, Brakebusch C, Rottner K. Cdc42 and phosphoinositide 3-kinase drive Rac-mediated actin polymerization downstream of c-Met in distinct and common pathways. Molecular and cellular biology. 2007;27(19):6615–6628. doi: 10.1128/MCB.00367-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez-Dominguez C, Barbieri AM, Beron W, Wandinger-Ness A, Stahl PD. Phagocytosed live Listeria monocytogenes influences Rab5-regulated in vitro phagosome-endosome fusion. The Journal of biological chemistry. 1996;271(23):13834–13843. doi: 10.1074/jbc.271.23.13834. [DOI] [PubMed] [Google Scholar]
- 21.Prada-Delgado A, Carrasco-Marin E, Pena-Macarro C, Del Cerro-Vadillo E, Fresno-Escudero M, Leyva-Cobian F, Alvarez-Dominguez C. Inhibition of Rab5a exchange activity is a key step for Listeria monocytogenes survival. Traffic. 2005;6(3):252–265. doi: 10.1111/j.1600-0854.2005.00265.x. [DOI] [PubMed] [Google Scholar]
- 22.Dramsi S, Cossart P. Listeriolysin O: a genuine cytolysin optimized for an intracellular parasite. The Journal of cell biology. 2002;156(6):943–946. doi: 10.1083/jcb.200202121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welch MD, Iwamatsu A, Mitchison TJ. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997;385(6613):265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- 24.Robbins JR, Barth AI, Marquis H, de Hostos EL, Nelson WJ, Theriot JA. Listeria monocytogenes exploits normal host cell processes to spread from cell to cell. The Journal of cell biology. 1999;146(6):1333–1350. doi: 10.1083/jcb.146.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarez-Dominguez C, Madrazo-Toca F, Fernandez-Prieto L, Vandekerckhove J, Pareja E, Tobes R, Gomez-Lopez MT, Del Cerro-Vadillo E, Fresno M, Leyva-Cobian F, Carrasco-Marin E. Characterization of a Listeria monocytogenes protein interfering with Rab5a. Traffic. 2008;9(3):325–337. doi: 10.1111/j.1600-0854.2007.00683.x. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Waldron RT, Dhaka A, Patel A, Riley MM, Rozengurt E, Colicelli J. The RAS effector RIN1 directly competes with RAF and is regulated by 14-3-3 proteins. Molecular and cellular biology. 2002;22(3):916–926. doi: 10.1128/MCB.22.3.916-926.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balaji K, Mooser C, Janson CM, Bliss JM, Hojjat H, Colicelli J. RIN1 orchestrates the activation of RAB5 GTPases and ABL tyrosine kinases to determine the fate of EGFR. J Cell Sci. 2012;125(Pt 23):5887–5896. doi: 10.1242/jcs.113688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deininger K, Eder M, Kramer ER, Zieglgansberger W, Dodt HU, Dornmair K, Colicelli J, Klein R. The Rab5 guanylate exchange factor Rin1 regulates endocytosis of the EphA4 receptor in mature excitatory neurons. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(34):12539–12544. doi: 10.1073/pnas.0801174105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tall GB, MA, Stahl PD, Horazdovsky BF. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev Cell. 2001;1(1):73–82. doi: 10.1016/s1534-5807(01)00008-9. [DOI] [PubMed] [Google Scholar]
- 30.Kong C, Su X, Chen PI, Stahl PD. Rin1 interacts with signal-transducing adaptor molecule (STAM) and mediates epidermal growth factor receptor trafficking and degradation. J Biol Chem. 2007;282(20):15294–15301. doi: 10.1074/jbc.M611538200. [DOI] [PubMed] [Google Scholar]
- 31.Hu H, Bliss JM, Wang Y, Colicelli J. RIN1 is an ABL tyrosine kinase activator and a regulator of epithelial-cell adhesion and migration. Current biology : CB. 2005;15(9):815–823. doi: 10.1016/j.cub.2005.03.049. [DOI] [PubMed] [Google Scholar]
- 32.Balaji K, Colicelli J. RIN1 regulates cell migration through RAB5 GTPases and ABL tyrosine kinases. Commun Integr Biol. 2013 doi: 10.4161/cib.25421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu L, Lubkov V, Taylor LJ, Bar-Sagi D. Feedback regulation of Ras signaling by Rabex-5-mediated ubiquitination. Current biology : CB. 2010;20(15):1372–1377. doi: 10.1016/j.cub.2010.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen Y, Naujokas M, Park M, Ireton K. InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell. 2000;103(3):501–510. doi: 10.1016/s0092-8674(00)00141-0. [DOI] [PubMed] [Google Scholar]
- 35.Vincentini O, Ciotta C, Bignami M, Stammati A, Zucco F. Normal rat intestinal cells IEC-18: characterization and transfection with immortalizing oncogenes. Cytotechnology. 1996;21(1):11–19. doi: 10.1007/BF00364833. [DOI] [PubMed] [Google Scholar]
- 36.Gao N, Kaestner KH. Cdx2 regulates endolysosomal function and epithelial cell polarity. Genes & development. 2010;24(12):1295–1305. doi: 10.1101/gad.1921510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu H, Milstein M, Bliss J, Thai M, Malhotra G, Huynh L, Colicelli J. Integration of transforming growth factor beta and RAS signaling silences a RAB5 guanine nucleotide exchange factor and enhances growth factor-directed cell migration. Moll Cell Biol. 2008;28(5):1573–1583. doi: 10.1128/MCB.01087-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monk IR, Casey PG, Hill C, Gahan CG. Directed evolution and targeted mutagenesis to murinize Listeria monocytogenes internalin A for enhanced infectivity in the murine oral infection model. BMC microbiology. 2010;10:318. doi: 10.1186/1471-2180-10-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nature cell biology. 2013;15(7):713–720. doi: 10.1038/ncb2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dortet L, Mostowy S, Cossart P. Listeria and autophagy escape: involvement of InlK, an internalin-like protein. Autophagy. 2012;8(1):132–134. doi: 10.4161/auto.8.1.18218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dortet L, Mostowy S, Samba-Louaka A, Gouin E, Nahori MA, Wiemer EA, Dussurget O, Cossart P. Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLoS pathogens. 2011;7(8):e1002168. doi: 10.1371/journal.ppat.1002168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coyne CB, Shen L, Turner JR, Bergelson JM. Coxsackievirus entry across epithelial tight junctions requires occludin and the small GTPases Rab34 and Rab5. Cell host & microbe. 2007;2(3):181–192. doi: 10.1016/j.chom.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pizarro-Cerda J, Payrastre B, Wang YJ, Veiga E, Yin HL, Cossart P. Type II phosphatidylinositol 4-kinases promote Listeria monocytogenes entry into target cells. Cellular microbiology. 2007;9(10):2381–2390. doi: 10.1111/j.1462-5822.2007.00967.x. [DOI] [PubMed] [Google Scholar]
- 44.Mustafi S, Rivero N, Olson JC, Stahl PD, Barbieri MA. Regulation of Rab5 function during phagocytosis of live Pseudomonas aeruginosa in macrophages. Infection and immunity. 2013;81(7):2426–2436. doi: 10.1128/IAI.00387-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gouin E, Adib-Conquy M, Balestrino D, Nahori MA, Villiers V, Colland F, Dramsi S, Dussurget O, Cossart P. The Listeria monocytogenes InlC protein interferes with innate immune responses by targeting the I{kappa}B kinase subunit IKK{alpha} Proceedings of the National Academy of Sciences of the United States of America. 2010;107(40):17333–17338. doi: 10.1073/pnas.1007765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greiffenberg L, Goebel W, Kim KS, Daniels J, Kuhn M. Interaction of Listeria monocytogenes with human brain microvascular endothelial cells: an electron microscopic study. Infection and immunity. 2000;68(6):3275–3279. doi: 10.1128/iai.68.6.3275-3279.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bulgin R, Raymond B, Garnett JA, Frankel G, Crepin VF, Berger CN, Arbeloa A. Bacterial guanine nucleotide exchange factors SopE-like and WxxxE effectors. Infection and immunity. 2010;78(4):1417–1425. doi: 10.1128/IAI.01250-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mateus T, Silva J, Maia RL, Teixeira P. Listeriosis during Pregnancy: A Public Health Concern. ISRN obstetrics and gynecology. 2013;2013:851712. doi: 10.1155/2013/851712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Starks H, Bruhn KW, Shen H, Barry RA, Dubensky TW, Brockstedt D, Hinrichs DJ, Higgins DE, Miller JF, Giedlin M, Bouwer HG. Listeria monocytogenes as a vaccine vector: virulence attenuation or existing antivector immunity does not diminish therapeutic efficacy. Journal of immunology. 2004;173(1):420–427. doi: 10.4049/jimmunol.173.1.420. [DOI] [PubMed] [Google Scholar]
- 50.Milstein M, Mooser CK, Hu H, Fejzo M, Slamon D, Goodglick L, Dry S, Colicelli J. RIN1 is a breast tumor suppressor gene. Cancer research. 2007;67(24):11510–11516. doi: 10.1158/0008-5472.CAN-07-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thai M, Ting PY, McLaughlin J, Cheng D, Muschen M, Witte ON, Colicelli J. ABL fusion oncogene transformation and inhibitor sensitivity are mediated by the cellular regulator RIN1. Leukemia. 2011;25(2):290–300. doi: 10.1038/leu.2010.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental