MicroRNA expression profiling in human Barrett's carcinogenesis (original) (raw)
. Author manuscript; available in PMC: 2015 Jan 22.
Published in final edited form as: Int J Cancer. 2011 Mar 11;129(7):1661–1670. doi: 10.1002/ijc.25823
Abstract
Barrett's esophagus (BE) is characterized by the native stratified squamous epithelium (N) lining the esophagus being replaced by a columnar epithelium with intestinal differentiation (Barrett's mucosa; BM). BM is considered as the main risk factor for esophageal adenocarcinoma (Barrett's adenocarcinoma; BAc). MicroRNAs (miRNAs) are a class of small noncoding RNAs that control gene expression by targeting messenger RNAs and they are reportedly dysregulated in BM. To test the hypothesis that a specific miRNA expression signature characterizes BM development and progression, we performed miRNA microarray analysis comparing native esophageal mucosa with all the phenotypic lesions seen in the Barrett's carcinogenic process. Specimens were collected from 14 BE patients who had undergone esophagectomy, including: 14 with N, 14 with BM, 7 with low-grade intraepithelial neoplasia, 5 with high-grade intra-epithelial neoplasia and 11 with BAc. Microarray findings were further validated by quantitive real-time polymerase chain reaction and in situ hybridization analyses using a different series of consecutive cases (162 biopsy samples and 5 esophagectomies) of histologically proven, long-segment BE. We identified a miRNA signature of Barrett's carcinogenesis consisting of an increased expression of 6 miRNAs and a reduced expression of 7 miRNAs. To further support these results, we investigated target gene expression using the Oncomine database and/or immunohistochemical analysis. We found that target gene expression correlated significantly with miRNA dysregulation. Specific miRNAs are directly involved in BE progression to cancer. miRNA profiling significantly expands current knowledge on the molecular history of Barrett's carcinogenesis, also identifying molecular markers of cancer progression.
Keywords: miRNA, Barrett's esophagus, gene target, expression signature
Barrett's esophagus (BE) is defined as the replacement of the native esophageal squamous epithelium by a lining of columnar epithelium with intestinal differentiation [Barrett's mucosa (BM)].1,2 Epidemiological and clinico-pathological studies have consistently shown BM to be the initial event in a cascade of phenotypic changes that may lead to Barrett's adenocarcinoma (BAc).3–5 Very little is known as yet about the molecular mechanisms involved in neoplastic transformation. From a prognostic point of view, the histological diagnosis of dysplasia [defined as low- and high-grade intraepithelial neoplasia (IEN or NiN)] is currently the only bio-marker considered in the definition of high risk BE populations.6–8 Expanding on such biological information might lead to the identification of new prognostic markers and targeted therapies.
MicroRNAs (or miRNAs) are a class of small noncoding RNAs that control gene expression by targeting messenger RNAs (mRNAs).9–11 A growing number of reports point to the involvement of miRNAs in carcinogenesis and/or tumor progression, and miRNA expression profiles have been suggested as a promising new class of biomarkers for tumor diagnosis and prognosis, including the prediction of response to therapy.
MiRNAs are reportedly involved in the oncogenic process leading to the onset of both BM and BAc. The first published report by Feber et al.12 described the miRNA expression profiling of esophageal cancers (both adenocarcinoma and squamous cell carcinoma) and a small series of BM samples. After this seminal study, Dijckmeester et al. investigated miR-143 and miR-205 expression in neosquamous esophageal epithelium after Argon plasma ablation of BM.13 In our study, miR-143 was found significantly up-regulated in neosquamous mucosa by comparison with the normal squamous epithelium of control subjects.13 In human Barrett's carcino-genesis, miR-196a has been established as a potential marker of progression by targeting KRT5, SPRR2C and S100A9,14 whereas in BM/BAc-derived cell lines, miRNA expression profiling revealed that miR-106b-25 polycistron is involved in the neoplastic progression via CDKN1A and BCL2L11 suppression.15 In a large series of esophageal adenocarcinoma samples, BAc showed a specific miRNA expression profile by comparison with cancers unrelated to BE.16 In a series of paired, diseased and normal tissues, Yang et al.17 showed that specific miRNA expression signatures are associated with BM cancerization.
None of the abovementioned studies, however, provided comprehensive information on the miRNA profile at each step of the natural history of Barrett's carcinogenesis. We undertook to find a specific miRNA expression signature consistently associated with the whole spectrum of this oncogenic process, also investigating miRNA target gene expression by means of the Oncomine database and/or immunohistochemical (IHC) analyses. Our results strongly support the direct involvement of specific miRNAs in BE progression and their potential as a novel diagnostic tool in the characterization of BAc gene targets.
Material and Methods
Tissue samples
All cases of BE patients who had undergone total (R0, no residual tumor) esophagectomy for HG-NiN and BAc reported between 2005 and 2009 were retrieved from the archives of the surgical pathology and cytopathology unit of Padova University. Of the 158 cases initially found, 144 were subsequently excluded for the following reasons: (i) prior neoadjuvant chemotherapy, 51 cases; (ii) prior surgical or endoscopic treatments, 40 cases; and (iii) archival tumor samples inconsistent with the aims of the study, 49 cases. Informed consent from involved patients was obtained in all, but not in four cases (which were also ruled out). Thus, 14 patients (mean age: 63.±67.9 years; range: 52–81; 12 men, 2 women; all Caucasian) were considered, who had all been surgically treated at the same institution (Department of Gastroenterological and Surgical Sciences of Padova University). All cases were assessed by two pathologists (PP and MF); in cases where their opinions differed, a third GI expert pathologist (MR) was consulted. Two 2-mm cores were obtained from the paraffin blocks from: (i) the proximal native squamous esophageal mucosa (N = 14 cases); (ii) intestinal metaplasia (IM) positive esophageal mucosa (BM = 14 cases); (iii) low-grade intraepithelial neoplasia (LG = 7 cases); (iv) high-grade intraepithelial neoplasia (HG = 5 cases); and (v) BAc (11 cases) were collected and further analyzed in the microarray study. For the quantitative real-time polymerase chain reaction (qRT-PCR) and the IHC studies, tissue samples were retrospectively collected from the files of the Veneto Region's multicenter Barrett's Esophagus Registry (EBRA; Padova Unit),5 selecting consecutive cases of histologically proven long-segment BE. A total of 162 biopsy samples obtained from different biopsy sets were considered, including: N = 40 cases, BM = 40 cases, LG = 31 cases, HG = 26 cases and BAc = 25 cases. For the in situ hybridization (ISH) study, tissues were collected from five further BAc patients who had undergone esophagectomy. All the patients considered in our study gave their written informed consent.
miRNA microarray
Tissue samples were deparaffinized with xylene at 50°C for 3 minutes. Total RNA extraction was done using the Recover-All kit (Ambion, Austin, TX) according to manufacturer's instructions. RNA labeling and hybridization on miRNA microarray chips were performed as described elsewhere.10,18 Briefly, 5 μg of total RNA from each sample were reverse-transcribed using biotin end-labeled random-octamer oligo-nucleotide primer. Biotin-labeled complementary DNA was hybridized on an Ohio State University custom miRNA microarray chip (OSU_CCC version 4.0), which contains ~1100 miRNA probes, including 326 human and 249 mouse miRNA genes and 10 control genes, spotted in duplicate. The hybridized chips were washed and processed for biotin-containing transcript detection by streptavidin-Alexa 647 conjugate and scanned on an Axon 4000B microarray scanner (Axon Instruments, Sunnyvale, CA).
Statistical and bioinformatic analyses
Microarray images were analyzed using GENEPIX PRO 6.0 (Axon Instruments, Sunnyvale, CA). Average values of the replicate spots of each miRNA were background subtracted, normalized using quantiles enabling a comparison between chips19 and further analyzed. The microarray data are deposited in the Gene Expression Omnibus at the National Center for Biotechnology Information (GEO:GSE20099). The miRNAs that were differently expressed between different esophageal lesions were identified using a random-variance _t_-test, which is an improvement over the standard separate _t_-test because it enables information on within-class variation to be shared among genes without assuming that all genes have the same variance.20 Genes were considered statistically signifi-cant if their p value was less than 0.001; a stringent significance threshold was used to limit the number of false positive findings.21 Only mature miRNAs that were differently expressed are reported.
Quantitative real-time polymerase chain reaction
The NCode™ miRNA qRT-PCR method (Invitrogen, Carlsbad, CA) was used to detect and quantify mature miRNAs on Applied Biosystems RT-PCR instruments in accordance with manufacturer's instructions. Normalization was performed with the small nuclear RNA U6B (RNU6B; Invitrogen). All real-time reactions, including no-template controls and real-time minus controls, were run in a GeneAmp PCR 9700 thermocycler (Applied Biosystems, Foster City, CA). Gene expression levels were quantified using the ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Comparative RT-PCR was performed in triplicate, including no-template controls. The fold difference for each sample was obtained using the equation 2−dCt; is the threshold cycle, the cycle number at which the fluorescence generated within a reaction crosses the threshold; dCt = (Ct average sample gene) – (Ct average RNU6B). Total RNAs from 15 N, 15 BM, 15 LG, 15 HG and 15 BAc biopsy samples were used in the qRT-PCR analysis.
In situ hybridization
ISH was performed using the GenPoint™ catalyzed signal amplification system (DakoCytomation) following the manufacturer's protocol. Briefly, slides were incubated at 60°C for 30 min and deparaffinized as described.22 Sections were treated with Proteinase K (DakoCytomation) for 30 min at room temperature, rinsed several times with dH2O and immersed in 95% ethanol for 10 sec before air drying. Slides were prehybridized at 49–56°C for 1 hr with mRNA ISH buffer (Ambion) before incubation overnight at 49–56°C in buffer containing the 5′-biotin labeled _miR_-203, _miR_-205, _let_-7c miRCURY™ LNA detection probe (Exiqon, Woburn, MA) or the scrambled negative control probe (U6, Exiqon) at 200 nM final concentration. Slides were washed in both TBST washing buffer and GenPoint stringent wash solution (54°C for 30 min). Slides were then exposed to H2O2 blocking solution (DakoCytomation) for 20 min and further blocked in a blocking buffer (DakoCytomation) for 30 min before being exposed to primary Streptavidin-HRP antibody, biotinyl tyramide, secondary Streptavidin-HRP antibody and DAB chromogen solutions following the manufacturer's protocol. Slides were then briefly counterstained in hematoxylin and rinsed with both TBST and water before mounting. For the ISH study, samples obtained from 5 BAc esophagectomies were considered.
cDNA microarray analysis
The Oncomine database and gene microarray analysis tool, a repository for published cDNA microarray data (www.onco-mine.org),23,24 was explored (15th December 2009) for HMGA2, ZEB1, ZEB2 and CDH1 (E-cadherin) mRNA expression in esophageal native mucosa, BM and BAc samples. Oncomine algorithms, which enable multiple comparisons among different studies, were used for the statistical analysis of the differences in mRNA expression between the aforementioned comparisons.23–25
Immunohistochemistry (IHC)
Staining was performed automatically (Ventana Benchmark XT system, Touchstone, AZ)26 for high mobility group A2 (HMGA2) (Biocheck, Foster City, CA; 1:100), ZEB1 (H-102; Santa Cruz Biotecnology, Santa Cruz, CA), SIP1 (ZEB2, H-260; Santa Cruz Biotecnology), and E-cadherin (Ventana, Touchstone, AZ; prediluted) according to the manufacturer's instructions. Sections were then lightly counterstained with hematoxylin. Appropriate positive and negative controls were run concurrently. The expression of each IHC marker was jointly scored by two pathologists (MR and MF), according to previous experiences with minor modifications.27,28 For HMGA2, ZEB1 and ZEB2 only nuclear immunostaining was considered. For ZEB1 and ZEB2 expression, cases were assessed as positive versus negative; HMGA2 immunostain was semiquantified using a three-tier scoring system based on extent of staining (0 = 0–5%; 1 = > 5–50% of positive cells and 2 = >50% of positive cells). For E-cadherin, positive membranous immunostaining was considered, semiquantified using a three-tier scoring system based on extent of staining (0 = 0–10%; 1 = > 10–90% of positive cells and 2 > 90% of positive cells). A total of 25 N, 25 BM, 16 LG, 11 HG and 10 BAc biopsy samples were analyzed in the IHC study.
Results
miRNAs are dysregulated during Barrett's carcinogenesis
To identify the miRNAs that are dysregulated in Barrett's carcinogenesis, we performed a miRNA microarray analysis on 51 macrodissected samples obtained from 14 BE patients who had undergone esophagectomy. The lesions considered were representative of the whole phenotypic spectrum of lesions observed in Barrett's carcinogenesis. The miRNA microarray analysis was performed using a custom microarray platform that had proved able to produce robust results, as validated by several studies.10,18,19,29,30 The analysis identified 13 miRNAs that were differently expressed during carcinogenesis (Fig. 1_a_ and Table 1, defined as the “progression” signature). Particularly, the signature included 6 miRNAs (hsa-miR-215, hsa-miR-560, hsa-miR-615-3p, hsa-miR-192, hsa-miR-326 and hsa-miR-147) with an increased expression and 7 miRNAs (hsa-miR-100, hsa-miR-23a, hsa-miR-605, hsa-miR-99a, hsa-miR-205, hsa-let-7c and hsa-miR-203) with a decreased expression.
Figure 1.
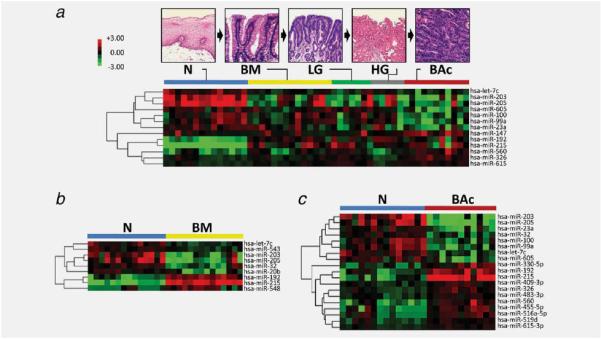
miRNAs differently expressed during human Barrett's carcinogenesis. Hierarchical clustering of the miRNA genes with a significantly different expression (p < 0.001) during Barrett's carcinogenesis (a), in Barrett's mucosa (BM versus normal squamous esophageal epithelium; b) and in Barrett's adenocarcinoma (BAc versus normal squamous esophageal epithelium; c). Rows represent individual genes; columns represent individual tissue samples. Pseudo-colors indicate transcript levels below, equal to, or above the mean (green, black and red, respectively). The scale represents the intensity of gene expression (log2 scale ranges between −3 and 3).
Table 1.
Differently expressed miRNAs in Barrett's carcinogenesis and between diseased and normal esophageal tissues
| Cancer progression1 | Normal vs Barrett's mucosa | Normal vs Barrett'sadenocarcinoma | |||||||
|---|---|---|---|---|---|---|---|---|---|
| miRNA | Correlationcoefficient | p | FDR-adjusted p | Foldchange | p | FDR-adjusted_P_ | Foldchange | p | FDR-adjusted_p_ |
| hsa-miR-215 | 0.608 | 3.8e–06 | 6.71e–04 | 62.77 | <1e–07 | <1e–07 | 50.51 | <1e–07 | <1e–07 |
| hsa-miR-560 | 0.550 | 3.99e–05 | 2.06e–03 | 4.39 | 3.18e–04 | 0.008 | |||
| hsa-miR-615-3p | 0.548 | 4.15e–05 | 2.06e–03 | 1.57 | 2.9e–06 | 2.56e–04 | |||
| hsa-miR-192 | 0.545 | 4.66e–05 | 2.06e–03 | 6.34 | <1e–07 | <1e–07 | 4.92 | 1.85e–05 | 0.001 |
| hsa-miR-326 | 0.489 | 3.22e–04 | 0.010 | 1.87 | 2.3e–05 | 0.001 | |||
| hsa-miR-147 | 0.458 | 8.3e–04 | 0.024 | ||||||
| hsa-miR-100 | –0.452 | 9.85e–04 | 0.027 | 0.31 | 3.6e–04 | 0.008 | |||
| hsa-miR-23a | –0.536 | 6.53e–05 | 0.002 | 0.26 | 1.36e–04 | 0.004 | |||
| hsa-miR-605 | –0.538 | 6.06e–05 | 0.002 | 0.23 | 1.84e–04 | 0.005 | |||
| hsa-miR-99a | –0.561 | 2.56e–05 | 0.002 | 0.20 | 9.9e–06 | 6.99e–04 | |||
| hsa-miR-205 | –0.591 | 7.7e–06 | 6.79e–04 | 0.10 | 1.39e–05 | 0.002 | 0.05 | 1e–07 | 1.76e–05 |
| hsa-let-7c | –0.595 | 6.4e–06 | 6.79e–04 | 0.49 | 3.11e–05 | 0.002 | 0.33 | 6.77e–05 | 0.003 |
| hsa-miR-203 | –0.661 | 3e–07 | 1.06e–04 | 0.15 | 3.2e–05 | 0.002 | 0.03 | 7e–07 | 8.24e–05 |
| hsa-miR-543 | 0.59 | 6.06e–05 | 0.004 | ||||||
| hsa-miR548b-3p | 2.64 | 1.91e–04 | 0.010 | ||||||
| hsa-miR-32 | 0.39 | 2.8e–04 | 0.012 | 0.46 | 3.35e–04 | 0.008 | |||
| hsa-miR-20b | 0.32 | 3.61e–04 | 0.014 | ||||||
| hsa-miR-409-3p | 1.69 | 5.3e–05 | 0.002 | ||||||
| hsa-miR-355-5p | 3.50 | 1.36e–04 | 0.004 | ||||||
| hsa-miR-330-5p | 2.62 | 4.45e–04 | 0.010 | ||||||
| hsa-miR-516a-5p | 2.43 | 4.88e–04 | 0.010 | ||||||
| hsa-miR-483-3p | 1.53 | 5.26e–04 | 0.010 | ||||||
| hsa-miR-519d | 1.68 | 6.37e–04 | 0.012 |
Different miRNA expression profiles were identified by comparing normal versus BM and normal versus BAc (Figs. 1_b_ and 1_c_, and Table 1). Particularly, nine miRNAs were found dysregulated in BM versus N and 19 in BAc versus N (Table 1). Some of these miRNAs (5 and 12, respectively) were shared with the progression signature, as obtained when the whole carcinogenic process was considered (Fig. 1_a_ and Table 1). Five 5 miRNAs were consistently dysregulated in all three signatures considered (i.e., progression signature, BM signature and BAc signature): up-regulation of hsa-miR-215 and hsa-miR-192, coexisting with hsa-miR-205, hsa-let-7c and hsa-miR-203 down-regulation suggested that this signature was significantly associated with BE progression.
qRT-PCR and ISH validation
To confirm the results of microarray analysis, we performed qRT-PCR and ISH analyses on two independent series of endoscopic biopsies (qRT-PCR) and esophagectomy specimens (ISH). For the qRT-PCR study, five miRNA dysregulations (the “progression signature”) were validated in a series of 15 N, 15 BM, 15 LG, 15 HG and 15 BAc biopsy samples. The replication results were consistent with those of the initial microarray experiments. Particularly, hsa-miR-215 and hsamiR-192 were significantly over-expressed, whereas hsa-miR-205, hsa-miR-203 and hsa-let-7c were significantly under-expressed during BE progression (all p < 0.001, Fig. 2). To further confirm the microarray data, we investigated, for the first time, miRNA expression by ISH using tissue samples (5 N, 5 BM and 5 BAc) obtained from five esophagectomy specimens from BAc patients. Using probes corresponding to hsa-miR-203, hsa-miR-205 and hsa-let-7c, a significant under-expression of these three miRNAs in BM and BAc samples was consistently confirmed (Fig. 3). All normal specimens showed miRNA expression as granular brown cytoplasmic staining consistently expressed by basal squamous epithelia (Fig. 3).
Figure 2.
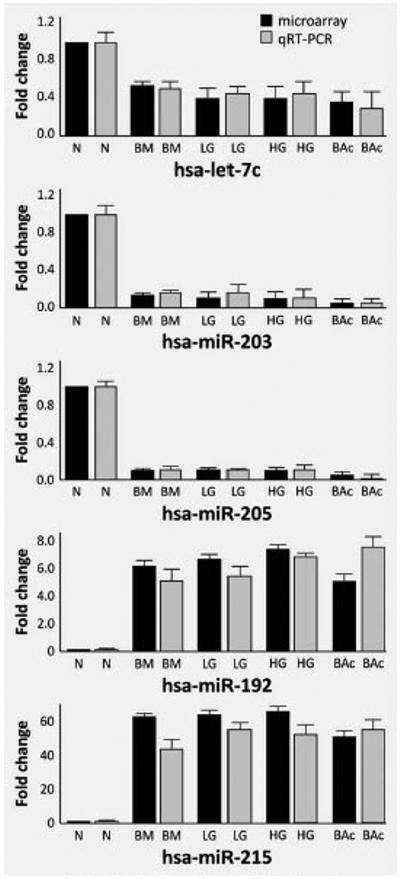
Quantitative RT-PCR analysis of miRNAs differently expressed during Barrett's carcinogenesis showing microarray and qRT-PCR results. The values for normal esophageal mucosa are set at 1 for both experiments. Results of qRT-PCR correlated closely with those of the microarrays. Both methods showed that let-7c, miR-203 and miR-205 are down-regulated and miR-192 and miR-215 are up-regulated in BM, LG, HG and BAc.
Figure 3.
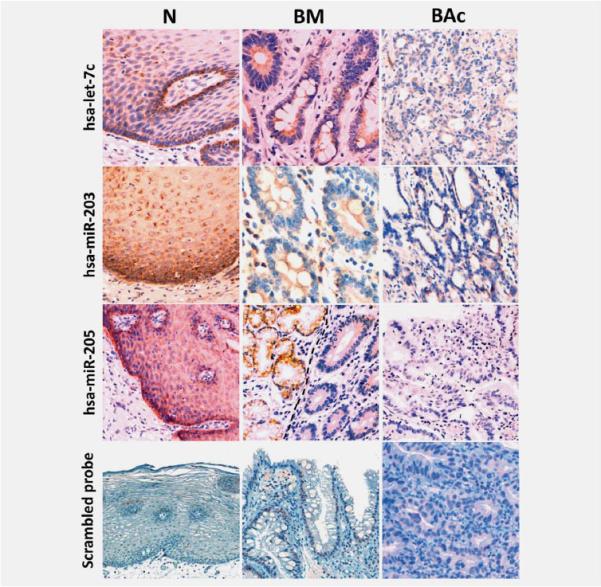
ISH analysis of miRNAs differently expressed in BM and BAc. Representative examples of let-7c, miR-203, miR-205 and the negative control (scrambled probe) in normal esophageal epithelium (N), BM and BAc. The presence of miRNAs is revealed by a grainy brown stain. In panel BM, note the different hsa-miR-205 expression between positive submucosal glands (left) and negative metaplastic epithelia (right). Original magnifications 40_X_ and 20_X_.
miRNA target gene expression during Barrett's carcinogenesis
A large body of evidence demonstrates that miRNAs modulate gene expression by binding to the 3′ untranslated region (UTR) of target mRNA, causing either mRNA degradation or translation inhibition.9,11 To demonstrate that miRNA analysis may be important in investigating BM- and BAc-specific genes, we correlate the expression of the five most significantly dysregulated miRNAs (i.e., hsa-miR215, hsa-miR-192, hsa-miR-205, hsa-let-7c and hsa-miR-203) with some of their previously-reported targets. To do so, we investigated target gene expression using the Oncomine database and/or IHC.
The Oncomine database and gene microarray data analysis tool enabled the meta-analysis of gene expression (mRNA) in the three available BAc microarray studies (Fig. 4).31–33 In the analysis, we considered the expression levels of HMGA2 (high mobility group 2, target of hsa-let-7c),34,35 ZEB1 and ZEB2 (targets of hsa-miR-205),36 as well as CDH1 (E-cadherin) mRNA expression. In fact, CDH1 is suppressed by ZEB1 and ZEB2, resulting in epithelial-mesenchymal transition (EMT), tumorigenesis and cancer metastasis.37
Figure 4.
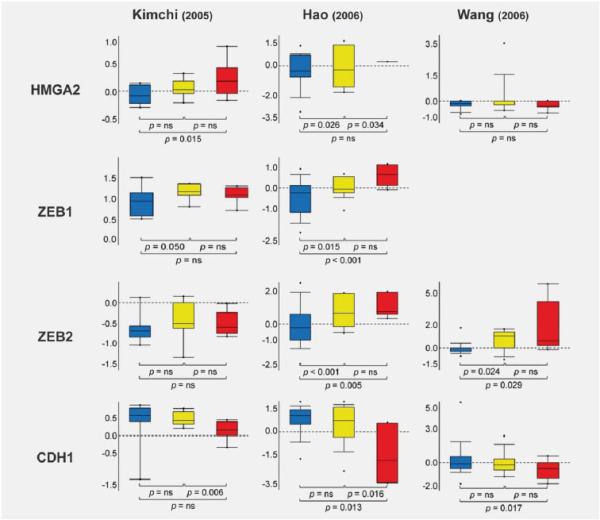
HMGA2, ZEB1, ZEB2 and CDH1 expression in BM and BAc. Expression (mRNA) array results of three Barrett's adenocarcinoma microarray data sets31–33 were analyzed, and statistical significance was calculated using the Oncomine website (www.oncomine.org). Box plots show differences in mRNA expression between normal esophageal mucosa (blue), BM (yellow) and BAc (red). The statistical significance is shown for comparisons of normal versus BM, BM versus BAc, and the sequence normal to BM to BAc.
HMGA2 is a small nonhistone chromosomal protein that modulates transcription by altering chromatin architecture.34,35 HMGA2 is abundantly expressed during embryogenesis, but undetectable in normal adult tissues, suggesting that it has a crucial role in cell proliferation/differentiation during embryonic development. On the other hand, HMGA2 over-expression is a hallmark of various benign and malignant human tumors, including gastric cancer. Given the significant down-regulation of hsa-let-7c expression during Barrett's carcinogen-esis, we performed an Oncomine study on HMGA2 mRNA expression. HMGA2 was found up-regulated in both BM and BAc in two of the three studies analyzed (but this up-regulation was only statistically significant in one; p = 0.015, Ref. 32; Fig. 4). To further support these findings, we investigated HMGA2 IHC expression in Barrett's carcinogenesis. As in gastric cancer, HMGA2 was significantly over-expressed in LG, HG and BAc samples (Figs. 5_a_–5_d_).
Figure 5.
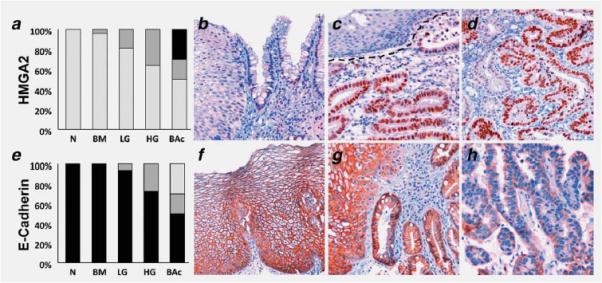
Representative examples of IHC expression of HMGA2 (a_–_d) and E-cadherin (e_–_h) in Barrett's carcinogenesis. The distribution of the IHC scores in the different histological lesions are also shown (a and e) [IHC scores: light gray = 0, dark grey = 1, black = 2; N = native squamous esophageal mucosa, BM = IM positive esophageal mucosa (Barrett's mucosa), LG = low-grade intra-epithelial neoplasia, HG = high grade intra-epithelial neoplasia, BAc = Barrett's adenocarcinoma]. (b) HMGA2 negative squamous and BM epithelia. (c) Well-differentiated HMGA2-positive BAc and its normal HMGA2-negative squamous counterpart. (d) BAc with scattered HMGA2 positive immunostaining. (f) Normal squamous epithelia featuring strong E-cadherin staining. (g) Strong membranous E-cadherin immunoreaction in squamous and BM epithelia. (h) Weak-to moderate E-cadherin expression in a HG sample (note the lack of membranous reinforcement). Original magnifications 40_X_ and 20_X_.
Given the significant down-regulation of hsa-miR-205 expression in Barrett's carcinogenesis, we also analyzed ZEB1 and ZEB2 expression using the Oncomine database (Fig. 4). ZEB1 and ZEB2 were found up-regulated in BM and BAc in two of two (one statistically significant; p < 0.001, Ref. 33) and two of three (both statistically significant, p = 0.005 and p = 0.025, Refs. 31 and 33) studies, respectively. Nuclear over-expression of both ZEB1 and ZEB2 was consistently observed in the BAc-associated stroma (data not shown). In contrast with the over-expression of mRNA levels, however, no positive nuclear immunostaining was documented in the epithelial compartment of preneoplastic/ neoplastic lesions.
As expected, CDH1 was found down-regulated in three of three studies (two statistically significant p = 0.013 and p = 0.017, Refs. 31 and 33). To further support the role of CDH1 in Barrett's carcinogenesis, we also investigated E-cadherin expression by IHC (Figs. 5_e_–5_h_): all N and BM samples showed strong membranous staining, whereas E-cadherin expression was down-regulated in 1/16 cases of LG, 3/11 cases of HG, and 5/10 cases of BAc.
Discussion
The incidence of BAc is increasing faster than that of any other solid tumor in the Western world.38 The phenotypic shift from BM to BAc involves a well-established sequence of histological lesions. The molecular profiling of each of these preneoplastic changes has produced inconsistent results.
Histological classification based on formalin-fixed, paraffin-embedded (FF-PE) tissue samples is a prerequisite for patients’ risk stratification, but in silico technologies are only partially applicable to FF-PE tissue samples and this significantly limits any efforts to associate molecular and histological profiles consistently.39
MicroRNA molecules are stable in FF-PP tissue samples40 and miRNA testing in previously histologically assessed lesions may consistently associate any target phenotype with its molecular profile. By using FF-PE tissues, aberrant miRNA expression patterns have been described in a variety of hematological and solid malignancies.9,11,41
In our study, a definite pattern of miRNA expression was consistently associated with the whole spectrum of the human Barrett's carcinogenic model (the “progression” profile); specific miRNA profiles were also obtained by comparing native esophageal mucosa with both BM and BAc. The miRNA signature of the whole BE carcinogenic sequence came closer to that of BAc than to that of BM (which had 12 and 5 miRNAs in common with the general signature, respectively; Table 1). In accordance with Yang et al., our results suggest that miRNAs play a more prominent part in the advanced than in the earlier steps of cancerous transformation.17
To expand on Yang's experience, however, we considered multiple tissue samples (representative of different precancerous/cancerous phenotypes) obtained from the same patients, enabling us to thoroughly explore the whole spectrum of the “cancerization field” in which BAc develops.
We identified five miRNAs significantly dysregulated in the progression of the disease (i.e., hsa-miR215 and hsa-miR-192 were up-regulated, whereas hsa-miR-205, hsa-let-7c and hsa-miR-203 were down-regulated), suggesting an important role for them in BE carcinogenesis. These results are consistent with previous findings in the three available miRNA microarray studies,12,15,17 in which hsa-let-7c, hsa-miR-203 and hsa-miR-205 were found significantly down-regulated in HG, BAc samples and cancer-derived cell lines,12,15,17 whereas hsa-miR-192 was up-regulated in BAc.12
Our oncomine and IHC studies confirmed previous reports of the down-regulation of E-cadherin in Barrett's carcinogenesis,42,43 suggesting its indirect miRNA-mediated dysregulation in BE.36,37 In tune with the previous findings on gastric carcinogenesis,34 we also obtained the first IHC images of HMGA2 protein in the metaplasia-dysplasia-carcinoma sequence occurring in BE.
In our series, the epithelial compartment of preneoplastic/ neoplastic lesions did not show any positive ZEB1 and/or ZEB2 nuclear immunostaining. This finding has been interpreted as related to the nature of the considered lesions: all the considered samples, in fact, consisted of preneoplastic lesions and/or well-differentiated BAc, which excluded those dedifferentiated cancers that are the most typically associated with EMT.
Unlike the case in the colon cancer model,44,45 we found hsa-miR-192 and hsa-miR-215 significantly up-regulated. These two miRNAs are reportedly induced by p53 activation, and they function as tumor suppressors through p21 accumulation and cell cycle arrest.44,45 In a similar series of samples, we observed a significant and progressive p53 inactivation (i.e., nuclear over-expression) along with the dedifferentiation of the considered lesions;46 further in vitro studies should specifically address the mechanisms involved in such an intriguing pattern of gene dysregulation.
Other miRNAs identified in our analysis have already been reported as down-regulated in BE carcinogenesis, i.e., hsa-miR-99a in HG and BAc samples,17 hsa-miR-100 in BAc and BAc-derived cells,12,15 hsa-miR20b and hsa-miR-23a in BAc-derived cells.15
An increased expression of hsa-miR-196a levels along with increased dysplasia was demonstrated by Maru et al.,14 while no such change in hsa-miR-196a expression was documented in our study; this difference could be reliably correlated with the significant interpatient and intrapatient variations in gene expression levels.47,48
In conclusion, our findings strongly support the role of miRNAs (and miRNA-related genes) in the molecular history of BAc. MicroRNA expression profiling may be a powerful tool for cancer diagnosis; regulating miRNA expression might be a novel strategy for the chemoprevention of human esophageal adenocarcinoma. Further, larger, multiinstitutional studies should investigate the prognostic significance of miRNA expression in BE patients.
Acknowledgements
The authors are grateful to Vincenza Guzzardo, Valentina Ferri, Cristiano Lanza and Vanni Lazzarin for their technical assistance. We wish to acknowledge the continuous support of the “G. Berlucchi” Foundation. The microarray data are deposited in the Gene Expression Omnibus at the National Center for Biotechnology Information (GEO:GSE20099).
Abbreviations
BAc
Barrett's adenocarcinoma
BE
Barrett's esophagus
BM
Barrett's mucosa
HG
high-grade intra-epithelial neoplasia
IHC
immunohistochemistry
IM
intestinal metaplasia
ISH
in situ hybridization
LG
low-grade intra-epithelial neoplasia
miRNA
microRNA
N
native stratified squamous epithelium
Footnotes
All authors of this research paper participated directly in the planning and execution of the study, and in the analysis of the results.
The authors have no competing interests to declare.
References
- 1.Sampliner RE. Update guidelines for the diagnosis, surveillance, and therapy of Barrett's esophagus. Am J Gastroenterol. 2002;97:1888–95. doi: 10.1111/j.1572-0241.2002.05910.x. [DOI] [PubMed] [Google Scholar]
- 2.Enzinger PC, Mayer RJ. Esophageal cancer. NEJM. 2003;349:2241–52. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 3.Paulson TG, Reid BJ. Focus on Barrett's esophagus and esophageal adenocarcinoma. Cancer Cell. 2004;6:11–6. doi: 10.1016/j.ccr.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgerald RC. Molecular basis of Barrett's oesophagus and oesophageal adenocarcinoma. Gut. 2006;55:1810–20. doi: 10.1136/gut.2005.089144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaninotto G, Minnei F, Guirroli E, Ceolin M, Battaglia G, Bellumat A, Betetto G, Bozzola L, Cassaro M, Cataudella G, Dal Bò N, Farinati F, et al. The Veneto Region's Barrett's Oesophagus Registry: aims, methods, preliminary results. Dig Liver Dis. 2007;39:18–25. doi: 10.1016/j.dld.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 6.Jankowski JA, Wright NA, Meltzer SJ, Triadafilopoulos G, Geboes K, Casson AG, Kerr D, Young LS. Molecular evolution of the metaplasia-dysplasia-adenocarcinoma sequence in the esophagus. Am J Pathol. 1999;154:965–73. doi: 10.1016/S0002-9440(10)65346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jankowski JA, Harrison RF, Perry I, Balkwill F, Tselepis C. Barrett's metaplasia. Lancet. 2000;356:2079–85. doi: 10.1016/S0140-6736(00)03411-5. [DOI] [PubMed] [Google Scholar]
- 8.Conio M, Blanchi S, Lapertosa G, Ferraris R, Sablich R, Marchi S, D'Onofrio V, Lacchin T, Iaquinto G, Missale G, Ravelli P, Cestari R, et al. Long-term endoscopic surveillance of patients with Barrett's esophagus. Incidence of dysplasia and adenocarcinoma: a prospective study. Am J Gastroenterol. 2003;98:1931–9. doi: 10.1111/j.1572-0241.2003.07666.x. [DOI] [PubMed] [Google Scholar]
- 9.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baffa R, Fassan M, Volinia S, O'Hara B, Liu CG, Palazzo JP, Gardiman M, Rugge M, Gomella LG, Croce CM, Rosenberg A. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–21. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 11.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–56. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feber A, Xi L, Luketich JD, Pennathur A, Landreneau RJ, Wu M, Swanson SJ, Godfrey TE, Litle VR. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135:255–60. doi: 10.1016/j.jtcvs.2007.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dijckmeester WA, Wijnhoven BPL, Watson DI, Leong MP, Michael MZ, Mayne GC, Bright T, Astill D, Hussey DJ. MicroRNA-143 and -205 expression in neosquamous esophageal epithelium following argon plasma ablation of Barrett's esophagus. J Gastrointest Surg. 2009;13:846–53. doi: 10.1007/s11605-009-0799-5. [DOI] [PubMed] [Google Scholar]
- 14.Maru DM, Singh RR, Hannah C, Albarracin CT, Li YX, Abraham R, Romans AM, Yao H, Luthra MG, Anandasabapathy S, Swisher SG, Hofstetter WL, et al. MicroRNA-196a is a potential marker of progression during Barrett's metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus. Am J Pathol. 2009;174:1940–8. doi: 10.2353/ajpath.2009.080718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kan T, Sato F, Ito T, Matsumura N, David S, Cheng Y, Agarwal R, Paun BC, Jin Z, Olaru AV, Selaru FM, Hamilton JP, et al. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136:1689–700. doi: 10.1053/j.gastro.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathe EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, Schetter AJ, Braun R, Reimers M, Kumamoto K, Hughes D, Altorki NK, Casson AG, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15:6192–200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang H, Gu J, Wang KK, Zhang W, Xing J, Chen Z, Ajani JA, Wu X. MicroRNA expression signatures in Barrett's esophagus and esophageal adenocarcinoma. Clin Cancer Res. 2009;15:5744–52. doi: 10.1158/1078-0432.CCR-09-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–8. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 19.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, Alder H, Bullrich F, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. PNAS. 2004;101:9740–4. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–55. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 21.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. PNAS. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamichi N, Shimomura R, Inada K, Sakurai K, Haraguchi T, Ozaki Y, Fujita S, Mizutani T, Furukawa C, Fujishiro M, Ichinose M, Shiogama K, et al. Locked nucleic acid in situ hybridization analysis of miR-21 expression during colorectal cancer development. Clin Cancer Res. 2009;15:4009–16. doi: 10.1158/1078-0432.CCR-08-3257. [DOI] [PubMed] [Google Scholar]
- 23.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale metaanalysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. PNAS. 2004;101:9309–14. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xin W, Rhodes DR, Ingold C, Chinnaivan AM, Rubin MA. Dysregulation of the annexin family protein family is associated with prostate cancer progression. Am J Pathol. 2003;162:255–61. doi: 10.1016/S0002-9440(10)63816-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rugge M, Fassan M, Clemente R, Rizzardi G, Giacomelli L, Pennelli G, Mescoli C, Segat D, Rea F. Bronchopulmonary carcinoid: phenotype and long-term outcome in a single-institution series of Italian patients. Clin Cancer Res. 2008;14:149–54. doi: 10.1158/1078-0432.CCR-07-1631. [DOI] [PubMed] [Google Scholar]
- 27.Mu G, Liu H, Zhou F, Xu X, Jiang H, Wang Y, Qu Y. Correlation of overexpression of HMGA1 and HMGA2 with poor tumor differentiation, invasion, and proliferation associated with let-7 down-regulation in retinoblastomas. Hum Pathol. 2010;41:493–502. doi: 10.1016/j.humpath.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 28.Jian WG, Darnton SJ, Jenner K, Billingham LJ, Matthews HR. Expression of E-cadherin in oesophageal carcinomas from the UK and China: disparities in prognostic significance. J Clin Pathol. 1997;50:640–4. doi: 10.1136/jcp.50.8.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, Gomella LG, Croce CM, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–92. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 30.Fassan M, Baffa R, Palazzo JP, Lloyd J, Crosariol M, Liu CG, Volinia S, Alder H, Rugge M, Croce CM, Rosenberg A. MicroRNA expression profiling of male breast cancer. Breast Cancer Res. 2009;11:R58. doi: 10.1186/bcr2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, Zhan M, Yin J, Abraham JM, Mori Y, Sato F, Xu Y, Olaru A, Berki AT, Li H, Schulmann K, Kan T, et al. Transcriptional profiling suggests that Barrett's metaplasia is an early intermediate stage in esophageal adenocarcinogenesis. Oncogene. 2006;25:3346–56. doi: 10.1038/sj.onc.1209357. [DOI] [PubMed] [Google Scholar]
- 32.Kimchi ET, Posner MC, Park JO, Darga TE, Kocherginsky M, Karrison T, Hart J, Smith KD, Mezhir JJ, Weichselbaum RR, Khodarev NN. Progression of Barrett's metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentation. Cancer Res. 2005;65:3146–54. doi: 10.1158/0008-5472.CAN-04-2490. [DOI] [PubMed] [Google Scholar]
- 33.Hao Y, Triadafilopoulos G, Sahbaie P, Young HS, Omary MB, Lowe AW. Gene expression profiling reveals stromal genes expressed in common between Barrett's esophagus and adenocarcinoma. Gastroenterology. 2006;131:925–33. doi: 10.1053/j.gastro.2006.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Motoyama K, Inoue H, Nakamura Y, Uetake H, Sugihara K, Mori M. Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clin Cancer Res. 2008;14:2334–40. doi: 10.1158/1078-0432.CCR-07-4667. [DOI] [PubMed] [Google Scholar]
- 35.Peng Y, Laser J, Shi G, Mittal K, Melamed J, Lee P, Wei JJ. Antiproliferative effects by Let-7 repression of high-mobility group A2 in uterine leiomyoma. Mol Cancer Res. 2008;6:663–73. doi: 10.1158/1541-7786.MCR-07-0370. [DOI] [PubMed] [Google Scholar]
- 36.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 37.Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008;5:115–9. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 39.Doleshal M, Magotra AA, Choudhury B, Cannon BD, Labourier E, Szafranska AE. Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2008;10:203–11. doi: 10.2353/jmoldx.2008.070153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, Tavarè S, Caldas C, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumour subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. PNAS. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darlavoix T, Seelentag W, Yan P, Bachmann A, Bosman FT. Altered expression of CD44 and DKK1 in the progression of Barrett's esophagus to esophageal adenocarcinoma. Virchows Arch. 2009;454:629–37. doi: 10.1007/s00428-009-0769-z. [DOI] [PubMed] [Google Scholar]
- 43.Feith M, Stein HJ, Mueller J, Siewert JR. Malignant degeneration of Barrett's esophagus: the role of the Ki-67 proliferation fraction, expression of E-cadherin and p53. Dis Esophagus. 2004;17:322–7. doi: 10.1111/j.1442-2050.2004.00434.x. [DOI] [PubMed] [Google Scholar]
- 44.Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, Jackson AL, Carleton MO, Linsley PS, Cleary MA, Chau BN. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68:10105–12. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- 45.Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Orntoft TF, Andersen CL, Dobbelstein M. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rugge M, Fassan M, Zaninotto G, Pizzi M, Giacomelli L, Battaglia G, Rizzetto C, Parente P, Ancona E. Aurora kinase A in Barrett's carcinogenesis. Hum Pathol. 2010;41:1380–6. doi: 10.1016/j.humpath.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 47.Xu JZ, Wong CW. Hunting for robust gene signature from cancer profiling data: sources of variability, different interpretations, and recent methodological developments. Cancer Lett. 2010;296:9–16. doi: 10.1016/j.canlet.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 48.Hennig EE, Mikula M, Orlowska J, Jarosz D, Bielasik A, Regula J, Ostrowski J. Large intra- and inter-individual variability of genes expression levels limits potential predictive value of molecular diagnosis of dysplasia in Barrett's esophagus. J Mol Med. 2008;86:233–42. doi: 10.1007/s00109-007-0271-5. [DOI] [PubMed] [Google Scholar]