5-Hydroxymethylcytosine is a predominantly stable DNA modification (original) (raw)
. Author manuscript; available in PMC: 2015 Apr 2.
Published in final edited form as: Nat Chem. 2014 Sep 21;6(12):1049–1055. doi: 10.1038/nchem.2064
Abstract
5-Hydroxymethylcytosine (hmC) is an oxidation product of 5-methylcytosine (mC) present in DNA of most mammalian cells. Reduction of hmC levels in DNA is a hallmark of cancers. Elucidating the dynamics of this oxidation reaction and the lifetime of hmC in DNA is fundamental to understanding hmC function. Using stable isotope labeling of cytosine derivatives in the DNA of mammalian cells and ultrasensitive tandem liquid-chromatography mass spectrometry (LCMS), we show that the majority of hmC is a stable modification, as opposed to a transient intermediate. In contrast with DNA methylation, which occurs immediately during replication, hmC forms slowly over the first 30 h following DNA synthesis. Isotopic labeling of DNA in mouse tissues confirmed the stability of hmC in vivo and demonstrated a relationship between global levels of hmC and cell proliferation. These insights have important implications for understanding the states of chemically modified DNA bases in health and disease.
Methylation of cytosine (C) at C-5 to form 5-methylcytosine (mC), by DNA methyltransferase enzymes, is an important epigenetic DNA modification that is essential for development, normal function and disease in all mammals1. In 2009, it was robustly demonstrated that 5mC could be enzymatically oxidized to 5-hydroxymethylcytosine (hmC)2,3. The initial discoveries were made in genomic DNA isolated from mouse brain and embryonic stem (mES) cell DNA, but hmC has subsequently been detected in all mammalian tissues4. In contrast with global DNA methylation levels which are stable across tissues, the levels of hmC are highly tissue-specific, ranging between 0.03 % of all cytosines in the spleen and 0.7 % in the brain4, and are reduced up to 8-fold in cancer tissues relative to healthy ones5-8. The oxidation of mC to hmC is carried out by the ten-eleven translocation (TET) enzymes3,9, iron and 2-oxoglutarate dependent dioxygenases, which are also able to further oxidize hmC to 5-formylcytosine (fC) and 5-carboxycytosine (caC)10,11. A number of pathways for the removal of the methyl group from mC via hmC have been suggested and validated in vitro, thus proposing hmC as an intermediate of DNA demethylation12. However, given that hmC binds to specific regulatory proteins13-15 and that it is mainly present at actively transcribed genes13,16-19, it appears that hmC may convey regulatory functions and be an epigenetic mark in its own right.
While the study of hmC has been rapidly growing during the past 5 years12 many fundamentally important questions remain unanswered20. Herein we have focused on the metabolism of C to mC and hmC in genomic DNA to specifically elucidate whether hmC is exclusively derived from mC, how hmC levels are maintained in proliferating and non-dividing cells, and what are the temporal dynamics between C, mC and hmC in DNA. To address these questions in mammalian cells and in an animal, we have used chemical isotopic labeling of genomic DNA coupled with ultra-sensitive analytical liquid chromatography-tandem mass spectrometry (LCMS).
Results and Discussion
Global hmC levels do not change during the cell cycle
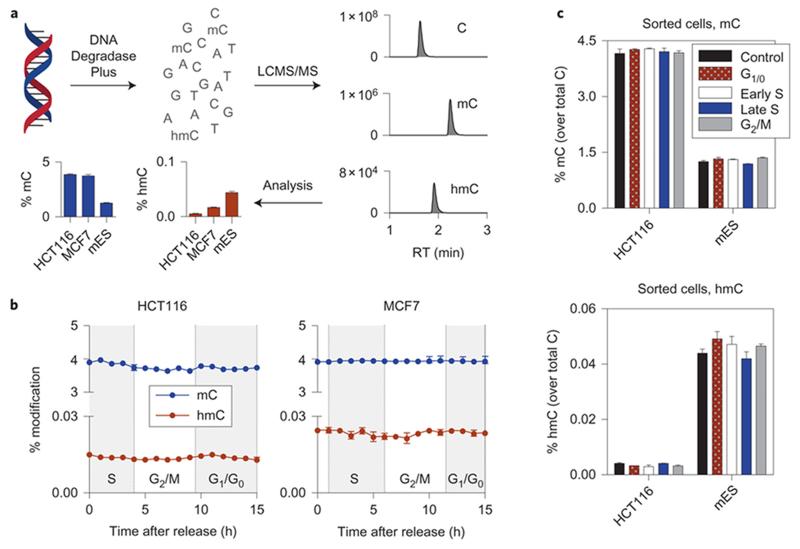
Our simple premise for these experiments was that if no hmC is produced during DNA replication (the S phase), when the total amount of DNA per cell doubles, then a two-fold drop in global hmC levels would be observed. To explore the timing of hmC formation, relative to replication, we set out to quantify hmC levels in genomic DNA in a synchronized population of cultured cells at defined points during the cell cycle. A robust LCMS assay (Fig. 1a and Supplementary Fig. S1) was used to quantify global levels of mC and hmC in the DNA of colorectal cancer (HCT116) and breast cancer (MCF7) cells. The asynchronous cell population was treated with a high dose of thymidine, arresting the cells in G1/S interphase, released into a thymidine-free medium and allowed to progress through one cell cycle (double-thymidine synchronization21) (Supplementary Fig. S2). In agreement with previous studies22,23, we measured mC levels to be constant throughout the cell cycle, consistent with restoration of the correct methylation pattern on the nascent strand during DNA replication (Fig. 1b). We found that hmC content is also uniform throughout one cell cycle in synchronized HCT116 and MCF7 cells. Both cell lines unexpectedly showed an increase in global hmC levels immediately after thymidine treatment (2.7- and 1.5-fold, P = 0.0003 and 0.001 for HCT116 and MCF7, respectively) (Supplementary Fig. S3), which is consistent with observations reported by Otani et al. in mES cells24. As a complementary approach to establish cell-cycle dependence of hmC, HCT116 and mES cells were fixed and stained with propidium iodide, then sorted by fluorescence activated cell sorting (FACS), according to the DNA content, into G1, early S, late S and G2/M phases. This confirmed that there are no major changes in global hmC levels during the cell cycle (Fig. 1c; Supplementary Fig. S4). Given there was not a two-fold decrease in hmC levels after replication, these data suggested that mC oxidation may be occurring during DNA replication (S phase) on either the nascent or the template strand.
Figure 1. Global levels of mC and hmC do not change during the cell cycle.
(a) An overview of the LCMS method for quantification of modified cytosines. Genomic DNA, isolated from cells, is digested in a single step with a mixture of enzymes to generate 2′-deoxynucleosides, which are separated on an ultra high performance liquid chromatography (UHPLC) column and quantified using tandem mass spectrometry. The levels of mC and hmC are expressed as a percentage of total cytosines. For calibration curves see Supplementary Figure S1. (b) Global levels of mC and hmC present in DNA isolated from HCT116 and MCF7 cells arrested in G1/S interphase and allowed to progress through one cell cycle. The shaded background indicates in which phase the majority of cells was found by fluorescence activated cell sorting (FACS) analysis (Supplementary Fig. S2). Shown are mean ± SEM of 3 and 2 biological replicates for HCT116 and MCF7, respectively, and at least 2 technical replicates per sample. (c) Global mC and hmC levels in HCT116 and mES cells sorted by the DNA content into G1/0, early S, late S and G2/M phases. Shown are mean ± SEM of 2 biological and 2 technical replicates per sample. See also Supplementary Figure S4.
hmC is generated on a newly synthesized DNA strand with a time delay
To address the timing of hmC formation on nascent DNA we set out to isolate DNA at different time points post-replication. We initially attempted to incorporate 5-bromo-2′-deoxyuridine (BrdU) during replication followed by isolation of newly replicated DNA with an anti-BrdU antibody.25,26 However, this approach failed as the anti-BrdU antibody also enriches for hmC (13-fold in a negative control containing no BrdU) (Supplementary Fig. S5). We therefore designed a strategy based on feeding cells with L-methionine-(_methyl_-13C,d3), the major methyl donor to mC via _S_-adenosylmethionine (SAM)-mediated alkylation by a DNA methyltransferase enzyme, to label newly synthesized mC and enable monitoring the kinetics of hmC formation in genomic DNA by LCMS (Fig. 2a). The light and heavy molecular species are clearly distinguishable in the LCMS analysis, allowing accurate and reproducible quantification of the labeling ratio (i.e. measured ratio of labeled hmC to total hmC (% hmC*)) (Fig. 2b; Supplementary Table S1). Firstly, HCT116, MCF7 and mES cells were cultured in heavy medium until a labeling plateau was obtained for both cytosine modifications. The measured ratio of labeled hmC to total hmC (% hmC*) was smaller than the ratio of labeled mC to total mC (% mC*) until a plateau was reached (Fig. 2c), which is consistent with label first being incorporated into mC that is then transformed directly into hmC by oxidation. We also observed a delay in hmC labeling in all cell lines (e.g. 10-20% hmC* when mC reaches 50% labeling). Thus, there is a lag in generating hmC from newly formed (labeled) mC. We further confirmed this lag using an alternative labeling system with stable isotopes on the cytosine ring and not the methyl group (Supplementary Fig. S6). Proliferation was not affected in labeled cells as judged from growth curves generated by live cell imaging (Supplementary Fig. S7), therefore we saw no evidence of a kinetic isotope effect associated with cleavage of the C-D bond.
Figure 2. DNA methylation and mC oxidation activities occur with a marked time difference.
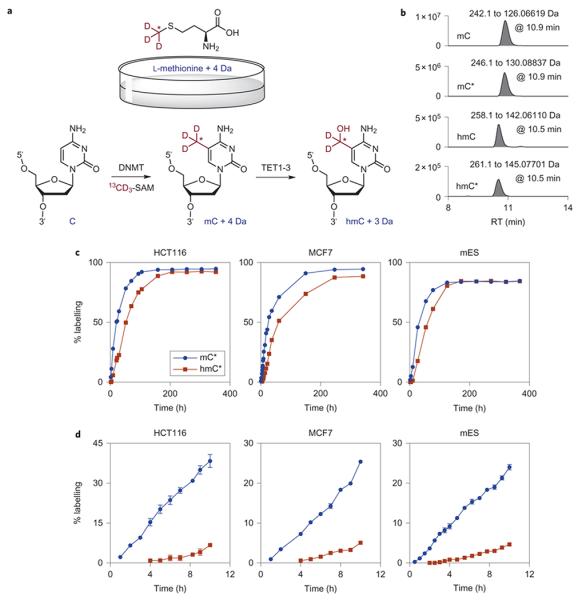
(a) An overview of the labeling strategy for measuring the timing of mC oxidation. Cells were grown in a medium containing L-methionine-(_methyl_-13C,d3), which leads to incorporation of stable isotopes into newly formed mCs in genomic DNA via _S_-adenosylmethionine (SAM). Subsequent LCMS analysis using accurate masses of labeled and unlabeled species provides mC and hmC labeling ratios (% mC* and hmC*). This in turn allows following the formation of hmC on a newly methylated DNA. (b) An example of extracted ion chromatograms from a DNA digest from mES cells labeled with L-methionine-(_methyl_-13C,d3) showing all analytes (heavy and light mC and hmC), and their corresponding retention times and mass transitions. The coefficient of variation for 3 technical replicates is typically less than 2%. MS signal intensity (arbitrary units) is shown on the y axis. (c) HCT116, MCF7 and mES cells were grown in the heavy medium for more than 14 days. The gap between mC and hmC labeling curves reveals that there is a timing difference between mC formation and its oxidation to hmC. (d) Detailed analysis of the first 10 h of labeling shows that in cancer cell lines and mES cells, it takes at least 4 h and 2 h, respectively, to begin oxidizing newly methylated DNA. Shown are mean and S.E.M. from 2 biological replicates (HCT116 and mES) or 2 technical replicates (MCF7). See also Supplementary Figures S8 and S9 for the detection limits of our LCMS, and more biological replicates for HCT116 and MCF7 cells.
To obtain the exact difference in timing between DNA methylation and the first oxidation of mC to hmC in newly replicated DNA, we looked for the initial emergence of labeled hmC species in DNA from cells grown with L-methionine-(_methyl_-13C,d3) for up to 10 h. The detection limit of our LCMS setup is in the low attomole range, which corresponds to 1 in 107 bases or about 1 in 500 hmCs in HCT116, 1 in 2000 hmCs in MCF7 and 1 in 6000 hmCs in mES cells when 100 ng of DNA is digested (Supplementary Fig. S8). We found that, in the human cancer cells and mES cells, it takes more than 4 h and 2 h, respectively, for the first detectable labeled hmC to appear in replicated DNA, and that this species initially builds up very slowly (Fig. 2d; Supplementary Fig. S9). This indicates that maintenance of hmC differs from mC in dividing cells. Most mC oxidation therefore does not occur on the nascent strand during DNA synthesis, and hemi-hydroxymethylated sites must form in the double-stranded DNA produced immediately after replication.
The majority of hmC in genomic DNA is stable in cultured cells and in vivo
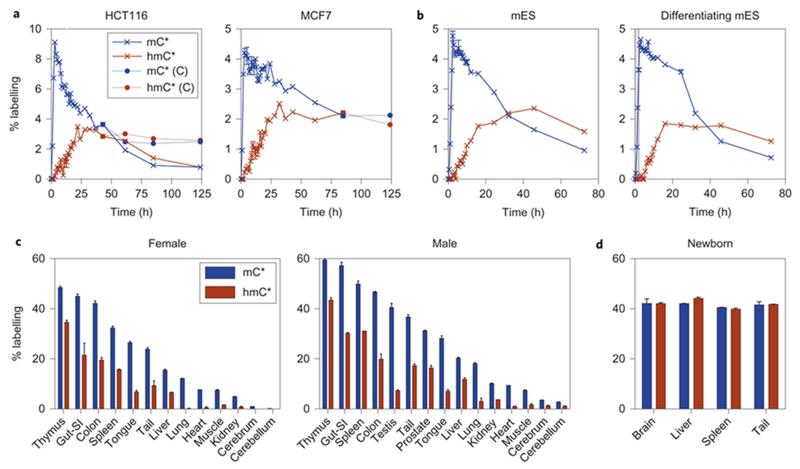
To measure how long mC and hmC persist in DNA and if they are removed via enzymatic transformation, we cultured HCT116 and MCF7 cells in the heavy (L-methionine-(_methyl_-13C,d3)) medium for 2 h to label a small population of mC (4-9%), and continued the culture in light medium for several days. In both studied cell lines, the hmC labeling curve is the steepest between 10-25 h, showing that the majority of hmC is formed from mC that is on average 10-25 h old (Fig. 3a, Supplementary Fig. S10). Such timing overlaps with the S phase of the next cell cycle, and therefore shows that there must be some DNA oxidation activity on the template strand. This effect will also be responsible for hiding any global hmC variations in synchronized cells, as during the release from G1/S there will still be oxidation activity on the DNA synthesized prior to the second thymidine block, or in sorted cells as shown in Figure 1 and by Otani et al.24.
Figure 3. The majority of genomic hmC is stable.
(a) Asynchronous HCT116 and MCF7 cells were labeled with L-methionine-(_methyl_-13C,d3) for 2 h and then grown in a light medium for several days. The bulk of hmC is derived from mC that is 15-25 h old, and persists in the genome until the labeling ratio gets diluted due to proliferation (i.e. generation of more unlabeled hmC). Dotted lines represent mC and hmC labeling in HCT116 cells that grew to confluency and stopped dividing, showing how stable the majority of hmC is in DNA. Shown are mean ± SEM of 2-3 technical replicates between 3-14 h of the experiment. The coefficient of variation between technical replicates is typically less than 2%. See also Supplementary Figure S10 for 6 and 10 h pulses in HCT116. (b) Labeling of undifferentiated and differentiating mES cells shows similar mC and hmC labeling profiles as for the differentiated cancer cells in (a). Shown are mean ± SEM of 2 technical replicates. Single replicate is shown for hmC in undifferentiated mES cells. (c) A breeding pair of wild-type animals was fed with a diet containing L-methionine-(_methyl_-13C,d3) for 117 days, and a range of tissues was analyzed by LCMS. The low labeling efficiency in slowly dividing and non-dividing tissues indicates that the majority of both mC and hmC must be stable in vivo. Gut-SI = small intestine. Shown are mean ± SEM of 2 technical replicates. See also Supplementary Figure S12. (d) mC and hmC labeling ratios in newborns (1 d old, parents labeled for 52 d before fertilization). Shown are mean ± SEM of 2 pups.
After this initial DNA oxidation activity, both heavy mC and hmC species remain present in DNA for more than 5 days, and the labeling ratios (%mC* and %hmC*) only decrease due to cell proliferation (i.e. adding more unlabeled mC and hmC into the population). We cannot completely rule out that small quantities of heavy hmC are not gradually generated from the relatively large pool of heavy mC, and subsequently removed. However, as cells become confluent leading to growth arrest and no more DNA is produced in the system, the labeling curves flatten (e.g. HCT116 past t = 60 h), strongly suggesting that the majority of both mC and hmC is long-lived or static in DNA of cultured differentiated cells (Fig. 3a and Supplementary Fig. S10). We have observed the same labeling profiles in undifferentiated mES cells, and also in mES cells which were allowed to differentiate at the start of labeling (Fig. 3b).
To explore the timing of mC oxidation and the stability of hmC in vivo, we generated mice comprising isotopically labeled mC and hmC in their genomic DNA. We fed a breeding pair of wild-type mice with a custom diet in which all L-methionine was replaced with L-methionine-(_methyl_-13C,d3). After 4 months (117 d), we measured the labeling efficiency of mC and hmC in the genomic DNA across a range of tissues by LCMS. We obtained a high labeling percentage (40-60% mC*) in the fast proliferating thymus and gut, lower levels (5-10% mC*) in kidney and heart, and only 0.2% and 2.9% mC* in the female and male cerebellum, respectively (Fig. 3c). It is noteworthy that brain DNA has the highest global hmC level of up to 0.7% of total cytosines4, and therefore the low levels of newly incorporated labeling in hmC (and mC) provide the first evidence that most of these modifications are stable in DNA in vivo. Similar to the cell culture experiments described in Figure 2, % mC* was higher than % hmC* in DNA from all adult tissues (Fig. 3c). If hmC were a transient intermediate, its labeling ratio would have to be equal to the labeling ratio of mC (the system would be in a steady state). Therefore, the majority of hmC must be stable also in these remaining tissues.
We did not observe any differences in mC and hmC labeling in developing tissues of pups that were born during the course of the above experiment (Fig. 3d and Supplementary Fig. S11). This indicates that the pool of labeled and unlabeled methyl donors in the female blood has reached equilibrium prior to fertilization and development of the pups. Consequently, the difference between % mC* and hmC* in proliferating adult tissues can only be explained by the presence of a non-proliferating (and therefore unlabeled) cell population with high hmC content, alongside a proliferating (labeled) cell population with relatively lower hmC content (Supplementary Fig. S12). This is consistent with our findings about the relationship between global hmC levels and proliferation (see below).
TET1 has been found to play a role in active DNA demethylation and memory formation in adult mouse brain27,28, therefore we also generated labeled mice deficient in one or both copies of the gene encoding the TET1 dioxygenase to determine the influence of TET1 on the lifetime of hmC. We fed a breeding pair of heterozygous Tet1tm1.1Jae mice29, each carrying one functional and one non-functional Tet1 allele, starting from the last week of pregnancy. After birth (a litter of 3 wild-type (WT, normal TET1), 4 heterozygous (Het, one functional copy of TET1) and 3 homozygous (Hom, no functional TET1) pups), the family was fed for 6 more days before measuring the labeling efficiency of mC and hmC in the genomic DNA across a range of tissues (Supplementary Figure S13).
Consistent with published work29, TET1 Hom pups had a slightly reduced weight relative to their WT littermates (1.3-fold, P = 0.01), and they incorporated correspondingly fewer isotopic labels into mC in all studied tissues (Supplementary Figs S14 and S15). Importantly, our data shows that the labeling ratio of hmC (% hmC*) or global levels of hmC do not significantly differ between WT and Hom pups (Supplementary Figs S15 and S16). This result indicates that the absence of TET1 does not impair the conversion of mC to hmC in any of the studied tissues, during the last and first week of pre- and postnatal development.
The observations about hmC stability are consistent with the studies in the early mouse development, where some hmC formed on the paternal genome in the zygote has been shown to be stable for several cell divisions and to disappear only due to the lack of maintenance during DNA replication30-34. However, this type of DNA demethylation, also called “active modification – passive removal”, differs from our observations in adult and developing tissues, where hmC is actively placed on new DNA with a time delay, and then remains stable, and therefore the removal of mC (demethylation) cannot be its sole function in DNA.
Cell proliferation is a major determinant of global hmC levels in vivo
We next plotted the global levels of hmC in genomic DNA against proliferation of the corresponding tissue, as determined by the incorporation of stable isotopes from L-methionine-(_methyl_-13C,d3) to mC, and found a linear correlation in all examined WT adult tissues apart from the brain (Fig. 4a). The brain is mostly a non-dividing tissue and therefore cannot follow this relationship. Antibody staining for hmC and the proliferation marker Ki67 in formalin-fixed paraffin-embedded tissue sections confirmed that hmC is abundant in the differentiated, non-proliferating cell types of the brain, liver or intestinal villi, and is low in proliferating Ki67 positive cells present in the splenic germinal centres or the intestinal crypts (Fig. 4b).
Figure 4. Isotopic labeling of DNA in vivo confirms the stability of hmC and reveals a relationship between global hmC levels and proliferation.
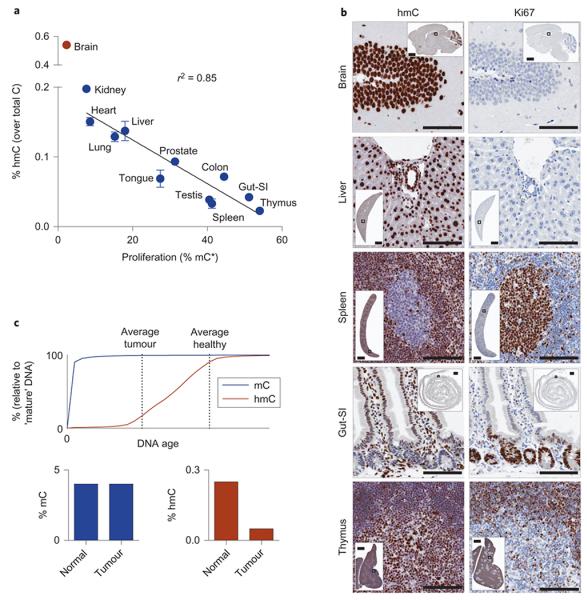
(a) Linear correlation between proliferation rate (estimated from % mC*) and global levels of hmC in tissues from WT adult mice labeled for 117 d (male and female). Brain outlier (red) was omitted from the correlation coefficient. Shown are mean ± SEM of 2 animals on the y axis (except for prostate and testis), and a mean %mC* was used on the x axis. (b) Immunohistochemistry for hmC and the proliferation marker Ki67. Shown are details of the dentate gyrus in the hippocampus, hepatic triad, splenic germinal centre, crypts and villi in the small intestine, and the cortico-medullary junction in the thymus. hmC is abundant in the differentiated, non-proliferating cell types and low in proliferating Ki67 positive cells. Horizontal bars indicate 100 μm (1 mm in inset image). (c) A model explaining the reduction of global hmC levels in faster proliferating cells and tissues compared to slowly proliferating ones (e.g. tumour vs. healthy tissue) based on our findings about the timing of DNA oxidation and the persistence of hmC in the genome. According to this model, the average age of DNA in a population of differentiated cells will be the major determining factor of global hmC content.
The central findings of our study show that the majority of hmC in genomic DNA is stable and that it takes a longer time to establish hmC on a newly synthesized DNA strand, in contrast with DNA methylation activity, which occurs immediately during replication. On the basis of these results, we propose that the average age of DNA (post replicative) in a population (and therefore the proliferation rate) is the major factor governing the global levels of hmC in both differentiated and undifferentiated cell types (Fig. 4c). This model explains the tissue specific levels of hmC (Fig. 4a), the increased global hmC levels in cells treated with either thymidine (Supplementary Fig. S3 and Otani et al.24) or a high dose of ascorbic acid (Supplementary Fig S17, Minor et al.35 and Blaschke et al.36), the high hmC level found in ‘immortal’ DNA strands37 and the reduced levels of hmC reported in all studied cancers5-7.
Conclusions
Since the discovery of hmC in mammalian DNA there has been an emphasis on the role of hmC as an intermediate in the mechanism of active demethylation. Herein we have presented the first evidence that the majority of hmC is actually stable in the genomic DNA of cultured cells and in vivo, rather than existing merely as a transient species. Given that hmC is particularly enriched in promoters and gene bodies of actively transcribing genes13,16-19, we propose that hmC is an epigenetic mark that is distinct from mC and therefore likely to have a different regulatory function. Contrary to mC, hmC is produced on a new DNA strand slowly and gradually over 30 h in cultured cells. Given the temporal stability of hmC in DNA, it will be vital in future studies to use methods that distinguish mC from hmC such as oxBS-seq38 when profiling DNA methylation in the genome.
We propose that the timing of mC oxidation during cellular proliferation is responsible for reduced levels of hmC in genomic DNA, such as those observed during tumorigenesis.
Methods
Cell culture
HCT116 cells were maintained in McCoy’s 5A medium supplemented with 10% FBS, penicillin and streptomycin. MCF7 cells were maintained in a DMEM medium supplemented with 10% FBS, penicillin and streptomycin. mES cells were derived by Dr Xiangang Zou in the CRUK Cambridge Institute from a C57BL/6 mouse and cultured on a gelatin-coated plate in a DMEM-KO medium (Invitrogen) supplemented with 10% FCS, MEM non-essential amino acids, glutamine, sodium pyruvate, penicillin, streptomycin, mouse leukemia inhibitory factor (mLIF) and 2i as described by Ying et al.39 For isotopic labeling experiments, all cells were maintained in a custom L-methionine-free DMEM-KO medium (Invitrogen) supplemented with 30 mg/L of either L-methionine (Sigma) or L-methionine-(_methyl_-13C,d3) (Cambridge Isotope), and the respective components above. For HCT116 and MCF7 cells, 3 g/L of D-(+)-glucose was added.
Animals
All in vivo experiments were performed under the terms of a UK Home Office license. C57BL/6 and B6;129S4-Tet1tm1.1Jae/J mice29 (Jackson Labs) were bred and housed according to UK Home Office guidelines. Custom L-methionine-free mouse diet supplemented with L-methionine-(_methyl_-13C,d3) (Sigma) was manufactured by TestDiet.
Genomic DNA extraction
Cells and tissues were resuspended in lysis buffer (100 mM Tris, pH 5.5, 5 mM EDTA, 200 mM NaCl, 0.2% SDS) supplemented with 400 μg/ml proteinase K (Invitrogen) and 200 μg/ml RNase A (Qiagen), and were incubated at 55°C overnight. DNA was purified using phenol:chloroform:isoamyl alcohol (25:24:1, Sigma) and Phase Lock Gel (5 Prime), precipitated from 70% ethanol and resuspended in ultrapure HPLC-grade water.
DNA degradation to 2′-deoxynucleosides and LCMS analysis
Up to 500 ng of DNA was incubated with 5 U of DNA Degradase Plus (Zymo Research) for 4 h at 37°C. Samples were filtered through a pre-washed Amicon 10 kDa centrifugal filter unit (Millipore) before LCMS analysis (filtration step for the Q-Exactive instrument only, see below).
LCMS analysis of global mC and hmC levels
Analysis of global levels of mC and hmC was performed on an AB Sciex Triple Quad 6500 mass spectrometer fitted with an Agilent Infinity 1290 LC system and an Acquity UHPLC HSS T3 column (50 × 2.1 mm, 1.8 μm particle size) at a flow rate of 300 μl/min, and a gradient of 0.1% formic acid in water and acetonitrile. Calibration curves were generated using a mixture of synthetic standards 2′-deoxycytidine (Sigma), 5-methyl- and 5-hydroxymethyl-2′-deoxycytidine (Berry&Associates), in the ranges of 0.01 – 100 μM, 0.0005 – 5 μM and 0.0001 – 1 μM for C, mC and hmC respectively. Samples and synthetic standards were spiked with an isotopically labeled mix containing 1 μM of 2′-deoxycytidine-(15N,d2) (synthesis and characterization in Supplementary Methods), 5-methyl-2′-deoxycytidine-(d3) and 5-hydroxymethyl-2′-deoxycytidine-(d3) (both Toronto Research Chemicals). The mass spectrometer was operated in multiple reaction monitoring (MRM) with transitions 228-112, 242-126, 258-142, 231-115, 245-129, 261-145 for C, mC, and hmC and their internal standards, respectively. The ion source was electrospray in positive mode. Results are expressed as a % of total cytosines.
LCMS analysis of isotope incorporation into genomic DNA
Analysis of isotope incorporation into DNA was performed on a Q-Exactive mass spectrometer (Thermo) fitted with an UltiMate 3000 RSLCnano HPLC (Dionex) and a self-packed hypercarb column (20 mm × 75 μm, 3 μm particle size) at a flow rate of 0.75 μl/min, and a gradient of 0.1% formic acid in water and acetonitrile. For labeling work with L-methionine-(_methyl_-13C,d3), ions of masses 228.1, 242.1, 246.1, 258.1 and 261.1 (corresponding to C, mC, mC*, hmC and hmC*, respectively) were fragmented in a positive ion mode at 10% normalized collision energy, and full scans (50 - 300 Da) were acquired for each channel. Extracted ion chromatograms of base fragments (112.0505, 126.0661, 130.0884, 142.0611 and 145.0770 +/− 5 ppm, respectively) were used for quantification. Results are expressed as % labeling (e.g. % mC* stands for labeled mC over total mC).
Supplementary Methods
Cell synchronization experiments, vitamin C treatments, FACS analysis of cell cycle, cell sorting by the DNA content, BrdU immunoprecipitation, immunohistochemical stainings of mouse tissue sections and chemical synthesis are described in Supplementary Methods.
Supplementary Material
Supplementary data
Acknowledgments
We would like to acknowledge the CRUK CI Flow Cytometry and Histopathology/ISH core facilities for their contributions, David Oxley, Clive d’Santos and Donna Michelle-Smith for their support with mass spectrometry, Xiangang Zou for his help with mES cells and David Tannahill for critical reading of the manuscript. This work was funded by Cancer Research UK (all authors) and the Wellcome Trust Senior Investigator Award (S.B.).
Footnotes
Competing financial interests
S.B. is a founder and shareholder in Cambridge Epigenetix, Ltd., and advisor and shareholder in Illumina, Inc.
References
- 1.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–30. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Globisch D, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haffner MC, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2:627–37. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin SG, et al. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011;71:7360–5. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lian CG, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–46. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kraus TF, et al. Low values of 5-hydroxymethylcytosine (5hmC), the “sixth base,” are associated with anaplasia in human brain tumors. Int. J. Cancer. 2012;131:1577–90. doi: 10.1002/ijc.27429. [DOI] [PubMed] [Google Scholar]
- 9.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–33. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–7. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–3. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song CX, He C. Potential functional roles of DNA demethylation intermediates. Trends Biochem. Sci. 2013;38:480–4. doi: 10.1016/j.tibs.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–30. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iurlaro M, et al. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 2013;14:R119. doi: 10.1186/gb-2013-14-10-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spruijt CG, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–59. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Yu M, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–80. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–8. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–84. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell. 2011;42:451–64. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen L, Zhang Y. 5-Hydroxymethylcytosine: generation, fate, and genomic distribution. Curr. Opin. Cell Biol. 2013;25:289–96. doi: 10.1016/j.ceb.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merrill GF. Cell synchronization. Methods Cell Biol. 1998;57:229–49. [PubMed] [Google Scholar]
- 22.Kappler JW. The kinetics of DNA methylation in cultures of a mouse adrenal cell line. J. Cell. Physiol. 1970;75:21–31. doi: 10.1002/jcp.1040750104. [DOI] [PubMed] [Google Scholar]
- 23.Woodcock DM, et al. Delayed DNA methylation is an integral feature of DNA replication in mammalian cells. Exp. Cell Res. 1986;166:103–12. doi: 10.1016/0014-4827(86)90511-2. [DOI] [PubMed] [Google Scholar]
- 24.Otani J, et al. Cell cycle-dependent turnover of 5-hydroxymethyl cytosine in mouse embryonic stem cells. PLoS One. 2013;8:e82961. doi: 10.1371/journal.pone.0082961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen RS, et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl. Acad. Sci. U. S. A. 2010;107:139–44. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vassilev L, Russev G. Purification of nascent DNA chains by immunoprecipitation with anti-BrdU antibodies. Nucleic Acids Res. 1988;16:10397. doi: 10.1093/nar/16.21.10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–34. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang RR, et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell. 2013;13:237–45. doi: 10.1016/j.stem.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dawlaty MM, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–75. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3642–7. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salvaing J, et al. 5-Methylcytosine and 5-hydroxymethylcytosine spatiotemporal profiles in the mouse zygote. PLoS One. 2012;7:e38156. doi: 10.1371/journal.pone.0038156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–9. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, et al. Programming and inheritance of parental DNA methylomes in mammals. Cell. 2014;157:979–91. doi: 10.1016/j.cell.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minor EA, Court BL, Young JI, Wang G. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J. Biol. Chem. 2013;288:13669–74. doi: 10.1074/jbc.C113.464800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blaschke K, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–6. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huh YH, Cohen J, Sherley JL. Higher 5-hydroxymethylcytosine identifies immortal DNA strand chromosomes in asymmetrically self-renewing distributed stem cells. Proc. Natl. Acad. Sci. U. S. A. 2013;110:16862–7. doi: 10.1073/pnas.1310323110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Booth MJ, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–7. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 39.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–23. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data