PHD Domain-Mediated E3 Ligase Activity Directs Intramolecular Sumoylation of an Adjacent Bromodomain which is Required for Gene Silencing (original) (raw)
. Author manuscript; available in PMC: 2015 Mar 4.
SUMMARY
Tandem PHD and bromodomains are often found in chromatin-associated proteins and have been shown to cooperate in gene silencing. Each domain can bind specifically modified histones: the mechanisms of cooperation between these domains are unknown. We show that the PHD domain of the KAP1 corepressor functions as an intramolecular E3 ligase for sumoylation of the adjacent bromodomain. The RING finger-like structure of the PHD domain is required for both Ubc9 binding and sumoylation and directs modification to specific lysine residues in the bromodomain. Sumoylation is required for KAP1-mediated gene silencing and functions by directly recruiting the SETDB1 histone methyltransferase and the CHD3/Mi2 component of the NuRD complex via SUMO interacting motifs. Sumoylated KAP1 stimulates the histone methyltransferase activity of SETDB1. These data provide a mechanistic explanation for the cooperation of PHD and bromodomains in gene regulation and describe a new function of the PHD domain as an intramolecular E3 SUMO ligase.
INTRODUCTION
Eukaryotic gene regulation requires the coordination of multiple enzymatic activities that target nucleosomal histones for post-translational modification. Protein domains which recognize specific histone modifications are key components of this mechanism (Seet et al., 2006). The bromodomain recognizes acetylated lysines in histone H3 and H4 (Mujtaba et al., 2007). The PHD domain of ING2 and BPTF binds methylated histone H3 (Li et al., 2006; Shi et al., 2006; Wysocka et al., 2006). The PHD and bromodomain are often found adjacent to each other suggesting that they may form a cooperative module used to interpret combinatorial modifications of histones. Cooperation of the PHD and the bromodomain occurs in p300, TIP5, BPTF and KAP1 proteins (Ragvin et al., 2004; Schultz et al., 2001; Wysocka et al., 2006; Zhou and Grummt, 2005).
KAP1/TIF1 beta is a universal corepressor for the largest family of transcriptional silencers encoded in the human genome, KRAB domain-containing zinc finger proteins (ZFPs) (Huntley et al., 2006). The KAP1 N-terminal RBCC region is responsible for KRAB domain binding (Friedman et al., 1996; Peng et al., 2000), while the C-terminal HP1 binding domain and tandem PHD-bromodomain are required for gene silencing. The KAP1 PHD-bromodomain module is necessary for recruitment of the H3-K9 specific HMTase SETDB1 and NuRD complex protein CHD3 to the promoter regions of KRAB modulated genes (Schultz et al., 2002; Schultz et al., 2001; Sripathy et al., 2006). Mutations in either the PHD or bromodomain compromise KAP1 interaction with these partner proteins and relieve repression, suggesting that they act cooperatively. The PHD domain of KAP1 is highly related to the RING finger (Capili et al., 2001), often found in ubiquitin and SUMO E3 ligases (Joazeiro and Weissman, 2000).
SUMO family proteins are conjugated to target lysines via a cascade of E1 activating, E2 transfer and E3 ligase enzymes (Johnson, 2004). The SUMO E2 protein Ubc9 often recognizes the consensus sequence ψKxE/D (ψ - hydrophobic) in the target protein and catalyzes SUMO conjugation (Sampson et al., 2001) leading to alterations in protein localization and function. A family of deconjugation enzymes, SENPs, is responsible for rapid removal of SUMO from target lysines (Mukhopadhyay and Dasso, 2007) which accounts for the very temporal nature of this modification. Sumoylation is emerging as a key posttranslational modification involved in transcriptional repression (Gill, 2005), however, the detailed mechanisms of its action remain obscure. SUMO modification can enhance recruitment of repression machinery, including HDACs (Girdwood et al., 2003; Yang and Sharrocks, 2004) and may also act to preclude other lysine modifications leading to impaired protein-protein interactions.
Here, we show that the KAP1 PHD domain binds to Ubc9 and directs SUMO conjugation of an adjacent bromodomain, and that sumoylation is required for KRAB-KAP1 mediated repression. The PHD domain of KAP1 functions as an intramolecular SUMO E3 ligase. SETDB1 and CHD3 encode functional SIM motifs and bind SUMO-modified KAP1. Thus, sumoylation-mediated repression occurs via direct recruitment of H3-K9 HMTase and HDAC activities.
RESULTS
KAP1 is Sumoylated In Vivo and In Vitro
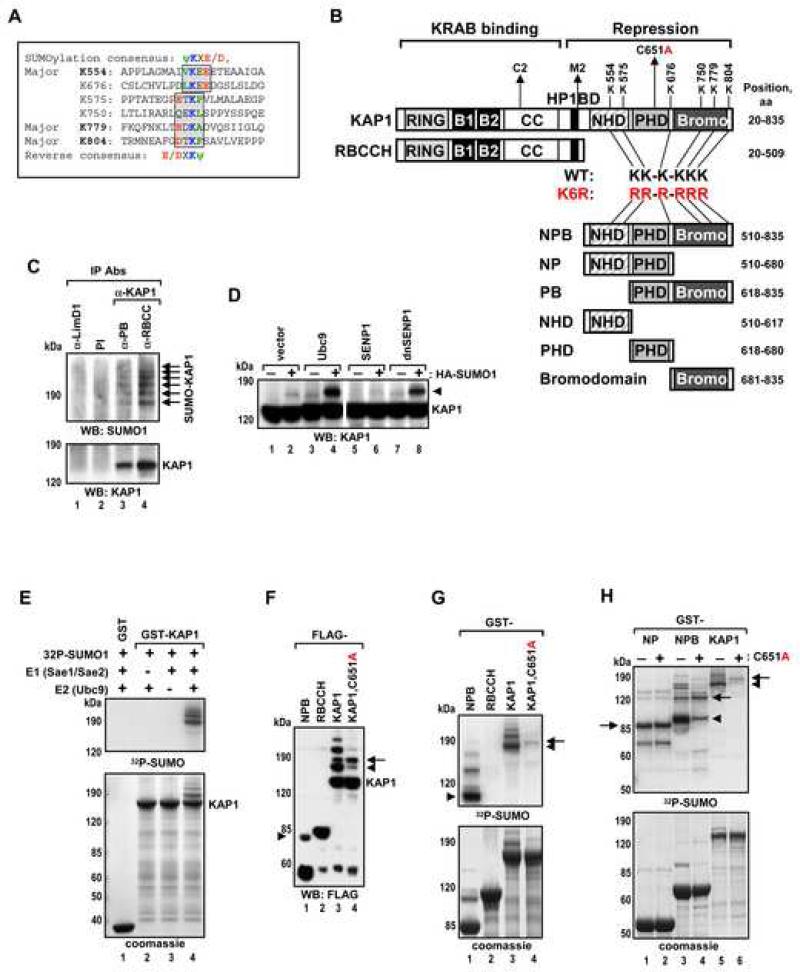
KAP1 contains six lysines conforming to the consensus ψKxE/D: canonical sites at K554 and K676, and four reverse sites at K575, K750, K779 and K804 (Figure 1A,B). Immunoprecipitation of endogenous KAP1 under conditions preserving the SUMO modification revealed a ladder of SUMO1-modified KAP1 species (Figure 1C). Expression of SUMO1, Ubc9 or a dominant negative SENP1 greatly enhanced KAP1-SUMO1 conjugation, whereas expression of wild type SENP1 prevented it (Figure 1D). KAP1 sumoylation was also efficiently reconstituted in vitro with purified proteins: Reactions containing all components produced a ladder of slower migrating KAP1 forms consistent with conjugation of multiple SUMO1 moieties. Reactions that lacked either E1 or Ubc9 did not show sumoylation products (Figure 1E). Thus, KAP1 is conjugated with SUMO on multiple sites, both in vivo and in vitro.
Figure 1. KAP1 is Sumoylated in Vivo and in Vitro in a PHD-Dependent Manner.
(A) Sequence alignment of six KAP1 sumoylation sites. Forward and reverse sumoylation consensuses are shown and boxed. Major sumoylation lysines are in bold.
(B) Schematic representation of KAP1 structural domains. B1 – box 1, B2 – box 2, CC – coiled-coil, HP1BD – HP1 binding domain. Amino acid positions of KAP1 sumoylation site lysines are indicated. Truncations of KAP1 are abbreviated with the first letters of each domain (PB = PHDBromodomain). Sumoylation deficient mutant with all six sumoylation site lysines substituted to arginines is designated RR-R-RRR or K6R.
(C) Proteins were immunoprecipitated from H1299 cells with pre-immune serum or antibodies specific for different regions of KAP1: PB or RBCC; or an unrelated LimD1 antibody. A SUMO1 antibody was used in Western blot. Sumoylated forms of KAP1 are indicated with arrows.
(D) HEK293 cells were transfected with vector, Ubc9, SENP1 or dnSENP1 plasmids with or without HA-SUMO1 as indicated. Proteins were detected by Western blot with KAP1 antibody. SUMO-KAP1 is indicated with an arrowhead.
(E) In vitro sumoylation of GST-KAP1. Immobilized GST and GST-KAP1 were incubated in the presence or absence of recombinant E1, Ubc9, and 32P-SUMO1 as indicated. All in vitro sumoylation reactions described below were analyzed by both Coomassie blue staining (bottom panel) and autoradiography (top panel).
(F) HEK293 cells were transfected with the indicated FLAG-KAP1 and T7-SUMO1 plasmids. Proteins were analyzed by Western blot with FLAG antibody.
(G-H) In vitro sumoylation of indicated GST-KAP1 proteins. The major mono-sumoylated forms of KAP1 are indicated with arrowheads.
Mutation of the PHD Domain Inhibits KAP1 Sumoylation
SUMO modification of KAP1 occurs only in the KAP1 C-terminus (NPB fragment, Figure 1F,G) overlapping the known repression domains NHD (**N-**CoR2 Homology Domain, see Figure S1A), the PHD and the bromodomain. The KAP1 PHD domain structure showed it to be highly related to the RING finger (Capili et al., 2001), often found in ubiquitin and SUMO E3 (Joazeiro and Weissman, 2000). The RING domain in PIAS proteins is responsible for E2 binding and sumoylation ((Kotaja et al., 2002) and Figure 3F). Few E3 SUMO ligses have been identified. To test the hypothesis that the PHD is a SUMO ligase, we introduced a C651A mutation, which disrupts its structure. SUMO modification of the C651A KAP1 protein was drastically reduced (Figure 1F,G, lane 4). However, the intact PHD was required for sumoylation of many, but not all sites in KAP1 (Figure 1F,G, arrow): Sumoylation of the NP fragment which lacks the bromodomain was unaffected by the C651A mutation (Figure 1H). Therefore, mutation in the PHD affected sumoylation of the bromodomain, but not of the adjacent NHD domain or the PHD itself. These results suggest that the PHD domain plays a key role in KAP1 sumoylation and it is required to target the adjacent bromodomain.
Figure 3. The KAP1 PHD Binds to Ubc9 with High Selectivity and Directs Sumoylation of the Adjacent Bromodomain.
(A) In vitro sumoylation of GST-PB wild type, indicated sumoylation site mutants and C651A mutant proteins. Arrowheads and arrows indicate the positions of mono- and di-sumoylated forms, respectively.
(B) HEK293 cells were transfected with indicated LexA-PB plasmids together with 4xLexA-TK-luc plasmid. Data are the mean ± SD of at least two experiments performed in duplicate.
(C-D) GST and GST fusions of SUMO1, Ubc9, Sae1 and Sae2 were immobilized on glutathione beads and incubated with recombinant KAP1 PHD. Bound proteins were analyzed by Coomassie blue staining. Lane 1 (C) was loaded with a protein marker in addition to an input sample of the PHD protein.
(E-G) In vitro sumoylation of GST- (E) PB modules from KAP1, TIF1α, TIF1γ, Sp100C; (F) RING or RING with adjacent domain (RR) from PIAS1 (aa 140-320), PHD or PB from KAP1; and (G) KAP1 PB and BP chimera.
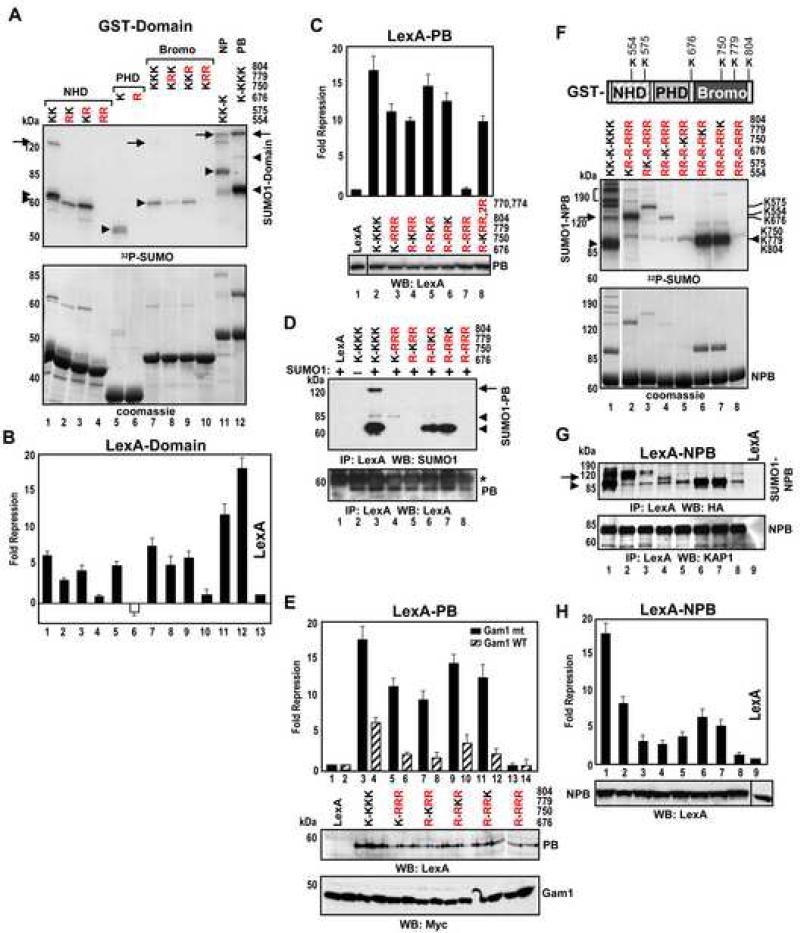
Selective Utilization of Sumoylation Sites and Their Contribution to KAP1 Repression Activity
An extensive lysine-to-arginine mutation analysis defined the contribution of sumoylation to the repression function of KAP1. Mutation of the canonical consensus, K554 in the NHD reduced sumoylation and repression (Figure 2A,B, lane 2). Mutation of K575 modestly reduced, while the double mutation abolished, both sumoylation and repression (lane 4). Mutation of K676 in the PHD domain had a similar effect (lane 6). Simultaneous mutation of K779 and K804 in the bromodomain completely abrogated sumoylation and repression (lanes 8-10). Introduction of triple substitutions in the PB (PHD-Bromodomain) revealed a cryptic acceptor, K750 in the bromodomain which was not detected in the absence of the PHD (lane 10) and occurs in a non-canonical motif, QEKL. Additional inactivation of this fourth site completely abrogated repression (Figure 2C, lane 7). Mutation of bromodomain lysines K770 and K774 had no effect (lane 8). Analysis of mutant PB proteins with only one wild type acceptor lysine (out of four) allowed us to evaluate relative contribution of each site to sumoylation efficiency and repression activity (Figure 2C,D). Lysines K779 and K804 were predominant. Though K676 and K750 sumoylation was much less efficient, these sites still contributed to repression.
Figure 2. Convergence of Sumoylation Sites and Repression Domains in KAP1.
(A) In vitro sumoylation of wild type or sumoylation site KAP1mutants. Arrowheads and arrows indicate the positions of mono- and di-sumoylated forms, respectively.
(B-C) HEK293 cells were transfected with plasmids expressing the KAP1 mutants as in (A) fused to LexA, together with a 4xLexA-TK-luc plasmid. LexA-PB plasmids were used in panel C. Data are the mean ± SD of at least two experiments performed in duplicate.
(D) HEK293 cells were transfected with T7-SUMO1 and LexA-PB plasmids. LexA-PB proteins were immunoprecipitated with LexA antibody and analyzed by Western blot with SUMO1 (top) and LexA antibodies (bottom). Asterisk – IgGs.
(E) HEK293 cells were transfected with the indicated LexA-PB plasmids and wild type or mutant Myc-Gam1 together with 4xLexA-TK-luc plasmid. Protein expression of LexA-PB and Gam1 proteins was confirmed by Western blot (bottom). Data are the mean ± SD of at least two experiments performed in duplicate.
(F) In vitro sumoylation of GST-NPB KAP1 proteins.
(G) HEK293 cells were transfected with HA-SUMO1 and the indicated LexA-NPB plasmids. Proteins were immunoprecipitated with LexA antibody and analyzed by Western blot using HA (top) and KAP1 antibodies (bottom).
(H) HEK293 cells were transfected with the same LexA-NPB plasmids as in (G) with 4xLexATK-luc plasmid. The expression of LexA-NPB proteins was confirmed by Western blot (bottom). Data are the mean ± SD of at least two experiments performed in duplicate.
The Gam1 protein is a specific inhibitor of the endogenous sumoylation machinery, and appears to target the SUMO E1 machinery for degradation (Boggio et al., 2004). When each KAP1 PB mutant was cotransfected with the wild type Gam1, the repression activity was strongly reduced (Figure 2E), suggesting that each lysine is a site for in vivo sumoylation and that, irrespective of other modifications which may occur on these lysines, sumoylation has a major contribution to repression.
Analysis of the NPB fragment of KAP1 containing all six acceptor lysines allowed us to establish a strict hierarchy in the site utilization. The most efficiently sumoylated sites were K779, K804 and K554 (Figure 2F,G). The difference in SDS-PAGE mobility of the SUMO-conjugated NPB mutants allowed us to assign specific bands to specific acceptor lysines. The predominant mono-sumoylated NPB protein (lane 1) migrated in an identical position to the SUMO conjugates at the bromodomain sites K779 or K804 (lanes 6,7; see also Figure S1B). Analysis of mutant NPB proteins with only one wild type acceptor lysine showed a clear correlation between the extent of sumoylation and repression activity (Figure 2F-H): mutation of all six acceptor lysines abolished sumoylation and repression (lane 8). Thus, sumoylation of KAP1 predominantly targets K779 and K804 in the bromodomain.
KAP1 PHD Binds to Ubc9 with High Selectivity and Directs Sumoylation of the Adjacent Bromodomain
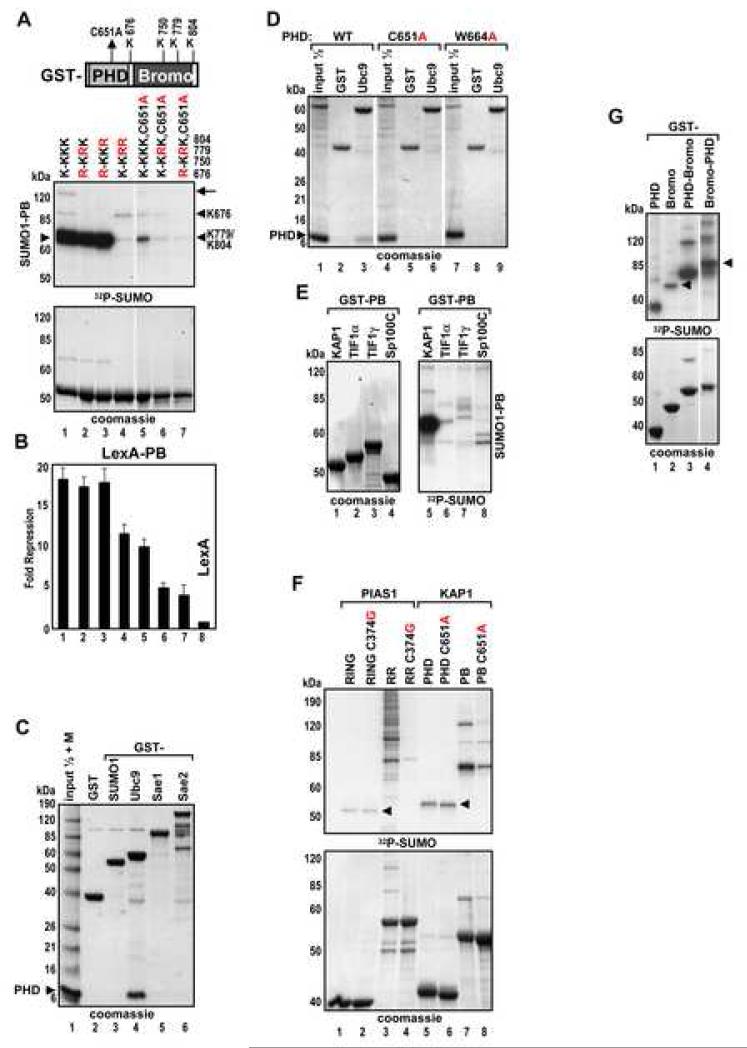
The PHD-bromodomain module (PB) was a >10-fold better substrate for sumoylation and better repressor than either domain alone (Figure 2A,B, compare lanes 5,7 and 12). Mutation of the PHD (C651A) greatly diminished SUMO conjugation to the bromodomain K779 and K804 and impaired repression (Figure 3A,B, lanes 5-7) implying that the PHD enhances bromodomain sumoylation. The SUMO E2 enzyme, Ubc9 bound strongly to recombinant PHD protein but did not bind to SUMO1, Sae1 or Sae2 (Figure 3C). Structural mutations C651A or W664A in the PHD abolished the binding (Figure 3D, lanes 3,6,9). The PHD domains from closely related TIF1 proteins and from Sp100C were unable to bind to Ubc9 (Figure S2A) and were very poor sumoylation substrates compared to the KAP1 PB (Figure 3E) despite the fact that they encode similar sumoylation sites (Figure S2B). Thus, the correlation between Ubc9 binding and sumoylation provides strong evidence that the KAP1 PHD-Ubc9 interaction is critical for KAP1 SUMO modification.
We compared KAP1 with the known SUMO E3 ligase PIAS1 and analyzed their sumoylation in relation to the structural integrity of the Ubc9 binding determinants, PHD and RING domains, respectively. Both, the RING and PHD domains were poor substrates themselves (Figure 3F, lanes 1 and 5, arrowhead). However, they efficiently stimulated sumoylation of adjacent domains, the N-terminal globular domain in PIAS1 (RR) and bromodomain in KAP1 (PB), respectively (lanes 3 and 7). Structural mutations in both RING and PHD domains, which disrupt Ubc9 binding, drastically reduced PIAS1 and KAP1 sumoylation (lanes 4,8) suggesting that the KAP1 PHD functions similarly to the PIAS1 RING domain and acts as an intramolecular SUMO E3 ligase.
To determine if the KAP1 PHD can function as SUMO ligase in-trans, we added increasing amounts of PHD protein to the isolated bromodomain or PB with a mutated PHD domain. This did not result in increased SUMO conjugation (Figure S2C). Fusion of the PHD domain to the C-terminus of the bromodomain (Bromo-PHD chimera) enhanced sumoylation (Figure 3G, compare lanes 2 and 4) indicating that a covalent link between the two domains is required for proper SUMO ligation.
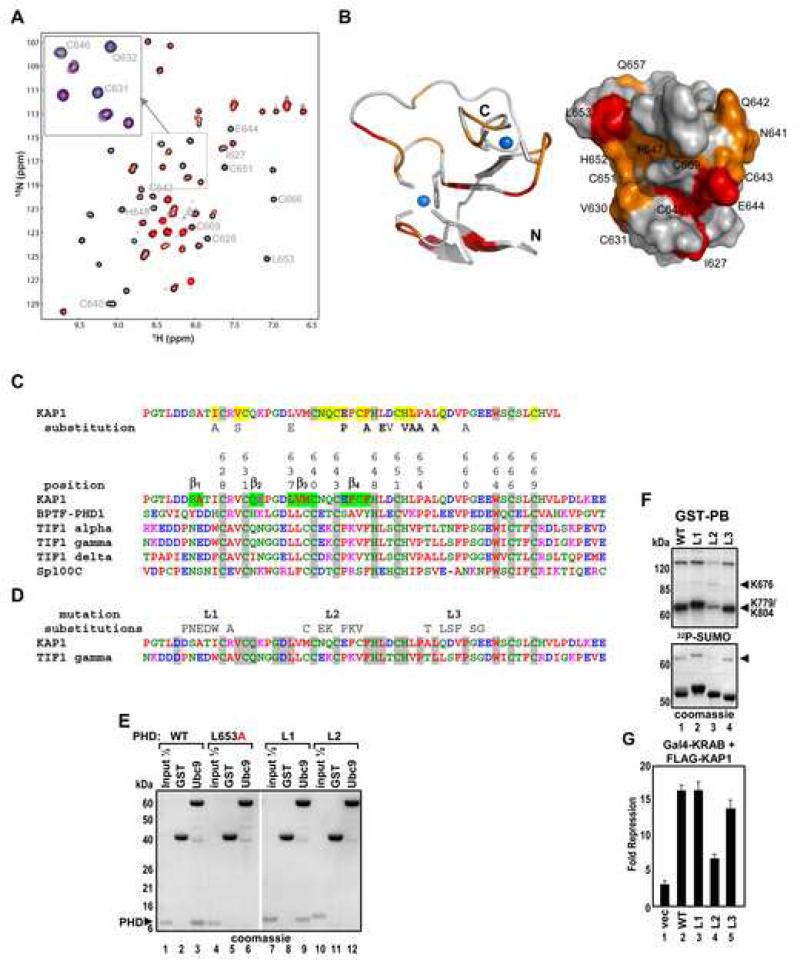
Mapping of Ubc9 Binding Site on the KAP1 PHD
NMR spectra of the 15N-labeled KAP1 PHD revealed that upon addition of Ubc9 many amino acid residues of the PHD protein exhibited significant chemical shift perturbations - predominantly line-broadening effects, indicative of direct PHD-Ubc9 binding (Figure 4A and S3). The ligand binding-induced line-broadening effect likely results from the dissociation rate of the PHD-Ubc9 complex on the millisecond NMR timescale, suggesting a low micromolar binding affinity of the two proteins. NMR resonance assignments (Figure S3) allowed mapping of the Ubc9 binding site on the PHD domain: the most perturbed amino acid residues on the surface representation model of the PHD molecule were color-coded as shown (Figure 4B). An exhaustive mutation analysis of the KAP1 PHD sequence revealed a very good correlation between decreased in vitro sumoylation efficiency and NMR resonance perturbation upon Ubc9 titration (Figure S4 and 4C).
Figure 4. Mapping of the Ubc9 Binding Site on KAP1 PHD.
(A) 2D 1H-15N HSQC spectral comparison of 15N-labeled KAP1 PHD in the free form (black) and in the presence of Ubc9 (red). Molar ratio of KAP1 PHD to Ubc9 is 1:2.4. (Inset) Expansion of spectral region illustrating three peaks that undergo line broadening during Ubc9 titration. The blue peaks correspond to a mid-point of the titration, in which the PHD:Ubc9 molar ratio is 1:1.2.
(B) Ribbon and surface representations of the KAP1 PHD structure (PDB code 1FP0), highlighting the residues that exhibited major resonance perturbations upon Ubc9 binding. KAP1 residues corresponding to NMR peaks that undergo line broadening at a PHD:Ubc9 molar ratio of 1:1.2 are in red, and those affected at 1:1.8 are in orange.
(C) Sequence alignment of PHD domains from KAP1, TIF1s, BPTF and Sp100C. The KAP1 PHD residues perturbed in the NMR titration experiment are shaded in yellow. Substitutions which inhibit sumoylation (Figure S3) are indicated.
(D) Sequence alignment of PHD domains from KAP1 and TIF1γ. The 6 aa substitutions of predicted loop segments L1, L2 and L3 are shown above. Identical residues are shaded in grey.
(E) GST, GST-Ubc9 were immobilized on glutathione beads and incubated with the indicated recombinant KAP1 PHDs. Bound proteins were analyzed by Coomassie blue staining.
(F) In vitro sumoylation of GST-PB wild type or the indicated loop mutants.
(G) U2OS K4 cells were transfected and treated as in Fig.5D. Data are the mean ± SD of at least two experiments performed in duplicate.
Most of the Ubc9 perturbed residues are located on one side of the PHD molecule and belong to both zinc coordination sites, as well as to loop 1 between β1 and β2 strands, loop 2 between β3 and β4 strands and to an adjacent region containing H652 and L653 (Figure 4C) at the PHD/bromodomain interface (see accompanying paper, Zeng et al). The perturbed residues within loop 1 are well conserved among PHD domains of TIF1 family members, whereas the loop 2 and β4 strand residues and L653 are unique to KAP1 PHD (Figure 4D and S5A). We performed subdomain swaps of the most diverged contiguous amino acid sequences between the KAP1 and TIF1γ PHD domains where KAP1 PHD segments containing loops 1, 2 or 3 were replaced with corresponding TIF1γ sequences (6 aa substitutions) (Figure 4D). The respective mutants, L1, L2 or L3 were tested in GST pull-down, in vitro sumoylation and repression assays (Figures 4E-G) which confirmed that the L2 segment of the KAP1 PHD together with L653 contain the critical amino acid contacts/surfaces required for binding to Ubc9. We verified that the L653A and L2 substitutions did not cause any gross changes in the PHD structural fold that were observed for the C651A mutant (Figure S5B).
Sumoylation Is Required for KRAB Domain-Mediated Repression
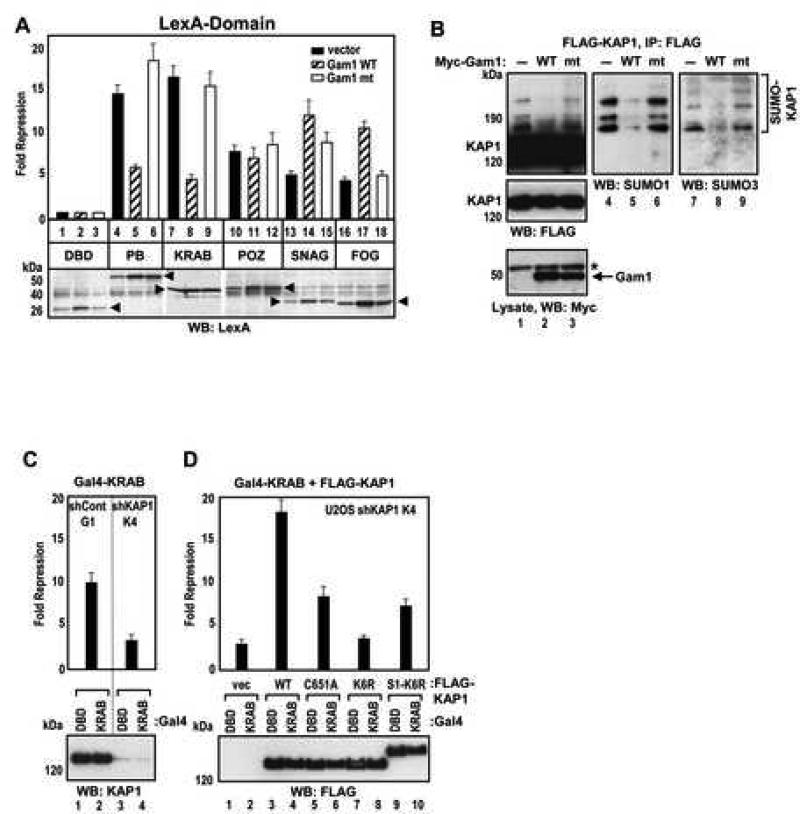
Since KAP1 is an obligate corepressor for the KRAB domain, we sought to determine if sumoylation is required for KRAB domain-mediated repression. As a comparison, several other well characterized repression domains were tested in addition to the KRAB domain. LexA fusions to KRAB, POZ, SNAG and FOG domains were cotransfected with Gam1. Each domain demonstrated potent repression in the absence of Gam1 (Figure 5A). However, the repression activity of LexA-KAP1 PB and LexA-KRAB was markedly reduced by wild type Gam1 but not the Gam1 mutant. The effect of Gam1 on repression was specific for the KRAB-KAP1 pathway since neither POZ, SNAG nor FOG domains showed significant relief of repression in the presence of Gam1. We confirmed that expression of Gam1 markedly decreased KAP1 sumoylation in cells (Figure 5B).
Figure 5. Role of KAP1 Sumoylation in KRAB Domain Mediated Repression.
(A) HEK293 cells were transfected with the indicated LexA fusions together with vector, wild type or mutant Gam1, and 4xLexA-TK-luc plasmid. The expression of the LexA fusion proteins (arrowheads) was confirmed by Western blot (bottom).
(B) HEK293 cells were transfected with FLAG-KAP1 and T7-SUMO1 plasmid together with vector, wild type or mutant Myc-Gam1 plasmids. Gam1 was detected with Myc antibody (bottom). Asterisk - non-specific band. KAP1 was immunoprecipitated with FLAG M2 antibody and Western blotted with FLAG (lanes 1-3), SUMO1 (lanes 4-6) and SUMO3 (lanes 7-9) antibodies.
(C) U2OS-G1 and -K4 cells were transfected with Gal4-DBD or Gal4-KRAB plasmids together with 5xGal4-TK-luc plasmid. Endogenous KAP1 protein expression level was verified by Western blot (bottom).
(D) U2OS-K4 cells were transfected and treated as in (C) except that vector, FLAG-KAP1 wild type or mutant plasmids were included in the transfection (top). FLAG-KAP1 protein expression was confirmed by Western blot (bottom).
(A,C,D) Data are the mean ± SD of at least two experiments performed in duplicate.
Sumoylated KAP1 is Required to Complement KRAB-mediated Repression in KAP1 Knockdown Cells
We created U2OS cell lines with a stable knockdown of KAP1 using vector-based shRNA technology (Wang et al., 2005). Clone U2OS-K4 showed a >95% knockdown of endogenous KAP1 (Figure S6A and 5C). Cotransfection of KRAB reporter plasmids into control U2OS cells yielded about 10-fold repression, whereas this repression activity in U2OS-K4 cells was significantly attenuated (Figure 5C). Complementation with a FLAG-KAP1 cDNA resistant to the action of the shRNA (Figure S6B) restored Gal4-KRAB mediated repression to 18-fold. In contrast, the sumoylation-deficient mutant of KAP1, K6R, which has each of the six SUMO acceptor lysines mutated to arginines, had no stimulatory effect on Gal4-KRAB repression activity (Figure 5D). Both wild type KAP1 and the mutant K6R protein were properly localized to pericentromeric heterochromatin (Figure S6C), indicating that the loss of repression was not due to defect in the K6R subnuclear targeting. The KAP1 PHD mutant C651A, which showed an intermediate level of sumoylation (Figure 1F,G) demonstrated poor ability to complement Gal4-KRAB-mediated repression. A fusion of the K6R protein to SUMO1 had a very modest ability to complement KRAB-mediated repression indicating that multiple sumoylation at naturally occurring sites in KAP1 is required for full activity.
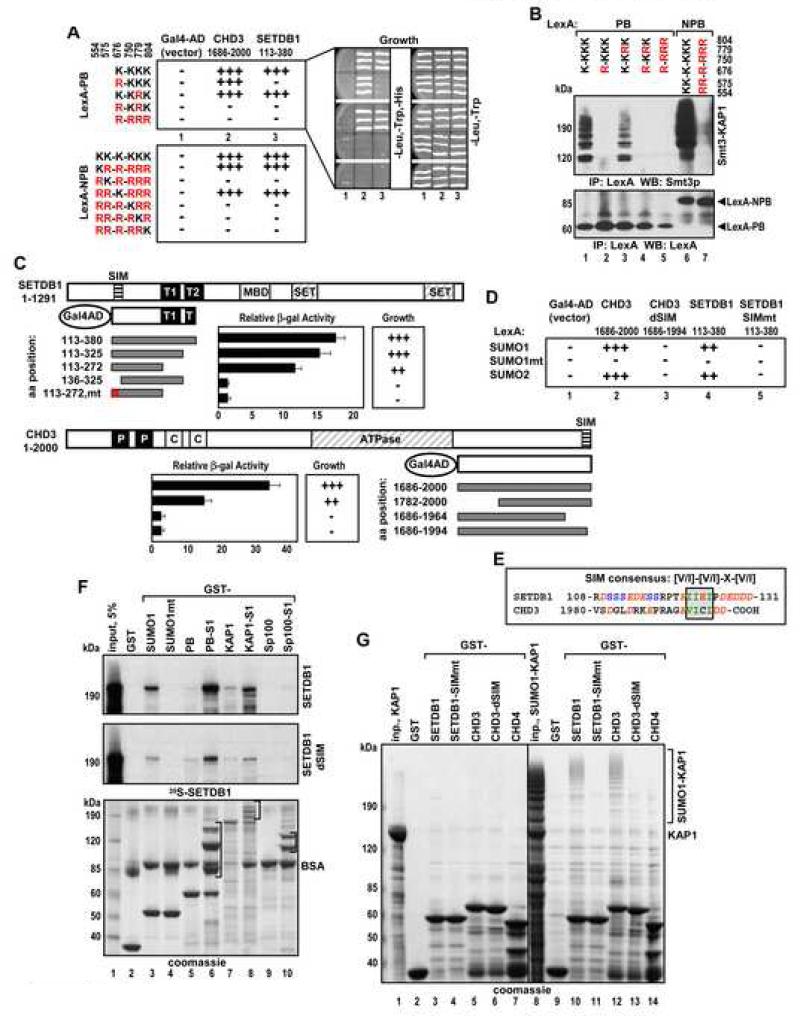
SETDB1 and CHD3 Interact Only with Sumoylated KAP1 in Yeast
Repression mediated by the KAP1 PHD-bromodomain (PB) module depends on its interaction with the H3-K9 histone methyltransferase SETDB1 and a component of histone deacetylase NuRD complex - chromatin remodeling factor CHD3 (Schultz et al., 2001; Sripathy et al., 2006). We asked whether these interactions are influenced by sumoylation of KAP1. Yeast two hybrid assay showed robust interactions of wild type LexA-KAP1 PB with Gal4 activation domain fusions to SETDB1 or CHD3 as indicated by yeast growth under the appropriate selective conditions (Figure 6A). However, LexA-PB with the K676R mutation interacted with CHD3 but not with SETDB1, and a double mutation of K676 and K779 abolished interaction with both. Immunoprecipitation and Western blot analysis of the KAP1 bait proteins showed that K676 and to a much lesser extent K779 were subject to sumoylation in yeast (Figure 6B, lanes 1-4). Unlike in mammalian cells, the yeast SUMO protein Smt3p formed poly-Smt3p chains on KAP1, consistent with published reports (Bylebyl et al., 2003). When the LexA-KAP1 NPB baits were used, only lysines K554 and K676 were required for association with SETDB1 and CHD3 (Figure 6A). Among the six identified sumoylation sites in KAP1, only these two sites conform to the canonical consensus (Figure 1A) suggesting that sumoylation in yeast preferentially occurs on canonical sites. These results emphasize a very different specificity in utilization of the KAP1 SUMO acceptor sites in yeast versus mammalian cells or in vitro, where K779 and K804 reverse consensus sites are the primary targets (Figures 2D,F,G).
Figure 6. SETDB1 and CHD3 Interact with KAP1 in Yeast and in Vitro in SIM Dependent Manner.
(A,C,D) Growth selection and quantitative β-galactosidase analyses of L40a yeast cells transformed with the indicated plasmids. The growth was scored on a three-point scale: (+++) - robust growth, (++) - slow growth and (−) – no growth on triple drop-out plates (right).
(B) L40a cells transformed with LexA-PB and -NPB plasmids as in (A) were grown in double drop-out media. LexA fusion proteins were immunoprecipitated with LexA antibody and analyzed by Western blot with Smt3p (top) and LexA antibodies (bottom).
(C) The domain structure of SETDB1 and CHD3: T1 and T2 – Tudor domains, P – PHD, C – chromodomain. SIM is indicated by a striped box. Data are the mean ± SD from at least three clones analyzed in duplicate.
(E) Sequence alignment of SUMO interacting motifs (SIM) from SETDB1 and CHD3. The SIM consensus is boxed and the flanking acidic residues are italicized.
(F) Indicated GST fusion proteins immobilized on glutathione beads were incubated with 35S-SETDB1 or SETDB1dSIM. Bound proteins were analyzed by Coomassie blue staining (bottom) and autoradiography (top). Brackets indicate positions of sumoylated isoforms.
(G) GST fusions of SETDB1 (aa 113-272) and CHD3 (aa 1782-2000) wild type and SIM mutants, and CHD4 (aa 1768-1912) immobilized on glutathione beads were incubated with recombinant unmodified (lanes 2-7) or sumoylated (lanes 9-14) full length KAP1. Bound proteins were analyzed by Coomassie blue staining. Inp. - input.
Identification of Functional SIM Motifs in SETDB1 and CHD3
We determined if binding of SETDB1 or CHD3 to SUMO is sufficient for the interaction with sumoylated KAP1 PB in yeast. SETDB1 and CHD3 interacted with SUMO1 and SUMO2, but not with SUMO1mt (Figure 6D). This SUMO mutant contains a well-characterized, two amino acid alteration (K37A,K39A) in the binding pocket of the molecule that abolishes its interaction with a SIM (SUMO Interacting Motif) found in proteins that bind SUMO. A core SIM with the consensus sequence V/I-X-V/I-V/I, (X - any amino acid) has been identified and structurally characterized (Chupreta et al., 2005; Hecker et al., 2006; Song et al., 2005). Using mutational analysis we identified functional SIM in both SETDB1 (aa 122-IIEI-125) and CHD3 (aa 1995-VICI-1998). Mutations of these sequences completely abrogated the interaction with KAP1 PB (Figure 6C-E).
We reconstituted KAP1–SIM interactions in vitro using _E.coli_-produced, sumoylated proteins (Figure 6F, brackets) (Uchimura et al., 2004). SETDB1 showed moderate binding to SUMO1 and this association was abolished by the K37A,K39A mutation in SUMO1 (SUMO1mt) (lane 4 and Figure S7A). SETDB1 bound poorly to unmodified PB but the interaction was dramatically stimulated when sumoylated PB (PB-S1) or sumoylated full length KAP1 (KAP1-S1) were used (lanes 6,8). SETDB1 which lacks the SIM (SETDB1dSIM) showed markedly reduced binding to all sumoylated proteins (middle). SETDB1 showed very little affinity for highly sumoylated Sp100A (Sp100-S1, lane 10) suggesting that the SUMO modification alone is not the sole determinant for interaction.
We observed similar results in reverse binding experiments (Figure 6G). Unmodified KAP1 did not bind to any of the GST-SETDB1 or GST-CHD3 resins (lanes 2-7). In contrast, sumoylated KAP1 was efficiently retained by wild type SETDB1 and CHD3, while mutation of their SIM sequences completely abolished binding (lanes 10-13). Together, these experiments identified bona fide SIM motifs in SETDB1 and CHD3 which have been highly conserved throughout evolution (Figure S7B).
Endogenous KAP1 binds to SETDB1 and CHD3 in a Sumoylation Dependent Manner
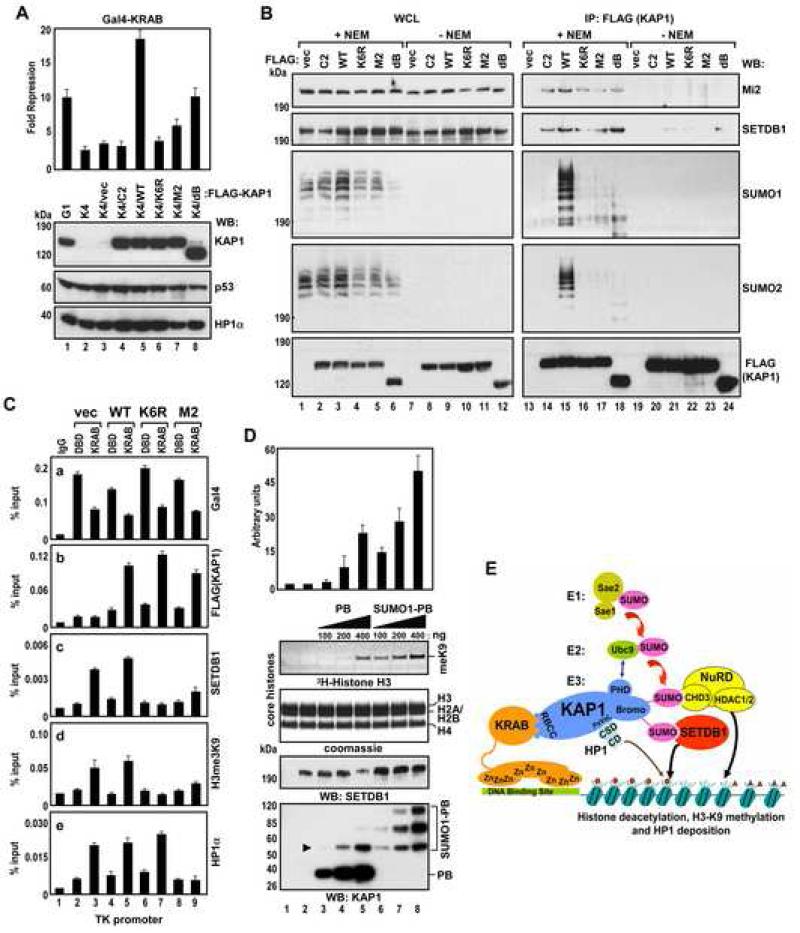
To assess the role of sumoylation for the KAP1-SETDB1/CHD3 interaction in vivo we reintroduced wild type or KAP1 mutants into U2OS-K4 KAP1 knockdown cells. We selected clonal cell lines expressing exogenous FLAG-KAP1 proteins at nearly physiological levels (Figure 7A, compare lanes 1 and 4-8). The reconstituted KAP1 mutants included C2 (abolishes trimerization/KRAB binding) (Peng et al., 2000), M2 (abolishes HP1 binding/subnuclear targeting) (Ryan et al., 1999), K6R (abolishes sumoylation) and dB (deletion of the bromodomain, decreases sumoylation). Cotransfection of a GAL4-TK-luciferase and Gal4-KRAB plasmids into K4/WT cells yielded about 17-fold repression, whereas this repression activity in control K4/vector and in K4/C2, K4/M2, K4/K6R and K4/dB cells was significantly attenuated (top). This is consistent with our previous data showing that KAP1 trimerization (abolished by the C2 mutation), HP1 binding/subnuclear targeting (abolished by the M2 mutation) and sumoylation (abolished by the K6R mutation, decreased by deletion of the bromodomain (dB)) (Figure 5D) are critical for KAP1 repression function. Surprisingly, in addition to the bona fide sumoylation-deficient mutant K6R, all the other KAP1 functional mutants showed dramatically reduced levels of SUMO modification in vivo by both SUMO1 and SUMO2 (Figure 7B, middle, lanes 14, 16-18) suggesting that impairment in sumoylation might be the major factor in the loss of their repression activity. We detected efficient coimmunoprecipitation of endogenous SETDB1 and Mi2/CHD3 with wild type KAP1 only when cells were lysed and processed in conditions preserving the SUMO modification (+NEM) (top, compare lanes 15 and 21). NEM is an inhibitor of desumoylating SENP enzymes, which deconjugate SUMO from substrate proteins upon cell lysis (lanes 7-12). Consistent with this, there was no or very little association between KAP1 and Mi2/CHD3 or SETDB1 in the absence of NEM (lanes 20-24). Moreover, markedly reduced sumoylation of the KAP1 mutants resulted in lower efficiency of SETDB1 and Mi2/CHD3 co-immunoprecipitation (lanes 14,16-18). Together these data suggest that KAP1 sumoylation plays a major role in its interaction with SETDB1 and CHD3 in vivo.
Figure 7. KAP1 Sumoylation Promotes Recruitment of SETDB1 to the Target Promoter and Stimulates Its Enzymatic Activity.
(A) U2OS-G1, -K4 and indicated reconstituted K4/FLAG-KAP1 cells were treated as in Fig.5C (top). The expression of KAP1, p53 and HP1α proteins was confirmed by Western blot (bottom). Data are the mean ± SD of at least two experiments performed in duplicate.
(B) Proteins from reconstituted K4/FLAG-KAP1 cells were immunoprecipitated with FLAG M2 antibodies and Western blotted with Mi2/CHD3, SETDB1, SUMO1, SUMO2 and FLAG antibodies.
(C) KAP1-mediated SETDB1 and HP1α recruitment to the TK promoter. U2OS-K4 cells were transfected with Gal4-DBD or Gal4-KRAB, vector, FLAG-KAP1 wild type or indicated mutants together with 5xGal4-TK-luc plasmid and subjected to ChIP analysis.
(D) Increasing amounts of unmodified or sumoylated KAP1 PB were mixed with 100 ng of SETDB1 along with 5 μg of core histones and subjected to an in vitro HMTase assay. The arrowhead indicates the position of mono-sumoylated PB. The autoradiograph shows the corresponding 3H-methyl-labeled histone H3. Data are the mean ± SD of three independent experiments (above).
(E) A model for sumoylation-dependent, KAP1–mediated gene silencing. The SUMO-conjugated bromodomain recruits the CHD3/NuRD complex and SETDB1 through SUMO-SIM interactions, which results in the deacetylation of histones and the methylation of histone H3-K9. KAP1-bound HP1 recognizes H3-K9 methylation via its chromodomain (CD). Small triangle – acetyl mark, small circle – H3-K9 trimethyl mark on histone tails.
KAP1 Sumoylation is Required for Recruitment of SETDB1 to the Target Promoter
To confirm this conclusion we tested the requirement of KAP1 sumoylation for the recruitment of SETDB1 to target promoter. We introduced GAL4-TK-luciferase, Gal4-DBD or Gal4-KRAB and various KAP1 variants into U2OS-K4 KAP1 knockdown cells and performed chromatin immunoprecipitation (ChIP) assay with primers specific to the TK promoter. As expected, all Gal4 fusion proteins bound to the promoter (Figure 7C, panel a). Consistently, the wild type (WT), K6R and M2 mutant KAP1 proteins were efficiently recruited to the promoter by Gal4-KRAB but not by Gal4-DBD (panel b, lanes 5,7,9). However, SETDB1 and the histone H3 K9-trimethyl mark were only enriched at the promoter in cells transfected with wild type KAP1 or control vector (panels c,d, lanes 3,5). Since HP1 binding to KAP1 does not directly depend on KAP1 sumoylation (Ryan et al., 1999), HP1α was recruited to the promoter by the K6R mutant as efficiently as by wild type KAP1, but it did not result in the appearance of the histone H3 K9-trimethyl mark and subsequent repression. The HP1 binding-deficient mutant, M2 did not recruit HP1 and neither SETDB1 nor the histone H3 K9-trimethyl mark (panels c-e, lane 9), since it is also defective in sumoylation (Figure 7B, lane 17). Together, these results suggest that KAP1 sumoylation is required for SETDB1 recruitment to the target promoter and the establishment of repressive chromatin marks.
Sumoylated KAP1 Stimulates SETDB1 HMTase Activity in Vitro
To investigate whether sumoylated KAP1 influences SETDB1 activity in vitro, we performed a histone methyltransferase (HMT) assay in the presence of unmodified or sumoylated KAP1 PB module (see Figure S8A). Low levels (100 ng) of baculovirus-expressed SETDB1 were incubated with core histones in a standard HMTase reaction containing increasing amounts of KAP1 PB. SETDB1-mediated methylation of histone H3 was undetectable in the absence of KAP1 (Figure 7D, lane 2). (A twenty fold higher amount of SETDB1 (2 μg) was required to produced robust methylation of histone H3 (data not shown)). Unmodified KAP1 PB showed a low level of H3-K9 methylation (lane 5). However, the addition of sumoylated KAP1 PB resulted in a robust, dose-dependent increase in histone H3 methylation (lanes 6-8). Titration reactions with the SUMO1 protein alone did not stimulate SETDB1 (Figure S8B). These data suggest that sumoylated KAP1 can increase the intrinsic SETDB1 HMTase activity toward histone H3.
DISCUSSION
KAP1 PHD is an Intramolecular SUMO E3 Ligase for the Adjacent Bromodomain
Tandem PHD and bromodomains are often found in chromatin associated proteins and have been shown to cooperate in gene regulation. Based upon recent structural analyses, it has been proposed that tandem PHD and bromodomains can independently recognize modifications in histones, and thus may act combinatorially to interpret the histone code (Li et al., 2006). In this article we provide an alternative basis for the cooperativity of these domains in corepressor protein KAP1 by demonstrating that the PHD can function as a SUMO E3 ligase in-cis for the bromodomain.
The PHD domain of KAP1 functions in a similar way to the RING finger of the PIAS SUMO E3 ligases: It directly binds to the SUMO E2 enzyme Ubc9 and is necessary for KAP1 (auto)sumoylation and KRAB-KAP1 mediated repression. The immediate target of the KAP1 PHD ligase activity is the adjacent bromodomain, where the major K779 and K804 sumoylation sites are located. In accordance with this, importance of PIASy autosumoylation for its function has also been reported (Ihara et al., 2005). KAP1 may also function as a SUMO E3 not only intramolecularly but also in-trans, by enhancing sumoylation of other KAP1 interacting proteins. We did not detect KAP1 dependent stimulation of SETDB1 and CHD3 protein sumoylation implying that they are effector molecules for KAP1 but are not direct targets for the E2-E3 (Ubc9-PHD) cascade. Like other E3 enzymes, the KAP1 PHD binds directly to its target. The solution structure of the KAP1 tandem PHD-bromodomain confirms that these two domains physically interact and form a functional module: mutations, which disrupt the interdomain interaction, also inhibit bromodomain sumoylation and KAP1-mediated repression (Zeng et al., see accompanying paper).
Sumoylation Dependent Interaction of KAP1 with Its Effector Proteins
It has been demonstrated that the controlled recruitment of a Gal4-fusion of KAP1 to a chromatinized GAL4-TK-luciferase transgene results in targeted localization of SETDB1 to the promoter region, a local increase in histone H3-K9 trimethylation and repression. Transient knockdown of SETDB1 in these conditions compromises repression and thus provides evidence for a functional link between KAP1 repression activity and SETDB1 recruitment (Sripathy et al., 2006). Here we provide direct evidence that the interaction between KAP1 and SETDB1 or CHD3 depends on KAP1 sumoylation and is a function of their specific SIM sequences. We show that SETDB1 and CHD3 are able to interact with SUMO1 and SUMO2. Consistent with this finding, SETDB1 was recently identified among other bound proteins in GST-SUMO2 affinity chromatography purifications (Rosendorff et al., 2006). It appears that specific recognition of SUMO-conjugated transcriptional regulators by individual components of repression machinery could be a general phenomenon. It has recently been shown that another partner protein of SETDB1, MCAF1, contains a SIM which facilitates its association with sumoylated MBD1 (Uchimura et al., 2006). Similarly, recruitment of the DAXX protein by SUMO-modified glucocorticoid receptor is dependent on its SIM (Lin et al., 2006).
A schematic model for KRAB-KAP1 mediated repression (Figure 7D) favors the hypothesis that sumoylation of KAP1 occurs at specific sites in chromatin likely due to KRAB-ZFP-KAP1 interaction at specific promoters (Mascle et al., 2007). KAP1 sumoylation clearly occurs on several sites simultaneously: the major ones being K779 and K804, located in the bromodomain. The PHD-Ubc9 interaction is required for this modification and presumably is regulated as well. Once modified by SUMO, the KAP1 bromodomain serves as a scaffold and recruits repression machinery through the recognition of the conjugated SUMO moieties by the SIM motifs of CHD3 and SETDB1 and their associated proteins. Intriguingly, sumoylated KAP1 stimulates the HMTase activity of SETDB1. The KAP1 scaffold is further exploited through recruitment of HP1 which recognizes the H3-K9 methyl mark and establishes a repressive chromatin state. Ultimately, the concerted action of these effector molecules leads to chromatin reorganization at the promoter region resulting in silent chromatin.
We provide conclusive evidence that the KAP1 PHD finger acts as an intramolecular SUMO E3 ligase for the adjacent bromodomain and that sumoylation of KAP1 is a major regulatory switch required for KRAB-KAP1 mediated repression.
EXPERIMENTAL PROCEDURES
Plasmids, Yeast Two-Hybrid Assay and NMR
For details of plasmids, yeast two-hybrid interaction assays and NMR, see Supplemental Data.
Cell Culture, Luciferase Assay and Infection
HEK293 and U2OS cells were cultured in DMEM supplemented with 10% FBS. Transfections were performed using CalPhos Kit (Takara) and FuGene6 (Roche) according to manufacturer's instructions. pCMV−β-gal or pTK−β-gal plasmids were included in all transfections. Fourty to 70 hrs after transfection, cells were lysed in Tween 20 buffer (Klenova et al., 2002) and Luciferase Assay System (Promega) and β-Galactosidase Assay Reagent (Pierce) were used to measure respective enzymatic activities and subsequent normalization. Lentiviral vector-based KAP1 gene knockdown was performed as described (Wang et al., 2005).
Recombinant Proteins, In Vitro Methylation and In Vitro Sumoylation
Baculovirus-expressed full-length SETDB1 was used in methylation reactions with purified HeLa core histones as described (Schultz et al., 2002). GST fusion proteins were expressed in E.coli BL21(DE3) and purified according to standard procedures using BLB150 buffer: 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 10% glycerol, 5 mM DTT, and protease inhibitors. 6His-KAP1 PHD wild type and mutants were purified as previously described (Peng et al., 2000). Sumoylated KAP1, KAP1 PB and Sp100A proteins were produced in E.coli BL21(DE3) as described (Uchimura et al., 2004). In vitro sumoylation reactions were carried out at 30°C for 3 hr. Briefly, 2 μg of target GST fusion protein were loaded on glutathione beads (GE Healthcare) and incubated in reaction mix containing 0.6 μM UbcH9 (BostonBiochem), 0.3 μM human E1 (BostonBiochem), 8 μM of 32P-labeled SUMO1 and sumoylation buffer with ATP (LAE Biotech Int) in a reaction volume of 20 μl. After incubation beads were washed five times with 1 ml of BLB500 buffer. Proteins were eluted in 2xLaemli buffer, resolved by SDS-PAGE and visualized by Coomassie blue staining and subsequent autoradiography. 32P-labeled SUMO1 was prepared from GST-PKA-SUMO1 as described (Yurchenko et al., 2006).
Immunoprecipitation, Western Blotting and Antibodies
For direct Western blotting, mammalian cells were lysed in 10% TCA, 2 mM DTT; yeast cells were lysed in 20% TCA. The pellets were solubilized in 1xLaemli buffer. For immunoprecipitation, the cells were extracted in modified IP buffer (mIP): 150 mM NaCl, 50 mM Tris-HCl (pH 8.0), 5 mM EDTA, 0.1% DOC Na, 0.5% NP-40, 0.5% Triton X-100, protease inhibitors and 10 mM N-ethylmaleimide (NEM). Lysates were cleared, diluted 4 times with FA buffer and incubated with appropriate antibody-agarose conjugates, for 4-12 hrs at 4°C. After washing 4 times with the same buffer, the proteins were analyzed by Western blotting. For details of the antibodies used, see Supplemental Data.
In vitro Translation and GST Pull-Down Assays
_In vitro_-translated proteins were 35S-methionine-labeled using TnT System (Promega). The GST fusion proteins (2 μg) were loaded on glutathione beads, pre-blocked with 2% BSA for 30 min and incubated with recombinant PHDs, SETDB1, unmodified or sumoylated KAP1 proteins in mST100 buffer: 50 mM HEPES (pH 7.5), 100 mM NaCl, 5 mM MgCl2, 0.1% Triton X-100, 1 mM DTT and protease inhibitors; or SETDB1 and SETDB1dSIM proteins in BLB150 buffer, for 2 hr at room temp and 2 hr at 4°C. After washing, the bound proteins were eluted in 2xLaemli buffer and analyzed by Coomassie blue staining and subsequent autoradiography.
ChIP
ChIPs were performed as described in Supplemental Data. Immunoprecipitated DNA was purified using Qiaquick PCR purification kit (Qiagen) and analyzed by qPCR on the ABI Prizm 7000 Sequence Detection Platform (Applied Biosystems) using SYBR Green technology.
Supplementary Material
01
ACKNOWLEDGEMENTS
We thank Dr. S. Chiocca for pSG9-Gam1 and Dr. H. Saitoh for pTE1E2-S1/2 plasmids; Dr. R. Marmorstein and M. Holbert for purified mononucleosomes and core histones; Dr. E. Johnson for Smt3p antibody; Lisa Gibson and Soumya Kandi for excellent technical assistance. F.J.R. is supported by NIH grants CA095561, CA092088, and DAMD17-02-1-0631 and the Samuel Waxman Cancer Research Foundation. M.-M.Z. is supported, in part, by NIH grant CA87658 and NSF grant #0517352. G.M. is supported by NIH grant AI41136 and GM57599. M.S. is supported by NIH grant AI41711, and V.Y. is supported by NIH grant CA09173. K.L.Y. is supported by a Terry Fox Foundation fellowship from the National Cancer Institute of Canada. D.E.W. is supported by NIH grant CA09171. We acknowledge the NCI Supported-Wistar Institute Cancer Center Shared Facilities: Genomics, Protein Expression, Proteomics and Hybridoma and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data
The Supplemental Data includes 8 figures, supplemental Experimental Procedures and References, and can be found with this article online at:
REFERENCES
- Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S. A mechanism for inhibiting the SUMO pathway. Mol Cell. 2004;16:549–561. doi: 10.1016/j.molcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Bylebyl GR, Belichenko I, Johnson ES. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem. 2003;278:44113–44120. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- Capili AD, Schultz DC, Rauscher IF, Borden KL. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. Embo J. 2001;20:165–177. doi: 10.1093/emboj/20.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupreta S, Holmstrom S, Subramanian L, Iniguez-Lluhi JA. A small conserved surface in SUMO is the critical structural determinant of its transcriptional inhibitory properties. Mol Cell Biol. 2005;25:4272–4282. doi: 10.1128/MCB.25.10.4272-4282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ., 3rd KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Girdwood D, Bumpass D, Vaughan OA, Thain A, Anderson LA, Snowden AW, Garcia-Wilson E, Perkins ND, Hay RT. P300 transcriptional repression is mediated by SUMO modification. Mol Cell. 2003;11:1043–1054. doi: 10.1016/s1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]
- Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1-and SUMO2-interacting motifs. J Biol Chem. 2006;281:16117–16127. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- Huntley S, Baggott DM, Hamilton AT, Tran-Gyamfi M, Yang S, Kim J, Gordon L, Branscomb E, Stubbs L. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16:669–677. doi: 10.1101/gr.4842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara M, Yamamoto H, Kikuchi A. SUMO-1 modification of PIASy, an E3 ligase, is necessary for PIASy-dependent activation of Tcf-4. Mol Cell Biol. 2005;25:3506–3518. doi: 10.1128/MCB.25.9.3506-3518.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Klenova E, Chernukhin I, Inoue T, Shamsuddin S, Norton J. Immunoprecipitation techniques for the analysis of transcription factor complexes. Methods. 2002;26:254–259. doi: 10.1016/S1046-2023(02)00029-4. [DOI] [PubMed] [Google Scholar]
- Kotaja N, Karvonen U, Janne OA, Palvimo JJ. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol Cell Biol. 2002;22:5222–5234. doi: 10.1128/MCB.22.14.5222-5234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT, Ho CC, Chen YC, Lin TP, Fang HI, et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell. 2006;24:341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Mascle XH, Germain-Desprez D, Huynh P, Estephan P, Aubry M. Sumoylation of the transcriptional intermediary factor 1beta (TIF1beta), the Co-repressor of the KRAB Multifinger proteins, is required for its transcriptional activity and is modulated by the KRAB domain. J Biol Chem. 2007;282:10190–10202. doi: 10.1074/jbc.M611429200. [DOI] [PubMed] [Google Scholar]
- Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Peng H, Begg GE, Schultz DC, Friedman JR, Jensen DE, Speicher DW, Rauscher FJ., 3rd Reconstitution of the KRAB-KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J Mol Biol. 2000;295:1139–1162. doi: 10.1006/jmbi.1999.3402. [DOI] [PubMed] [Google Scholar]
- Ragvin A, Valvatne H, Erdal S, Arskog V, Tufteland KR, Breen K, AM OY, Eberharter A, Gibson TJ, Becker PB, Aasland R. Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. J Mol Biol. 2004;337:773–788. doi: 10.1016/j.jmb.2004.01.051. [DOI] [PubMed] [Google Scholar]
- Rosendorff A, Sakakibara S, Lu S, Kieff E, Xuan Y, DiBacco A, Shi Y, Shi Y, Gill G. NXP-2 association with SUMO-2 depends on lysines required for transcriptional repression. Proc Natl Acad Sci U S A. 2006;103:5308–5313. doi: 10.1073/pnas.0601066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ., 3rd KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ., 3rd Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhang Z, Hu W, Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J Biol Chem. 2005;280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- Sripathy SP, Stevens J, Schultz DC. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol. 2006;26:8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura Y, Ichimura T, Uwada J, Tachibana T, Sugahara S, Nakao M, Saitoh H. Involvement of SUMO modification in MBD1-and MCAF1-mediated heterochromatin formation. J Biol Chem. 2006 doi: 10.1074/jbc.M602280200. [DOI] [PubMed] [Google Scholar]
- Uchimura Y, Nakamura M, Sugasawa K, Nakao M, Saitoh H. Overproduction of eukaryotic SUMO-1-and SUMO-2-conjugated proteins in Escherichia coli. Anal Biochem. 2004;331:204–206. doi: 10.1016/j.ab.2004.04.034. [DOI] [PubMed] [Google Scholar]
- Wang C, Ivanov A, Chen L, Fredericks WJ, Seto E, Rauscher FJ, 3rd, Chen J. MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. Embo J. 2005;24:3279–3290. doi: 10.1038/sj.emboj.7600791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD. SUMO promotes HDAC-mediated transcriptional repression. Mol Cell. 2004;13:611–617. doi: 10.1016/s1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- Yurchenko V, Xue Z, Sadofsky MJ. SUMO modification of human XRCC4 regulates its localization and function in DNA double-strand break repair. Mol Cell Biol. 2006;26:1786–1794. doi: 10.1128/MCB.26.5.1786-1794.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Grummt I. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr Biol. 2005;15:1434–1438. doi: 10.1016/j.cub.2005.06.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01