Proto-Oncogenic Role of Mutant IDH2 in Leukemia Initiation and Maintenance (original) (raw)
. Author manuscript; available in PMC: 2015 Mar 31.
Published in final edited form as: Cell Stem Cell. 2014 Jan 16;14(3):329–341. doi: 10.1016/j.stem.2013.12.016
SUMMARY
Mutations in the metabolic enzymes isocitrate dehydrogenase-1 (IDH1) and IDH2 that produce the oncometabolite D-2-hydroxyglutarate (2-HG) occur frequently in human acute myeloid leukemia (AML). 2-HG modulates numerous biological pathways implicated in malignant transformation, but the contribution of mutant IDH proteins to maintenance and progression of AML in vivo is currently unknown. To answer this crucial question we have generated transgenic mice that express IDH2R140Q in an on/ off- and tissue-specific manner using a tetracycline-inducible system. We found that IDH2R140Q can cooperate with overexpression of HoxA9 and Meis1a and with mutations in FMS-like tyrosine kinase 3 (FLT3) to drive acute leukemia in vivo. Critically, we show that genetic deinduction of mutant IDH2 in leukemic cells in vivo has profound effects on their growth and/or maintenance. Our data demonstrate the proto-oncogenic role of mutant IDH2 and support its relevance as a therapeutic target for the treatment of human AML.
INTRODUCTION
The isocitrate dehydrogenase (IDH) family of enzymes catalyzes the oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG) and carbon dioxide. Mutations in active site arginines of IDH1 and IDH2 have recently been identified in ~20% of acute myeloid leukemias (AMLs) (Cancer Genome Atlas Research Network, 2013; Mardis et al., 2009), as well as in a range of other malignancies including glioblastoma, chondrosarcoma, and prostate cancer (Amary et al., 2011; Kang et al., 2009; Parsons et al., 2008). Mutations confer on the enzymes a novel ability to produce D-2-hydroxyglutarate (2-HG), a molecule that is structurally similar to α-KG and can act as a competitive inhibitor of α-KG-dependent dioxygenases that in turn regulate a wide array of biological processes including DNA and histone demethylation, collagen maturation, and the hypoxic response (Dang et al., 2009; Ward et al., 2010; Xu et al., 2011). In AML, patient IDH mutations are associated with a normal karyotype and often co-occur with other genetic lesions including internal tandem duplication in FMS-like tyrosine kinase 3 (Flt3ITD) and mutations that promote the cytoplasmic localization of Nucleo-phosmin 1 (NPMc+) (Patel et al., 2012).
Mutant IDH proteins have been proposed as attractive drug targets and molecules that block the production of 2-HG have recently been reported (Popovici-Muller et al., 2012; Wang et al., 2013). Expression of mutant IDH1 or IDH2 proteins is sufficient to block differentiation of hematopoietic cells in vitro as well as in vivo and can be reversed by inhibitor treatment (Figueroa et al., 2010; Losman et al., 2013; Sasaki et al., 2012; Wang et al., 2013). Retroviral transduction of IDH mutations in combination with additional oncogenes into primary mouse bone marrow cells followed by transplantation have been shown to drive leukemia development (Chaturvedi et al., 2013; Chen et al., 2013); however, the central question of whether mutant IDH1 and IDH2 proteins are required for leukemia maintenance in vivo remains yet to be answered. Herein we address this question through the generation and characterization of transgenic murine models where expression of IDH2R140Q is conditional and regulatable.
RESULTS
Generation and Characterization of IDH2R140Q Transgenic Mice
To develop a mouse model of IDH2R140Q mutation that has the capacity to be both tissue-specific and on/off-inducible, we used the tetracycline response element (TRE)/tetracycline trans-activator (tTA) system. Briefly, the IDH2R140Q cDNA was cloned downstream of a TRE followed by a protamine-1 polyA cassette (Fisher et al., 2001) and targeted into the mouse Collagen A1 locus of C2-embryonic stem cells (ESCs) using Flp-recombinase-mediated genomic integration (Beard et al., 2006) (Figure 1A and S1A available online). Chimeric mice harboring the inducible IDH2R140Q allele were backcrossed into the C57BL/6 background and then crossed to mice that constitutively express the M2 reverse tTA from the ROSA26 locus (ROSA26-M2) (Hochedlinger et al., 2005). Compound IDH2R140Q+/−; ROSA26-M2+/− (hereafter referred to as IDH2R140Qt) were born at Mendelian ratios (data not shown) and appeared normal and were compared to single transgenic (i.e., IDH2 R140Q+/− or ROSA26-M2+/−) or wild-type littermates (hereafter referred to as controls).
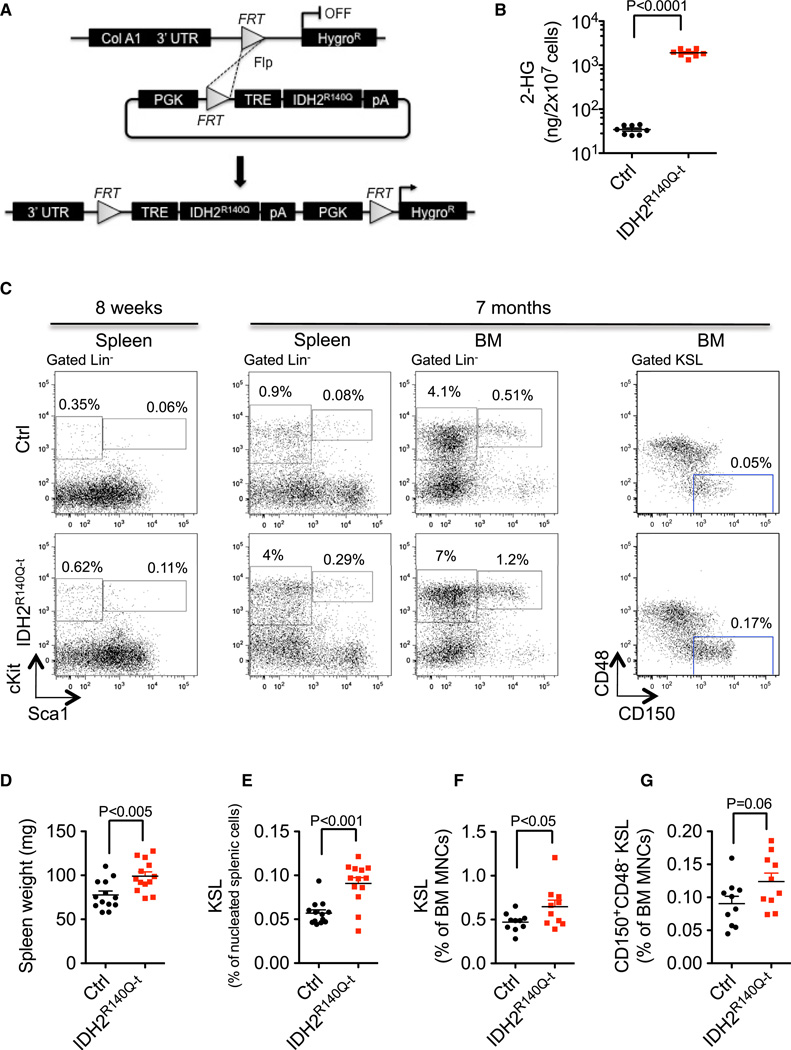
Figure 1. IDH2R140Q–t Mice Demonstrate Extramedullary Hematopoiesis Characterized by Spleen Enlargement and Expansion of Hematopoietic Stem/Progenitor Cells.
(A) Schematic depicting the strategy for generation of an inducible IDH2R140Q allele in the mouse. Following Flp-mediated recombination, the IDH2R140Q cDNA flanked by a tetracycline response element (TRE) and a protamine-1 poly-A cassette is integrated into the ColA1 locus. The PGK promoter drives hygromycin resistance, allowing selection of integrants.
(B) 2-HG in BM MNCs measured by LC-MS following 8 weeks of transgene induction.
(C) Representative flow cytometry analysis of bone marrow mononuclear cells and splenocytes from mice treated with doxycycline for 8 weeks or 7 months. Numbers indicate cells within the gate as a percentage of all living cells in the sample.
(D) Spleen weights of mice following 8 weeks of doxycycline treatment.
(E) Percentage of cKit+Sca1+Lin− (KSL) cells in the spleens of mice following 8 weeks of doxycycline treatment.
(F) Percentage of KSL cells in the bone marrow of mice following 7 months of doxycycline treatment.
(G) Percentage of CD48−CD150+KSL hematopoietic stem cells in the bone marrow of mice following 7 months of doxycycline treatment.
Error bars, mean ± SEM; p value calculated using a two-tailed unpaired Student’s t test. See also Figure S1.
Due to the documented frequency of IDH mutations in AML patients (Cancer Genome Atlas Research Network, 2013; Mardis et al., 2009; Patel et al., 2012), we chose to focus specifically on the hematopoietic system of our transgenic mice. IDH2R140Q expression was induced in animals at ~3 weeks of age by introducing doxycycline in the chow. Quantitative reverse-transcriptase PCR (qRT-PCR) using primers specific for the trans-gene confirmed IDH2R140Q mRNA expression in both mature and immature bone marrow cells from IDH2R140Qt animals (Figure S1C). Western blot analysis using an antibody that recognizes both the mutant IDH2R140Q and the endogenous wild-type IDH2 proteins revealed that the total levels of IDH2 in the bone marrow of IDH2R140Q-t animals were increased by approximately 2-fold in comparison with controls (Figure S1B). Because the level of endogenous Idh2 mRNA was unaffected by transgene expression (data not shown), these data suggest that the ratio of wild-type to mutant protein in our system is approximately 1:1.
The IDH2R140Q mutation, as well as other cancer-associated mutations in IDH1 and IDH2, has been shown to lead to the production of the oncometabolite 2-hydroxyglutarate (2-HG) (Dang et al., 2009; Ward et al., 2010). 2-HG accumulates in IDH mutant cancer cells at low mM concentrations and can also be detected in the serum of patients with mutant IDH AML (Gross et al., 2010). We used liquid chromatography-mass spectrometry (LC-MS) to quantify 2-HG in both serum (Figure S1E) and isolated bone marrow mononuclear cells (BM MNCs) (Figure 1B) from our mice. As expected, high levels of 2-HG were detected only in IDH2R140Q–t animals.
To confirm that transgene expression and 2-HG production could be reversed in vivo, mice that had been on a doxycycline diet for a period of 8 weeks were placed on a diet without doxycycline for 2 weeks. Following this period of deinduction, both transgene expression and serum 2-HG levels dropped significantly and returned almost to basal levels (Figures S1D and S1E). Together these data demonstrate that we have generated an inducible model of the cancer-associated IDH2R140Q mutation in which transgene expression and the consequent production of 2-HG can be regulated by administration of doxycycline.
Expression of IDH2R140Q in the Bone Marrow Results in Aberrant Hematopoiesis in Vivo
In order to establish the impact of IDH2R140Q expression on normal hematopoietic stem/progenitor cells (HSPCs) as well as lineage development in vivo, we undertook a detailed analysis of hematopoiesis in our transgenic animals. Eight weeks after transgene induction, IDH2R140Q–t and control peripheral blood cell counts showed no differences (Figures S1F–S1H). Likewise, there were no alterations in the numbers of hematopoietic stem cells, myeloid progenitors, or mature B, T, or myeloid cells in the bone marrow (Figures S1I–S1O). However, the spleens of IDH2R140Q–t mice were approximately 1.3-fold larger (99.05 ± 4.851 mg versus 77.69 ± 4.54 mg for the controls; p < 0.005, Student’s t test) (Figure 1D) and fluorescence-activated cell sorting (FACS) analysis revealed significant extramedullary hematopoiesis, as evidenced by increased numbers of cKit+Sca1+Lin− (KSL) HSPCs (Figures 1C and 1E).
To assess the longer-term effect of IDH2R140Q mutation, we analyzed mice following 7 months of continuous transgene induction. In addition to the changes observed at earlier time points, IDH2R140Q–t mice also had increased numbers of KSL cells in the bone marrow compared to controls (Figure 1C and 1F). Five out of ten mutants also displayed an expansion of the long-term hematopoietic stem cell (LT-HSC) compartment (CD48−CD150+ KSL), although overall this trend was not statistically significant (Figures 1C and 1G). To extend our findings we performed competitive bone marrow transplantation assays in which sorted KSL cells from IDH2R140Q–t and control mice (CD45.2+) were transplanted into sublethally irradiated recipients (CD45.1+). In this assay, IDH2R140Q–t HSPCs showed no difference in repopulation capacity compared to control HSPCs for at least 16 weeks after transplantation (Figures S1P–S1S).
IDH2R140Q Expression in HSPCs Drives a Block of Erythroid Differentiation in Vitro
Deregulated hematopoietic differentiation is frequently observed in AML and 2-HG has been shown to induce a partial block of differentiation in the erythroid leukemia cell line TF-1 (Losman et al., 2013; Wang et al., 2013). We therefore sought to investigate the effect of IDH2R140Q on differentiation of primary hematopoietic cells using a well-established methylcellulose differentiation assay. Following 8 weeks of transgene induction, we sorted KSL cells from IDH2R140Q–t and control mice and performed colony-forming unit (CFU) assays using semisolid medium with cytokines formulated to drive differentiation into the myeloid and erythroid lineages (MethoCult GF M3434). In the initial experiments, doxycycline was added to the medium to ensure continued transgene expression throughout the differentiation process. Strikingly, we observed a potent block of differentiation in IDH2R140Q–t HSPCs compared with controls (Figure 2A). Importantly, while the number of myeloid colonies (M, G/GM) was unaffected, the number of erythroid colonies was severely reduced (19.5 ± 1.3 in the controls versus 3.6 ± 0.9 in IDH2R140Q–t; p < 0.0001, Student’s t test). Notably, when cells from the first plating were collected and replated, IDH2R140Q–t cells formed more colonies in the second plating when compared with controls. IDH2R140Q–t cells could also be serially replated at least five times, whereas control cells no longer formed colonies after the third plating (Figure S2A).
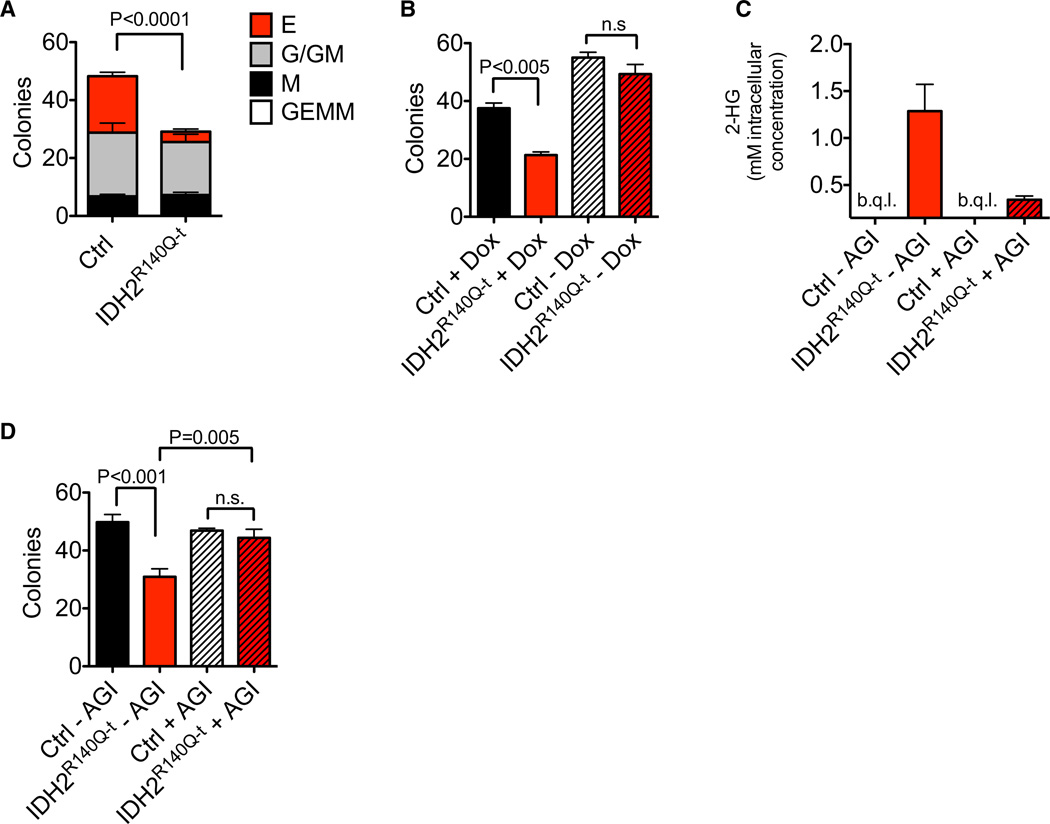
Figure 2. IDH2R140Q–t Induces a Block of Erythroid Differentiation in KSL Cells That Is Reversible by Genetic Deinduction of the Transgene and by a Specific Inhibitor of Mutant IDH2.
(A) Methylcellulose CFU-GEMM assay using KSL cells sorted from IDH2R140Q–t or control mice following 8 weeks of doxycycline treatment. Doxycycline was added to the media to maintain transgene expression throughout the differentiation process (n = 7 biological replicates/group).
(B) Methylcellulose CFU-GEMM assay done as in (A) but with or without adding doxycycline to the medium (n = 3 biological replicates/group).
(C and D) Methylcellulose CFU-GEMM assay done as in (A), with doxycycline in the medium and with or without the mutant IDH2 inhibitor AGI-6780. 2-HG was quantified by LC-MS in cells collected from the assay after 7 days of culture. b.q.l., below quantitative limit (C). Colonies were scored (n = 4 biological replicates/ group) in (D).
Error bars, mean ± SEM; p value calculated using a two-tailed unpaired Student’s t test. See also Figure S2.
To determine if the block of differentiation induced by IDH2 mutation in our model can be reversed, we performed a series of experiments using both genetic and pharmacological inactivation of IDH2R140Q. First, we compared differentiation of IDH2R140Q–t and control KSL cells in the presence or absence of doxycycline. We sorted cells from mice following 8 weeks of in vivo transgene induction and then cultured them in CFU assays either with or without doxycycline. We confirmed that within 72 hr of doxycycline-free culture, transgene expression was efficiently silenced (Figure S2B). Excitingly, upon silencing IDH2R140Q, differentiation of mutant cells was restored to wild-type levels, demonstrating that continued expression of the mutant protein is required for the block of differentiation (Figure 2B).
Next we used a specific inhibitor of IDH2R140Q (AGI-6780) that has recently been shown to block production of 2-HG by the mutant protein (Wang et al., 2013). We confirmed that in our system 5 µM of the compound was able to reduce 2-HG production in mutant cells by ~75% (1.29 ± 0.29 mM in untreated cells versus 0.34 ± 0.04 mM in AGI-6780-treated cells; p < 0.05, Student’s t test) (Figure 2C). Similar to genetic deinduction, AGI-6780 completely reversed the block of differentiation in IDH2R140Q–t KSL cells (Figure 2D). AGI-6780 also reversed aberrant colony formation in the second and third plating (Figure S2C). Taken together our data demonstrate that mutant IDH proteins are sufficient to induce defects in the differentiation of primary hematopoietic cells. Critically, we have shown that these effects are reversible by pharmacological inhibition of 2-HG production by the mutant protein.
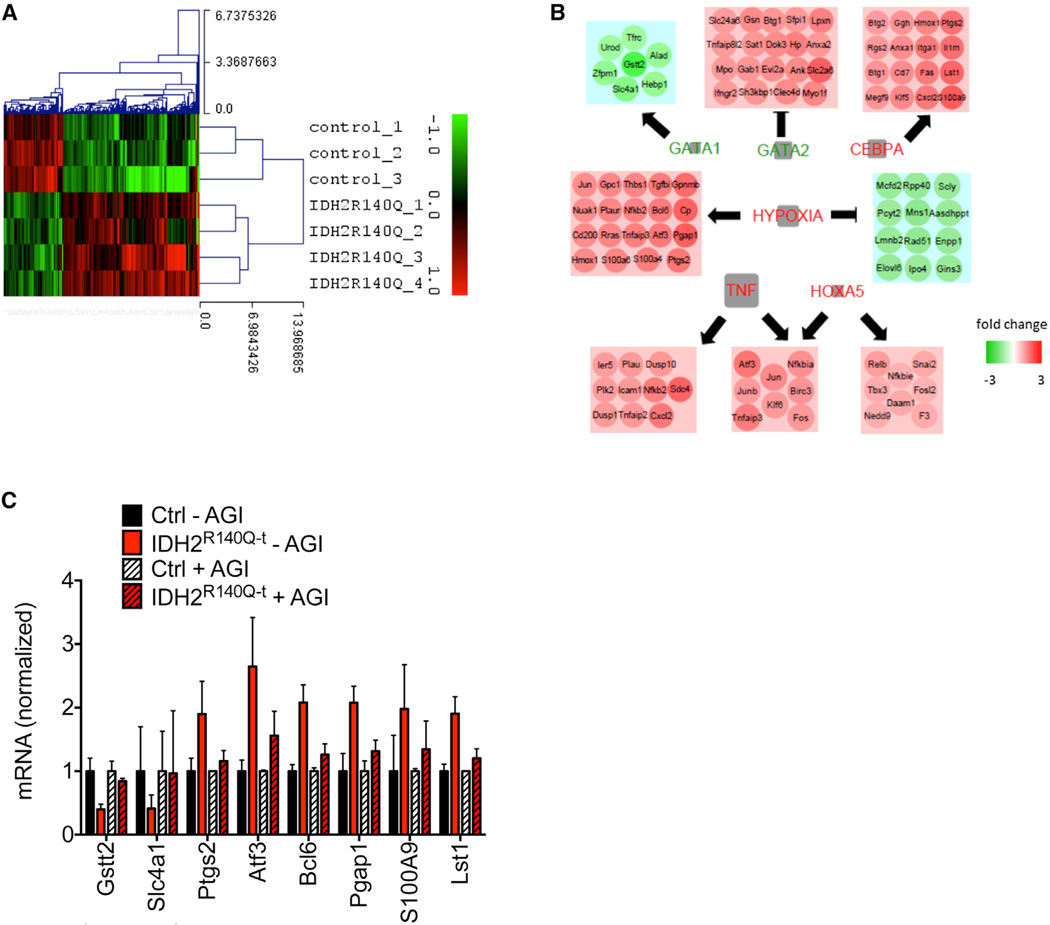
Mutant IDH proteins have been demonstrated to affect multiple biological pathways in a cell-context-dependent manner. We sought to explore the transcriptional signature that was associated with the block of erythroid differentiation and so we performed global gene expression microarray analysis. In order to identify genes that are responsible for the block of differentiation, rather than a consequence of it, we sorted KSL cells as previously, but cultured them in MethoCult GF M3434 for only 3 days prior to RNA extraction. At this stage, cells had undergone multiple rounds of division, but ~70% of cells retained an undifferentiated morphology (data not shown). Unsupervised hierarchical clustering revealed a clear separation of control and IDH2R140Q–t samples (Figures 3A and S3A) and gene set enrichment analysis identified multiple pathways that were deregulated in mutant cells (Figure 3B). In particular, IDH2R140Q–t cells displayed reduced levels of transcripts activated by the transcription factor GATA1, a master regulator of erythroid differentiation (Fujiwara et al., 1996). Mutant cells also displayed activation of the hypoxia and TNF-α pathways. To validate our microarray findings and to determine whether the transcriptional signature induced by mutant IDH2 is reversible, we performed qRT-PCR analysis on a subset of genes that were identified as being altered and included control and IDH2R140Q–t samples that were treated with AGI-6780 (Figure 3C). For each of the transcripts examined, AGI-6780 at least partially reversed the changes induced by IDH2R140Q. This data further confirms our earlier findings that 2-HG-dependent effects are rapidly reversible. We also sorted megakaryocyte-erythroid progenitors (MEPs) from control and IDH2R140Q–t animals and found that Bcl6, identified in our in vitro analysis, was also consistently upregulated in MEPs in vivo (Figure S3B).
Figure 3. Identification of a Transcriptional Signature Associated with the _IDH2R140Q_-Induced Block of Differentiation.
(A and B) Microarray analysis of cells cultured for 3 days in the methylcellulose CFU-GEMM assay (n ≥ 3 biological replicates/group). Hierarchical clustering of significantly altered genes (p < 0.05, fold change >1.5) is shown. Green, downregulated in IDH2R140Q–t cells; red, upregulated in IDH2R140Q–t cells (A). Pathway enrichment analysis shows genes that are downregulated in IDH2R140Q–t cells (green) or upregulated (red) in IDH2R140Q–t cells. The significance of the enrichment for each pathway (based on the false discovery rate q value) is proportional to the size of the gray box (i.e., the larger the gray box, the more significant the enrichment) (B).
(C) qRT-PCR analysis of cells cultured as in (A) and (B) but with or without AGI-6780. For each of the transcripts examined, pharmacological inhibition of 2-HG production at least partially reverses transcriptional changes in IDH2R140Q–t cells. Error bars, mean ± SEM.
See also Figure S3 and Tables S1, S2, and S3.
Continued Expression of IDH2R140Q Is Essential for Maintenance of _HoxA9/Meis1a_–Transformed Leukemic Cells
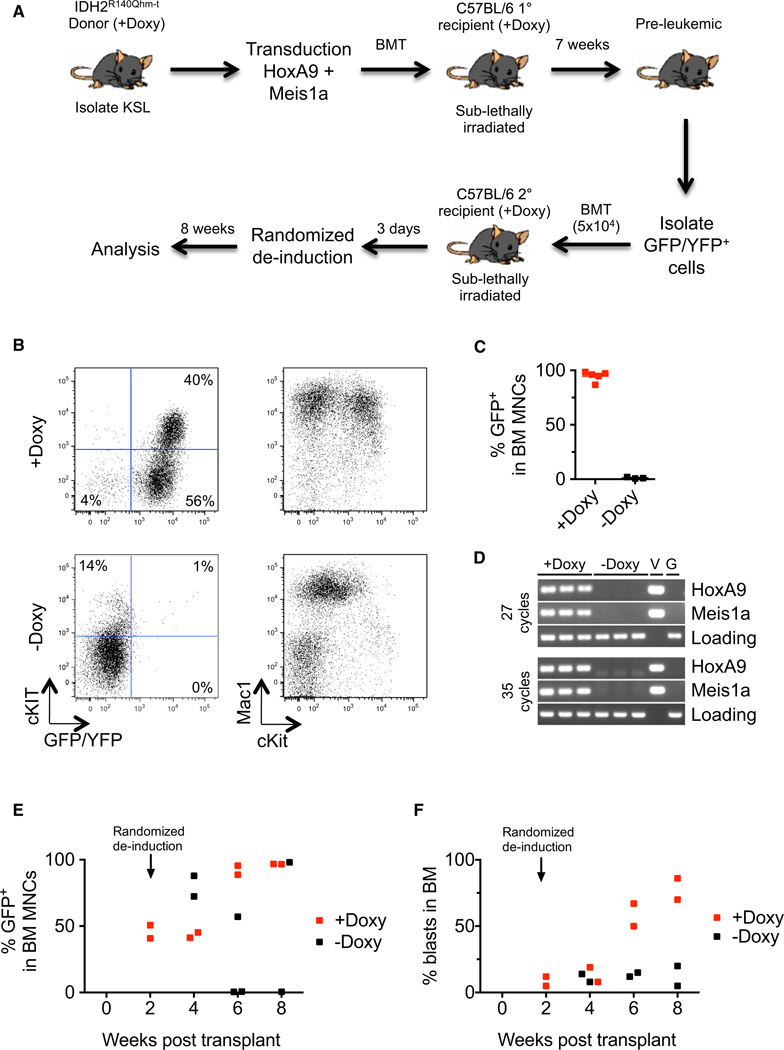
IDH mutations have been implicated in cancer but their ability to contribute to the maintenance of established disease in vivo has not been demonstrated. IDH2R140Q–t mice followed for 1 year of continuous doxycycline treatment did not develop leukemia, suggesting that additional genetic events are required for cancer development. This prompted us to investigate a well-established surrogate model of AML using retroviral overexpression of the homeobox proteins HoxA9 and Meis1a (Kroon et al., 1998). KSL cells isolated from IDH2R140Q+/+;ROSA26-M2+/− (hereafter referred to as IDH2R140Qhm-t) that had been treated with doxycycline were transduced with retroviruses encoding HoxA9 (MSCV-HoxA9-GFP) and Meis1a (MSCV-Meis1a–YFP) and transplanted into syngeneic sublethally irradiated recipients (Figure 4A). Following 7 weeks of administering continued doxycycline treatment, we observed an expansion of GFP/ YFP+ cells in the peripheral blood of primary recipients (not shown), indicating the establishment of an early leukemic or preleukemic state. These animals were then euthanized, GFP/ YFP+ bone marrow cells were sorted (Figure S4A), and 105 cells were retransplanted into secondary syngeneic recipients. After an initial 3 day period of administering doxycycline treatment to allow engraftment, we performed randomized deinduction with some of the recipients being taken off doxycycline food and thereby experiencing genetic inactivation of IDH2R140Q (Figure 4A).
Figure 4. Continued Expression of IDH2R140Q Is Required for Proliferation and/or Maintenance of _HoxA9/Meis1a_–Transformed Leukemic Cells.
(A) Schematic outlining the strategy for randomized deinduction experiments.
(B) Representative FACS plots of bone marrow from secondary recipients untreated or treated with doxycycline. Continued expression of IDH2R140Q is essential for proliferation and/or maintenance of GFP/YFP+ leukemic cells and the onset of leukemia.
(C) Percentage of GFP/YFP+ cells in the bone marrow of secondary recipients untreated or treated with doxycycline (n = 5 recipients on doxycycline, n = 3 recipients off doxycycline).
(D) PCR on genomic DNA isolated from BM MNCs of doxycycline-treated and -untreated recipients using primers specific for MSCV-HoxA9-GFP and MSCV-Meis1a–YFP. Vector (i.e., MSCV-HoxA9-GFP or MSCV-Meis1a–YFP, respectively) DNA (V) and wild-type C57BL/6 genomic DNA (G) were used as controls.
(E) Percentage of GFP/YFP+ cells in the bone marrow of secondary recipients untreated or treated with doxycycline at various time points following transplantation. Randomized deinduction was performed 2 weeks after transplantation (n = 2–3 recipients/time point/treatment).
(F) Percentage of blasts in the bone marrow of secondary recipients untreated or treated with doxycycline at various time-points following transplantation. Randomized deinduction was performed 2 weeks after transplantation (n = 2–3 recipients/time point/treatment). Blasts were scored on May-Grunwald-Giemsa-stained bone marrow cytospins.
See also Figure S4.
After 8 weeks, all (5/5) doxycycline-treated secondary recipients developed AML, with their bone marrow being composed almost exclusively of GFP/YFP+Mac1+ and GFP/YFP+Mac1 + cKit+ cells (Figure 4B, 4C, and S4B). All five recipients showed virtually identical disease development (Figure S4B), strongly suggesting that additional stochastic events that may influence the proliferative or immuno-phenotypic properties of the cells had not occurred after transplantation. We confirmed overexpression of HoxA9 and Meis1a in doxycycline-treated recipients using qRT-PCR (Figure S4D). Strikingly, all (3/3) secondary recipients that had been withdrawn from doxycycline had no clearly discernible GFP/YFP+ population and normal ratios of myeloid and lymphoid cells, demonstrating that the vast majority of leukemic blasts had been eliminated (Figure 4B, 4C, and S4B).
To determine if a small number of leukemia-initiating cells that was not detectable by FACS remained and could act as a source of disease relapse, we used a PCR assay to detect MSCV-HoxA9-GFP and MSCV-Meis1a–YFP with primers designed to span introns so as not to amplify endogenous HoxA9 and Meis1a from genomic DNA. Using this assay and following an extended cycling protocol, we could not detect a reservoir of transduced cells in the bone marrow of recipients that had been withdrawn from doxycycline (Figure 4D). These data strongly suggest that all, or at least the vast majority, of leukemic cells, including the leukemia-initiating cell population, were eliminated upon genetic deactivation of IDH2R140Q.
Next, we aimed to characterize the kinetics of disease regression upon IDH2R140Q deinduction. As before, sublethally irradiated C57BL/6 recipients were transplanted with 105 GFP/YFP+ cells sorted from a preleukemic IDH2R140Qhm-t/HoxA9/Meis1a primary donor. This time, however, all secondary recipients were treated with doxycycline for 14 days, at which point two recipients were sacrificed and FACS analysis revealed that their bone marrow comprised ~50% GFP/YFP+ cells (Figure 4E). We then performed randomized deinduction and sacrificed recipients for analysis every 2 weeks. Consistent with what has recently been reported in vitro upon pharmacological inhibition of IDH2R140Q in human and mouse AML cells (Chen et al., 2013; Wang et al., 2013), animals withdrawn from doxycycline treatment initially showed a hyperproliferation of GFP/YFP+ leukemic cells (Figure 4E; 4 week time point, 2 weeks off doxycycline food), followed by a reduction and elimination. In total, six of eight animals across the two independent experiments that had been withdrawn from doxycycline treatment for at least 4 weeks showed elimination of leukemic cells from the bone marrow and restoration of normal hematopoiesis (Figure S4C).
FACS, morphological, and transcriptional analyses of bone marrow at the 4 week time point suggested that upon IDH2R140Q deinduction, leukemic cells underwent differentiation. Despite having almost twice as many GFP/YFP+ cells in the bone marrow at this time point, recipients that had been withdrawn from doxycycline treatment had similar numbers of blasts to doxycycline-treated recipients (Figure 4F), implying a greatly increased ratio of morphologically differentiated to undifferentiated cells within the GFP/YFP+ fraction. Similarly, analysis of the stem/progenitor marker cKit showed a reduced ratio of cKit+ to cKit− cells upon deinduction (Figure S4E). Finally, we explored a panel of transcripts identified in our microarray analysis, and found that a subset of genes (Atf3, Ptgs2, and S100A9) that were upregulated in IDH2R140Q–t KSL cells undergoing differentiation in vitro were downregulated upon transgene deinduction in vivo in leukemic cells (Figure S4F). Taken together, our data demonstrate that HSPCs, which initially acquire an IDH2 mutation and subsequently become transformed by additional genetic events, at least in some contexts, are addicted to the continued function of the mutant IDH2 protein for maintenance.
Genetic Deinduction of IDH2R140Q Has Profound Effects on Leukemic Blasts in a Compound IDH2R140Q;Flt3ITD AML Model
We next sought to explore the function of mutant IDH2 in a system that more closely recapitulates the genetics of human AML. Internal tandem duplication in the receptor tyrosine kinase Flt3 is a frequent event in AML patients, is associated with poor prognosis, and co-occurs with mutations in IDH1 or IDH2 (Patel et al., 2012). Knockin Flt3ITD transgenic mice develop lethal meyloproliferative neoplasms (MPNs), but not overt leukemia (Chu et al., 2012; Li et al., 2008; Rau et al., 2013). Notably, combining the Flt3ITD allele with other mutations observed in AML patients can transform this MPN into full-blown acute leukemia (Rau et al., 2013).
To test whether the IDH2R140Q mutation can cooperate with Flt3ITD to drive disease progression, we generated compound IDH2R140Qt;Flt3ITD animals and treated them with doxycycline. Excitingly, and in contrast to Flt3ITD single transgenic littermate controls, compound animals developed acute leukemias (Figure 5A and 5B). In our cohort, IDH2R140Q–t;Flt3ITD animals displayed a significantly reduced overall survival compared with Flt3ITD controls (median survival of 229 days versus 352 days, respectively; p = 0.0299, Mantel-Cox test) (Figure S5A). Consistent with previous published observations (Chu et al., 2012; Li et al., 2008; Rau et al., 2013), the MPD in Flt3ITD animals was characterized by a hypercellular bone marrow with myeloid predominance and maturation (Figure 5A). The leukemias that developed in compound transgenic mice, on the other hand, were characterized by infiltration of large monomorphic blasts into hematopoietic (bone marrow, spleen, thymus) and non-hematopoietic (liver, lung, kidney) organs. We analyzed bone marrow cells, splenocytes, and thymocytes by FACS and found that the immuno-phenotype of cells from different animals, and occasionally from different organs within the same animal, varied (Figure 5B). We identified leukemias that displayed both myeloid (CD3ε−B220−Mac1lowcKit+) and lymphoid (Mac1− B220−CD3ε+CD8+CD4− and Mac1−B220−CD3ε−CD8+CD4+) markers (Figure 5B).
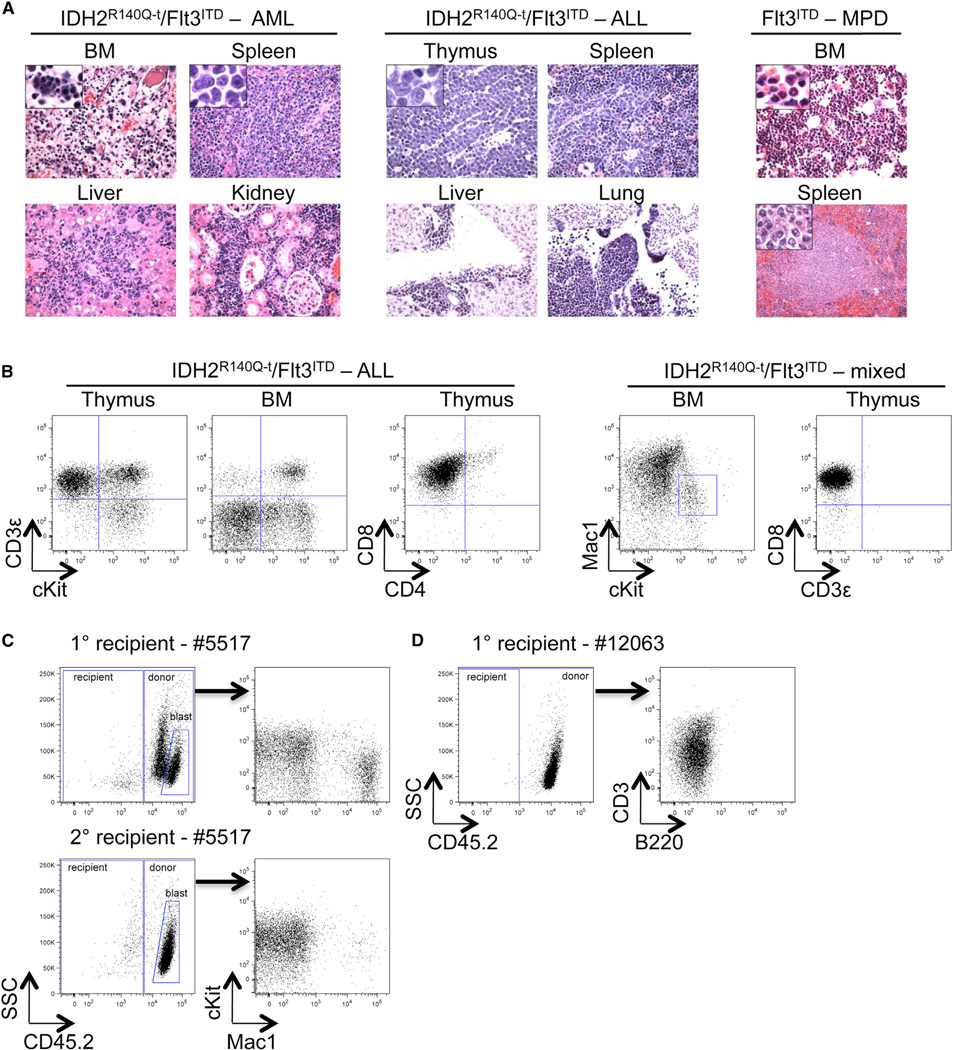
Figure 5. IDH2R140Q and Flt3ITD Cooperate in Leukemia Initiation.
(A) Representative H&E staining of tissue sections from IDH2 R140Q–t;Flt3ITD compound transgenic animals and an Flt3ITD control animal. Compound transgenic animals develop acute myeloid (AML-like) and lymphoid (ALL-like) malignancies whereas_Flt3ITD_ animals develop myeloproliferative disease (MPD). Left panels: a compound transgenic animal diagnosed with AML-like disease. Apoptotic debris and numerous tingible body macrophages (magnified in inset, upper left panel) characteristic of highly proliferative disease are evident throughout. Infiltration of intermediate-sized blast-like immature mononuclear cells (magnified in inset, upper right panel) is evident in the red pulp of the spleen, the liver, and kidney. Middle panels: a compound transgenic animal diagnosed with ALL-like disease. T lymphoblasts (confirmed by immunohistochemical staining using anti-CD3ε antibody, not shown) with high-grade morphology are present in the thymus, spleen, liver, and lung. Numerous mitoses are present, characteristic of a rapidly proliferative disease. Right panels: MPD in an Flt3ITD animal with a hypercellular bone marrow with myeloid maturation and increased myeloid cells in the spleen. Original magnification, 600×.
(B) Examples of FACS plots demonstrating the types of acute leukemia diagnosed in IDH2R140Q–t;Flt3ITD compound transgenic animals. Left panels: presence of CD3ε+cKit+ lymphoblasts in the thymus and bone marrow; cells in the thymus are exclusively CD8+CD4−. Right panels: cells in the bone marrow are almost exclusively Mac1+/low and a population of Mac1lowcKit+ immature cells is evident; in the thymus of the same animal, CD3ε−CD8+ lymphoblasts are present.
(C and D) Primary AML (BM MNCs from donor #5517) and ALL (thymocytes from donor #12063) cells recapitulate a lethal leukemia in transplanted recipients treated with doxycycline. FACS plots demonstrate the overwhelming majority of donor-derived (CD45.2+) cells in the bone marrow of moribund recipients and the loss of lineage markers (Mac1 and CD3ε, respectively) upon serial transplantation.
See also Figure S5.
Our data strongly suggest that IDH2R140Q is a bona fide cooperating oncogene in vivo. However, in order to further confirm the malignant nature of the disease and the diagnosis of acute leukemia that developed in IDH2R140Q–t;Flt3ITD animals, we performed transplantation experiments (Kogan et al., 2002). To this end, we transplanted leukemic cells into sublethally irradiated CD45.1+ syngeneic recipients and treated them with doxycycline. In this system, leukemic cells compete with normal recipient bone marrow cells in the niche and we were able to directly track their fate as they express the alternate surface antigen CD45.2. In two independent experiments we used BM MNCs or thymocytes derived from two different compound transgenic animals (#5517 and #12063, respectively). In both instances, the leukemic cells were able to recapitulate a lethal leukemia in transplanted recipients, and notably upon transplantation they mostly lost the expression of mature lineage markers (Mac1 for #5517 and CD3ε for #12063) (Figure 5C, 5D, and S5B–S5D). The disease derived from donor #5517 was used in serial transplantation experiments and became more aggressive and homogenous, and in secondary recipients the bone marrow was composed almost exclusively of CD45.2+ CD3ε−B220−Mac1− immature blasts.
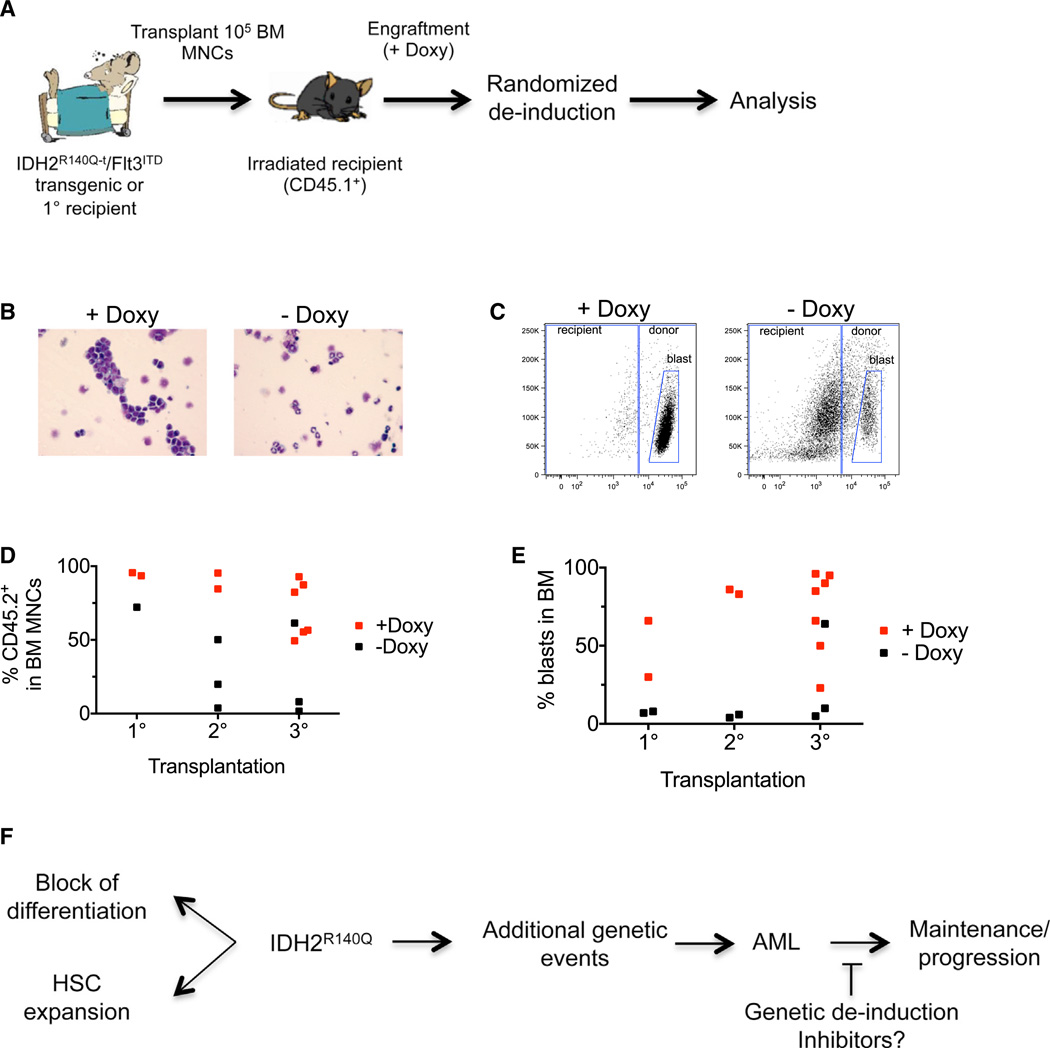
Having established a model where mutant IDH2 contributes to leukemia initiation, our next step was to determine whether genetic inactivation of IDH2R140Q in this context had an effect on leukemia maintenance and/or progression. Due to the variable onset of disease and the time required to generate large cohorts of compound transgenic animals, we focused our efforts on a transplantation-based “chimeric” model. We used primary leukemia cells from the bone marrow of donor #5517 to perform randomized deinduction experiments. As previously, leukemic cells were allowed to engraft for 3 days in the presence of doxycycline, and then half of the recipients were taken off doxycycline food (Figure 6A). We followed animals until they became moribund and had to be euthanized. We observed no clear differences in survival of doxycycline-treated and -untreated animals across primary, secondary, or tertiary transplantation experiments. Analysis of serum 2-HG levels confirmed that the metabolite returned to baseline levels following transgene deinduction (Figure S5E). All animals that were caught prior to succumbing to disease were subjected to a detailed histopathological analysis. Infiltration of leukemic cells into non-hematopoietic organs was similar between the two treatment cohorts (Figure S5F). In contrast, there was a clear difference with respect to bone marrow involvement. The bone marrow of animals treated with doxycycline was composed predominantly of blasts whereas animals in the untreated group displayed fewer blasts and marked differentiation (Figures 6B–6E). The morphological analysis was in agreement with the percentage chimerism as assessed by CD45.1/CD45.2 staining. Thus, inhibition of IDH2R140Q in acute leukemia in the context of a cooperating Flt3ITD mutation, at least in some cases, results in reduced proliferation and/or differentiation of leukemic blasts.
Figure 6. Mutant IDH2 Contributes to Leukemia Initiation and Maintenance in the Context of a Cooperating Flt3ITD Lesion.
(A) Schematic outlining the strategy for randomized deinduction experiments using AML cells from recipient #5517.
(B) May-Grunwald-Giemsa-stained bone marrow cytospins showing presence of leukemic blasts and differentiated myeloid cells in doxycycline-treated and -untreated recipients, respectively.
(C) Examples of FACS plots showing reduced donor chimerism in untreated versus doxycycline-treated recipients based on differential staining for the cell surface markers CD45.1/CD45.2.
(D and E) Genetic deinduction of IDH2R140Q expression by withdrawal of doxycycline has a profound effect on the proliferation and/or differentiation of leukemic blasts in vivo. Percentage of donor-derived (CD45.2+) cells (D) and blasts (E) in the bone marrow of primary, secondary, or tertiary recipients untreated or treated with doxycycline is shown. Blasts were scored on May-Grunwald-Giemsa-stained bone marrow cytospins. Randomized deinduction was performed 3 days after transplantation.
(F) Model for the role of _IDH2R140_Q in primary HSPCs and in the maintenance of leukemia.
See also Figure S5.
DISCUSSION
IDH mutations have been proposed as attractive targets for the development of novel anticancer therapeutics because specific small molecule inhibitors can directly block the neomorphic activity of the enzymes, namely the production of 2-HG (Popovici-Muller et al., 2012; Wang et al., 2013). However, the contribution of mutant IDH proteins to transformation in vivo remains poorly understood, and importantly, whether or not cancer cells are addicted to 2-HG in their native niche for growth and/or maintenance is yet to be determined.
We have generated a mouse model of IDH2R140Q, the most common IDH2 mutation observed in human AML. To our knowledge this is the first transgenic allele of mutant IDH2 reported to date. In our model, expression of the transgene and the consequent production of 2-HG is on/off inducible and comparable to that observed in AML patients, providing a valuable system for evaluating the biological effects of deinduction as well as the pharmacological efficacy of potential 2-HG inhibitors. Consistent with what was previously reported for a knockin model of IDH1R132H (Sasaki et al., 2012), we found that expression of mutant IDH2 resulted in alterations within the hematopoietic compartment characterized by an expansion of HSPCs within the spleen and bone marrow, but on its own it was not sufficient for leukemogenesis, at least within the time frame of our experiment.
In a well-established multilineage differentiation assay, IDH2R140Q–t KSL cells formed a greatly reduced number of erythroid colonies, but an equivalent number of myeloid colonies, in comparison with controls. We established a transcriptional signature associated with this block of erythroid differentiation and identified downregulation of GATA1 activity and upregulation of the hypoxia and TNF-α pathways as being potentially important in this process. Inhibition of GATA1 function was previously reported in mutant IDH AML patients (Figueroa et al., 2010). The effects of mutant IDH proteins on the hypoxia pathway have been disputed with some studies reporting hyper-activation (Xu et al., 2011) and others reporting inactivation (Koivunen et al., 2012; Losman et al., 2013), albeit in different systems. In our model, the observed transcriptional changes strongly argue in favor of activation of hypoxic signaling. Importantly, both the block of differentiation and the altered transcriptional profile in IDH2R140Q–t cells is reversible by the mutant IDH2 inhibitor AGI-6780. Our results confirm and expand on previous studies showing that mutant IDH proteins can induce a reversible block of differentiation in the erythroid leukemia line TF-1 (Losman et al., 2013; Wang et al., 2013). We conclude that at least under some conditions, production of 2-HG by mutant IDH proteins may not only block differentiation of hematopoietic cells, it may in fact bias it toward the myeloid lineage.
Though studies on the effects of IDH mutations on primary and leukemic cells in vitro or on HSPC function and lineage development in vivo provide important mechanistic insights, they fail to answer the most crucial question of whether mutant IDH2 is required for leukemia initiation and maintenance in vivo in the presence of a functional niche. To address this, we used two compound transgenic models in which IDH mutation was combined with activation of additional oncogenes. HoxA9 and Meis1a are downstream targets of numerous pathways deregulated in AML and forced coexpression of these two proteins is sufficient to transform murine HSPCs (Kroon et al., 1998; Wang et al., 2010). Notably, when this powerful combination was introduced into IDH2 mutant KSL cells, the proliferation and/or maintenance of the resultant leukemic cells were dependent on continued expression of IDH2R140Q. Within 2 weeks of genetic deinduction of mutant IDH, we observed evidence of differentiation, and 2 weeks later, 6/8 animals showed complete remission with elimination of any detectable leukemic cells. Interestingly, we found that Atf3, which has been implicated as a positive regulator of TGF-β signaling and was shown to enhance cancer-initiating cell activity in breast cancer cells, was downregulated upon IDH2R140Q inactivation (Yin et al., 2010). Our results are both surprising and encouraging in that we have modeled a situation where IDH mutation occurs as an early event and leukemic transformation occurs as a result of subsequent genetic “hits.” Indeed, IDH mutation is considered an early event in the evolution of AML as well as other cancer types including glioma (Kandoth et al., 2013; Watanabe et al., 2009). Thus our results strongly suggest that IDH mutation may establish a state of oncogene addiction even in a genetic setting where mutant IDH is not required for cancer initiation.
By generating compound IDH2R140Q;Flt3ITD transgenic mutants, we demonstrated that IDH2R140Q cooperates with Flt3ITD in leukemia initiation in vivo, strongly suggesting that IDH mutations are cooperative tumorigenic drivers in AML. Our results are coherent with two recent studies demonstrating that retroviral transduction of IDH mutations into mouse bone marrow cells can cooperate with other oncogenic stimuli in leukemogenesis (Chaturvedi et al., 2013; Chen et al., 2013). Retroviral models, though highly useful, are limited by numerous factors including artificial levels of transgene expression and random integration of the provirus into the genome. Our transgenic approach demonstrates the additional requirements and consequences of mutant IDH expression at physiologically relevant levels, as well as those of mutant IDH deinduction in vivo. Indeed the latency of disease development in IDH2R140Q;Flt3ITD mice suggests that additional mutations were required for full leukemic transformation. Recent sequencing efforts by the Cancer Genome Atlas have revealed that the median number of recurrent mutations in adult normal karyotype AML is approximately five (Cancer Genome Atlas Research Network, 2013). The spectrum of disease that developed in different transgenic animals in our model may be explained by the nature of additional mutations or by the type of cell in which they occurred. Indeed, numerous transgenic models develop different types of hematopoietic malignancies among animals of the same genotype (Rau et al., 2013; Yilmaz et al., 2006).
As with our HoxA9/Meis1a transduction model, we observed that genetic inactivation of mutant IDH2 in the context of a cooperating Flt3ITD lesion had an effect on disease progression, and though we were unable to show increased survival upon mutant IDH2 deinduction, we did demonstrate effects on the proliferation and/or differentiation of leukemic blasts. Analysis of leukemic infiltrates suggested that in this highly aggressive setting, mice that display a response in the bone marrow may nonetheless succumb to the disease due to damage of nonhematopoietic organs. Additionally, because a reduction in 2-HG levels may take some time (based on our measurements in nonleukemic mice 2 weeks after deinduction), leukemic cells may cause irreparable damage to the bone marrow niche and/or to other organs before the cells are eliminated. If that is the case, inhibition of mutant IDH proteins may synergize with chemotherapy or other drugs (in this case Flt3 inhibitors) that produce more immediate effects.
In summary, our findings, and those of other groups (Losman et al., 2013; Rohle et al., 2013; Wang et al., 2013), prove that at least some of the effects of IDH mutation are dependent on continued production of 2-HG. Critically, we extend previous work by demonstrating that those effects are reversible in vivo and addicted to the proto-oncogenic activity of the IDH mutant enzyme even in a genetic milieu where leukemia initiation is IDH mutant independent. We therefore validate mutant IDH proteins as very strong candidates for continued development of targeted anticancer therapeutics. Our model can serve as an invaluable tool for evaluating the biological effects of genetic deinduction of IDH mutations in various genetic settings as well as the pharmacological efficacy of potential mutant IDH2 inhibitors either alone or in combination with other compounds (Figure 6F).
EXPERIMENTAL PROCEDURES
Transgenic Animals
The tetracycline-inducible IDH2R140Q allele was created following the scheme shown in Figure 1A and backcrossed into the C57BL/6 background (see Supplemental Experimental Procedures for more information). ROSA26-M2 (Hochedlinger et al., 2005), C57BL/6, and CD45.1+ (B6.SJL-Ptprca Pepcb/ BoyJ) animals were purchased from Jackson Laboratories. Flt3ITD transgenic (Li et al., 2008) animals were kindly provided by Donald Small and Patrick Brown from Johns Hopkins University School of Medicine (Baltimore). To activate transgene expression in vivo, a doxycycline-containing chow (Harlan Laboratories) was administered to animals at the time of weaning (~3 weeks of age). To deactivate transgene expression, mice were taken off the doxycycline-containing diet and placed on a regular laboratory rodent diet. We obtained approval for all animal experiments from the Beth Israel Deaconess Medical Center IACUC Committee on Animal Research (Protocol #066–2011; Approved 08/15/2011).
CFU-GEMM Assays
CFU-GEMM assays were performed using MethoCult GF M3434 (StemCell Technologies) in accordance with the manufacturer’s instructions. Briefly, KSL cells from control and IDH2R140Q–t mice were isolated by FACS using a BD FACSAria IIu sorter (Becton Dickinson) (see Supplemental Experimental Procedures) and 900 cells were added to individual 3 ml aliquots of MethoCult GF M3434. One microgram per milliliter doxycycline and five micromolar AGI-6780 (Wang et al., 2013) were added as indicated. Cells were then plated at a density of 300 cells/dish in duplicate for each biological replicate and cultured for 7 days at 37°C and 5% CO2. Colonies were scored in a 2.5 cm × 2.5 cm area in the center of each dish using a grid. For the replating experiment, cells were collected in IMDM media supplemented with 2% fetal bovine serum (FBS), washed once in PBS, counted, and replated in MethoCult GF M3434 supplemented with 1 µg/ml doxycycline at a density of 5,000 cells/dish. Colonies were counted after 7–14 days of culture as above.
Generation of the HoxA9/Meis1 a Transduction Model
KSL cells were sorted (see Supplemental Experimental Procedures) from IDH2R140Qhm-t mice that had been treated with doxycycline for 6 weeks. Cells were cultured for 8 hr in StemSpan serum free expansion medium (StemCell Technologies) supplemented with murine SCF, TPO, IL-3, IL-6, and Flt3-L (PeproTech) on Retronectin-coated (Takara Bio) petri dishes. Cells were transduced overnight using MSCV-HoxA9-GFP and MSCV-Meis1a–YFP (kindly provided by Dr. Christian Bach). The next day, cells were harvested and washed twice with phosphate buffered saline (PBS). Viable cells were counted following trypan blue staining and 5,000 cells were injected retro-orbitally into sublethally irradiated (6.5Gr) C57BL/6 recipients. For the initial experiment, secondary transplantation and randomized deinduction was performed as outlined in Figure 4A. For the repeat experiment, all recipients were maintained on doxycycline food for 2 weeks, at which time 2 recipients were analyzed to confirm presence of GFP/YFP+ cells in the bone marrow. Randomized deinduction was then performed with recipients randomly assigned to doxycycline-treated or untreated groups. Mice were then analyzed every 2 weeks to monitor the kinetics of disease regression.
Serial Transplantation of IDH2R140Q–t/Flt3ITD Leukemic Cells
Bone marrow cells extracted from the femurs and tibias of a compound transgenic animal diagnosed with AML were subjected to red blood cell (RBC) lysis using ACK lysis buffer (GIBCO), washed twice with PBS supplemented with 2% FBS, passed through a 70 µM strainer, and frozen in FBS supplemented with 10% dimethyl sulfoxide (DMSO). On the day of transplantation, cells were thawed at 37°C and washed twice in PBS. Viable cells were counted following trypan blue staining. A hundred thousand donor cells were injected retro-orbitally into sublethally irradiated (6.5Gr) CD45.1+ recipients. Alternatively, 105 donor cells were mixed with 5 × 105 wild-type CD45.1 + competitor cells and injected retro-orbitally into lethally irradiated (9.5Gr) CD45.1+ recipients. For serial transplantations, the process was repeated using 105 cells from primary or secondary doxycycline-treated recipients.
Microarray Analysis
KSL cells from control and IDH2R140Q–t mice were cultured for 3 days in MethoCult GF M3434 (StemCell Technologies) supplemented with 1 µg/ml doxycycline. All cells were then collected and total RNA was extracted using Trizol (Ambion) with GlycoBlue (Life Technologies) as a carrier. Two-hundred nanograms of RNA was hybridized to Affymetrix Mouse Gene 2.1 ST arrays by the Beth Israel Deaconess Medical Center Genomics and Proteomics Core. The obtained raw intensity .cel files were normalized by robust multichip analysis (Bioconductor release 2.12) and differential expression was determined using the limma Bioconductor package by fitting a linear model. Gene set enrichment analysis was conducted with the gene sets from the Molecular Signatures Database (MolSigDB v3.1) (see Supplemental Experimental Procedures and Table S4 for more information).
Supplementary Material
Supplementary data
ACKNOWLEDGMENTS
We thank Dr. Robert Welner, Dr. Christian Bach, and members of the Pandolfi laboratory for critical discussion. L.M.K. was supported by an Overseas Postdoctoral Fellowship from the National Health and Research Council of Australia. M.R. was supported by the German Academy of Sciences Leopoldina (Leopoldina Research Fellowship grant number: LPDS 2009-27). R.T. was supported by a Marie Curie International Outgoing Fellowship for Career Development. K.Y., M.S., and K.S. are employees and shareholders of Agios Pharmaceuticals.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information for this article includes Supplemental Experimental Procedures, five figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.stem.2013.12.016.
REFERENCES
- Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, Pollock R, O’Donnell P, Grigoriadis A, Diss T, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J. Pathol. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi A, Araujo Cruz MM, Jyotsana N, Sharma A, Yun H, Görlich K, Wichmann M, Schwarzer A, Preller M, Thol F, et al. Mutant IDH1 promotes leukemogenesis in vivo and can be specifically targeted in human AML. Blood. 2013;122:2877–2887. doi: 10.1182/blood-2013-03-491571. [DOI] [PubMed] [Google Scholar]
- Chen C, Liu Y, Lu C, Cross JR, Morris JP, 4th, Shroff AS, Ward PS, Bradner JE, Thompson C, Lowe SW. Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes Dev. 2013;27:1974–1985. doi: 10.1101/gad.226613.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu SH, Heiser D, Li L, Kaplan I, Collector M, Huso D, Sharkis SJ, Civin C, Small D. FLT3-ITD knockin impairs hematopoietic stem cell quiescence/homeostasis, leading to myeloproliferative neoplasm. Cell Stem Cell. 2012;11:346–358. doi: 10.1016/j.stem.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher GH, Wellen SL, Klimstra D, Lenczowski JM, Tichelaar JW, Lizak MJ, Whitsett JA, Koretsky A, Varmus HE. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 2001;15:3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl. Acad. Sci. USA. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J. Exp. Med. 2010;207:339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Seo SI, Lee JY, Yoo NJ, Lee SH. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int. J. Cancer. 2009;125:353–355. doi: 10.1002/ijc.24379. [DOI] [PubMed] [Google Scholar]
- Kogan SC, Ward JM, Anver MR, Berman JJ, Brayton C, Cardiff RD, Carter JS, de Coronado S, Downing JR, Fredrickson TN, et al. Hematopathology subcommittee of the Mouse Models of Human Cancers Consortium Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100:238–245. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Piloto O, Nguyen HB, Greenberg K, Takamiya K, Racke F, Huso D, Small D. Knock-in of an internal tandem duplication mutation into murine FLT3 confers myeloproliferative disease in a mouse model. Blood. 2008;111:3849–3858. doi: 10.1182/blood-2007-08-109942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losman JA, Looper R, Koivunen P, Lee S, Schneider RK, McMahon C, Cowley G, Root D, Ebert BL, Kaelin WG. (R)-2-Hydroxyglutarate Is Sufficient to Promote Leukemogenesis and Its Effects Are Reversible. Science. 2013;339:1621–1625. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC-H, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu I-M, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, Van Vlierberghe P, Dolgalev I, Thomas S, Aminova O, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovici-Muller J, Saunders JO, Salituro FG, Travins JM, Yan S, Zhao F, Gross S, Dang L, Yen KE, Yang H, et al. Discovery of the First Potent Inhibitors of Mutant IDH1 That Lower Tumor 2-HG in Vivo. ACS Med. Chem. Lett. 2012;3:850–855. doi: 10.1021/ml300225h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau R, Magoon D, Greenblatt S, Li L, Annesley C, Duffield AS, Huso D, McIntyre E, Clohessy JG, Reschke M, et al. NPMc+ cooperates with Flt3/ITD mutations to cause acute leukemia recapitulating human disease. Exp. Hemat. 2013 doi: 10.1016/j.exphem.2013.10.005. in press Published online October 29, 2013. http://dx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brüstle A, Harris IS, Holmes R, Wakeham A, Haight J, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, Zon LI, Armstrong SA. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Travins J, DeLaBarre B, Penard-Lacronique V, Schalm S, Hansen E, Straley K, Kernytsky A, Liu W, Gliser C, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am. J. Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S-H, Ito S, Yang C, Wang P, Xiao M-T, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ¨ Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Yin X, Wolford CC, Chang Y-S, McConoughey SJ, Ramsey SA, Aderem A, Hai T. ATF3, an adaptive-response gene, enhances TGFβ signaling and cancer-initiating cell features in breast cancer cells. J. Cell Sci. 2010;123:3558–3565. doi: 10.1242/jcs.064915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data