Genetic and functional characterization of clonally derived adult human brown adipocytes (original) (raw)
. Author manuscript; available in PMC: 2015 May 11.
Published in final edited form as: Nat Med. 2015 Mar 16;21(4):389–394. doi: 10.1038/nm.3819
Abstract
Brown adipose tissue (BAT) acts in mammals as a natural defense system against hypothermia, and its activation to a state of increased energy expenditure is believed to protect against the development of obesity. Even though the existence of BAT in adult humans has been widely appreciated1–8, its cellular origin and molecular identity remain elusive largely because of high cellular heterogeneity within various adipose tissue depots. To understand the nature of adult human brown adipocytes at single cell resolution, we isolated clonally derived adipocytes from stromal vascular fractions of adult human BAT from two individuals and globally analyzed their molecular signatures. We used RNA sequencing followed by unbiased genome-wide expression analyses and found that a population of uncoupling protein 1 (UCP1)-positive human adipocytes possessed molecular signatures resembling those of a recruitable form of thermogenic adipocytes (that is, beige adipocytes). In addition, we identified molecular markers that were highly enriched in UCP1-positive human adipocytes, a set that included potassium channel K3 (KCNK3) and mitochondrial tumor suppressor 1 (MTUS1). Further, we functionally characterized these two markers using a loss-of-function approach and found that KCNK3 and MTUS1 were required for beige adipocyte differentiation and thermogenic function. The results of this study present new opportunities for human BAT research, such as facilitating cell-based disease modeling and unbiased screens for thermogenic regulators.
Recent studies using 18fluoro-2-deoxyglucose positron emission tomography (18FDG-PET) scanning demonstrated that the prevalence of adult human BAT is inversely correlated with body mass index (BMI), adiposity, and fasting plasma glucose level1–6, indicating that BAT is likely to play a role in human metabolism. Current evidence indicates that two types of UCP1-positive thermogenic adipocytes exist in rodents and humans: classical brown adipocytes and beige adipocytes (also known as brite adipocytes). Classical brown adipocytes arise from a subset of the dermomyotome during embryonic development9–12. They are found predominantly in adipose depots of rodents and infants that are mostly dedicated to BAT, such as those in the interscapular regions. Beige adipocytes, on the other hand, reside mainly in subcutaneous white adipose tissues (WAT), where they arise postnatally in response to certain external cues—such as chronic cold exposure or long-term treatment with agonists of peroxisome proliferator-activated receptor-γ (PPAR-γ)—a process often referred to as the ‘browning’ of WAT13–17.
Although previous studies have reported that adult human BAT possesses a molecular signature resembling that of mouse beige adipocytes16,18,19, more recent data imply that cultured adipocytes derived from adult human BAT express several classical brown adipocyte–selective markers that were originally found in mice20. This discrepancy appears to be primarily due to a few reasons. First, adult human BAT is a highly heterogeneous tissue compared to mouse BAT, consisting of UCP1-positive multilocular brown adipocytes, UCP1-negative unilocular white adipocytes, endothelial cells, stromal cells, and immune cells. Indeed, the gene-expression profile of human BAT in the neck region varies depending on the depth of the tissue21. Hence, molecular analyses of biopsied adult human BAT samples could be confounded by potential contamination from UCP1-negative cells such as white adipocytes and myocytes. Second, conclusions made in previous studies were entirely based on the mRNA-expression profiles of a few selected genes that were originally identified in mice. Global and unbiased molecular analyses in a homogeneous cell population are therefore warranted to clarify the nature of adult human BAT. The discrepancy regarding the cellular identity of adult human BAT needs to be critically assessed to make it possible to strategize future therapeutic interventions for anti-obesity treatment through targeting this tissue. Namely, the identification of human-specific BAT molecular markers will allow for the development of cell type–selective activators that are likely to act more effectively and more safely than non-specific activators in recruiting new thermogenic adipocytes, especially in subjects who do not possess appreciable levels of existing BAT. For example, even though non-selective pharmacological activation of the sympathetic nervous system via beta3-adrenoceptor agonists can activate both classical brown adipocytes and beige adipocytes, adverse effects on the cardiovascular system prevents the clinical use of these agonists22.
To this end, we have isolated a total of 65 clonally immortalized preadipocyte lines from stromal vascular fractions (SVFs) obtained from UCP1-positive supraclavicular BAT biopsies of two non-obese individuals20. We found seven clonal lines (10.8%) among the clones isolated from these biopsies that exhibited highly adipogenic properties. This result was similar to that of a previous report showing that approximately 6.5% of cloned SVFs (20 out of 305 clonal lines) displayed adipogenic properties in culture16. We subsequently analyzed three out of the seven adipogenic lines for RNA sequencing (RNA-seq) and bioinformatics analyses. We also examined UCP1 mRNA expression of the seven adipogenic clones in response to cAMP and to rosiglitazone (Supplementary Fig. 1a,b).
In order to study and identify the gene signatures of clonal brown adipocytes, we experimentally differentiated the three clonal preadipocyte cell lines into brown adipocytes under an adipogenic culture condition. We took this approach because it is not possible to immortalize clonally isolated brown adipocytes directly from human biopsy material because adipocytes are post-mitotic cells. These experimentally derived brown adipocytes cannot be formally construed as a cell line (that is, they cannot be serially passaged while maintaining their differentiated state), so we refer to them here as brown adipocyte cultures 1–3. Additionally, because matched WAT pairs from the same subjects were not available, we isolated 35 clonal preadipocyte lines from SVFs of subcutaneous WAT from a lean BMI-matched individual as a control, using the same immortalization protocol as for the brown preadipocyte lines to avoid potential contamination from SV40 large T antigen–mediated immortalization. The clonally derived preadipocyte lines from subcutaneous WAT were differentiated under the same cultured conditions as the clonal brown adipocyte cultures. We refer to them as white adipocyte cultures 1–3.
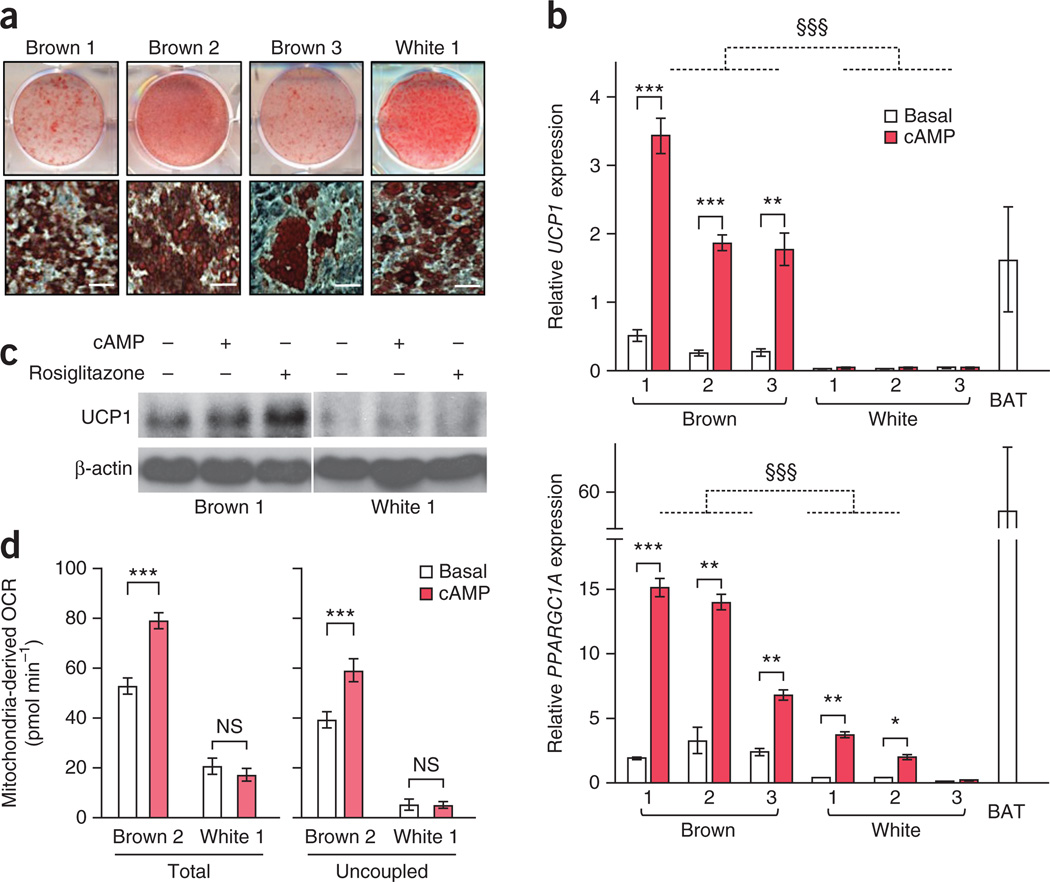
We found that these three clonal preadipocyte cell lines, upon differentiation, were highly adipogenic, as assessed by Oil-Red-O staining (Fig. 1a). We further found that after experimentally induced differentiation, brown adipocyte cultures 1–3 showed significantly (P < 0.001) higher expression of UCP1 and PPARGC1A (encoding PPAR-γ coactivator 1α) mRNA, both in the basal state and after a 4 h–long treatment with cAMP before harvesting, as compared to white adipocyte cultures 1–3 (Fig. 1b). The UCP1 mRNA expression in the three brown adipocyte cultures after cAMP treatment was similar to that found in biopsied BAT samples isolated from the supraclavicular region, whereas such induction was not seen in the white adipocyte cultures (Fig. 1b). The induction of PPARGC1A expression upon cAMP stimulation in the brown adipocyte cultures, however, did not reach the levels seen in the biopsied BAT control, although it was still potent (Fig. 1b).
Figure 1.
Isolation of clonal brown adipocytes from adult human BAT. (a) Representative Oil-Red-O staining of differentiated brown adipocyte cultures 1–3 and white adipocyte culture 1 at low magnification (top) and at high magnification (bottom). n = 3 for all groups. Scale bars, 50 µm. (b) Expression of UCP1 (top) and PPARGC1A (bottom) in the differentiated clonal brown adipocyte cultures 1–3 and white adipocyte cultures 1–3 treated with forskolin (cAMP) or vehicle (basal). BAT, biopsied human BAT from the supraclavicular regions (positive control). mRNA expression relative to expression of housekeeping gene TBP. n = 3 for all groups. §§§P < 0.001, brown versus white adipocyte lines; *P < 0.05, **P < 0.01, ***P < 0.001, brown or white (as indicated) versus basal by one-sided Welch’s _t_-test. The error bars for UCP1 in white adipocytes are 0.001, 0.002 and 0.001 in cultures 1, 2 and 3, respectively. (c) Western blot of UCP1 in differentiated brown adipocyte culture 1 and white adipocyte culture 1 treated with forskolin (cAMP) or rosiglitazone. β-actin, loading control. Data are representative of two experiments. (d) Total and uncoupled cellular respiration in differentiated brown adipocyte culture 2 and white adipocyte culture 1 treated with forskolin (cAMP) or vehicle (basal). OCR, oxygen consumption rate. n = 8 for all groups. ***P < 0.001 by one-sided Student’s _t_-test. NS, not significant. The variance was similar between basal and cAMP groups (P = 0.709). Data are expressed as means ± s.e.m. for all bar graphs.
UCP1 protein expression was also detected in the three brown adipocyte cultures, but not in the white adipocyte cultures. We observed higher UCP1 expression in brown adipocyte cultures treated with cAMP and rosiglitazonethan in the vehicle-treated cells (Fig. 1c). Notably, the differentiated clonal brown adipocyte cultures showed greater total and uncoupled cellular respiration in both the basal and the cAMP-stimulated states than in the clonal white adipocyte cultures with similar adipogenic properties (Fig. 1d). Induction of greater total and uncoupled cellular respiration was not observed in the brown preadipocyte cell lines (Supplementary Fig. 1c). Furthermore, the differentiated clonal brown adipocyte cultures possessed cAMP-induced lipolysis capacity, as glycerol release was higher in the cells treated with forskolin, a selective activator of the enzyme adenylyl cyclase, as well as in the cells treated with a cell-penetrant cAMP analog, dibutyryl-cAMP than in the vehicle-treated cells (Supplementary Fig. 1d). These data indicate that the three clonal human adipocyte cultures exhibited molecular and functional characteristics of thermogenic adipocytes.
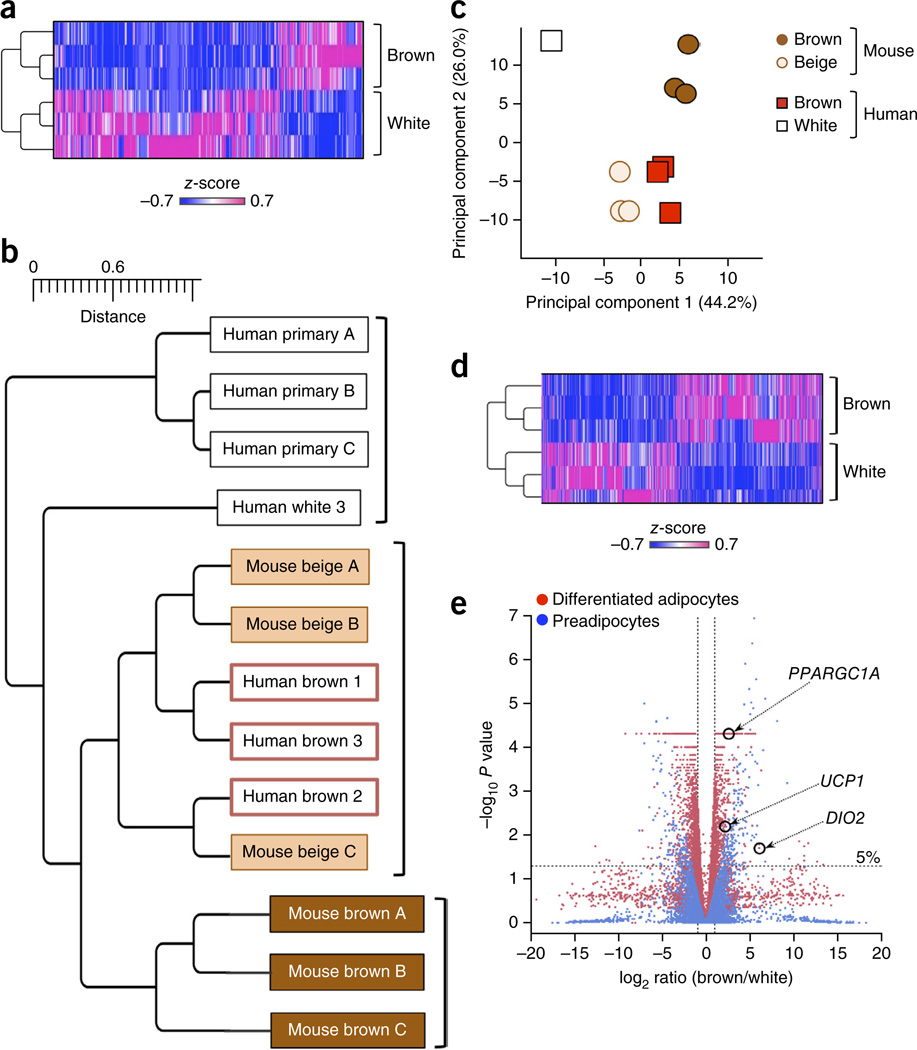
To identify gene signatures that were distinct between human brown and white adipocytes, we employed RNA-seq of clonal brown adipocyte cultures 1–3 and clonal white adipocyte cultures 1–3 (Fig. 2a). A complete list of the genes is provided in Supplementary Table 1. To determine if differentiated human brown adipocytes were similar or different to mouse classical brown adipocytes or beige adipocytes, we overlapped the top 800 differentiated human brown adipocyte–enriched genes with mouse orthologs that were enriched in differentiated classical brown and/or beige adipocytes as previously reported16 (Fig. 2a). Microarray data sets from clonally derived immortalized mouse beige and classical brown adipocyte cultures under a differentiated state were obtained from the Gene Expression Omnibus (GEO; accession no. GSE39562)16. In addition to our own human brown and white adipocyte RNA-seq data sets, we included publicly available microarray data sets of primary differentiated human adipocytes (referred to here as primaries A, B, and C) derived from subcutaneous WAT (GEO accession no. GSE41223; Fig. 2b). Hierarchical clustering revealed that the three differentiated clonal human brown adipocyte cultures belonged to a cluster of mouse beige adipocytes but were distinct from a cluster of mouse classical brown adipocytes (Fig. 2b). As an independent approach, principal component (PC) analysis suggested that the gene-expression profiles of the three differentiated clonal human brown adipocyte cultures were much closer to those of mouse beige adipocytes than to those of mouse classical brown adipocytes (relative Euclidean distance from human brown adipocytes to mouse beige and classical brown adipocytes was 9.4. and 18.1, respectively) (Fig. 2c). These results suggest that the molecular signatures of adult human brown adipocytes resemble those of mouse beige adipocytes rather than mouse classical brown adipocytes.
Figure 2.
Genome-wide gene expression analyses indicate a close relationship between human brown adipocytes and mouse beige adipocytes. (a) Expression profile and hierarchical clustering of the differentially expressed genes between differentiated clonal human brown adipocyte cultures 1–3 and differentiated clonal white adipocyte cultures 1–3 by two-fold or more. n = 3 for each cell type. The color scale shows _z_-scored FPKM (fragments per kilobase of exon per million fragments mapped) representing the mRNA level of each gene in a blue (low expression)-white-red (high expression) scheme. (b) Hierarchical clustering of human and mouse adipocytes as visualized by TreeGraph. The horizontal distance represents similarities among each cluster. (c) Principal component (PC) analysis of the transcriptome from human and mouse differentiated adipocytes. PC analysis was done using the same gene expression data set used in b, that is, the RNA-seq data set obtained from differentiated clonal human brown and white adipocyte cultures, and the microarray data set (GSE39562) from differentiated clonal mouse classical brown and beige adipocytes. Numbers in parentheses represent the proportion of data variance explained by each PC. (d) Expression profiles and hierarchical clustering of the differentially expressed genes between undifferentiated clonal human brown preadipocyte cell lines 1–3 and white preadipocyte cell lines 1–3 by two-fold or more. n = 3 for each cell type. The color scale is the same as in a. (e) Volcano plot of transcriptomes in the clonal differentiated human brown and white adipocyte cultures (red) and in the clonal undifferentiated human brown and white preadipocyte lines (blue). n = 3 for each cell type. The log-fold change between brown versus white is shown on the _x_-axis. The _y_-axis represents the −log10of the P values by delta method–based test. Previously defined BAT-enriched markers are shown.
Of note, RNA-seq analysis of the three clonal human preadipocyte cell lines before differentiation revealed that the molecular signatures of brown and white preadipocytes were distinct and formed separate clusters (Fig. 2d). Consistent with a recent report23, we found that the expression of contactin-associated protein-like 3 (CNTNAP3), leucine-rich repeat-containing 17 (LRRC17), regulator of G-protein signaling 7 binding protein (RGS7BP), phosphodiesterase 5A, cGMP-specific (PDE5A), and ATP-binding cassette, subfamily A, member 9 (ABCA9) was higher in the clonal brown preadipocyte lines than that in the clonal white preadipocyte lines (Supplementary Table 2). This separation at the gene-expression level was further validated by volcano plot analysis, which showed that the degree of difference in brown and white adipocyte transcriptomes was comparable between undifferentiated and differentiated states (Fig. 2e). These results indicate that the cellular fate of brown versus white adipocytes is largely determined at the precursor cell stage in adult humans.
From a developmental standpoint, it has been reported that mouse beige adipocytes, but not classical brown adipocytes, express smooth muscle lineage–selective genes and that a subset of beige adipocytes arises from smooth muscle myosin, heavy chain 11, (Myh11)-positive smooth muscle–like precursors24. Indeed, the isolated clonal human brown preadipocyte lines abundantly expressed smooth muscle lineage–selective genes, including ACTA2, TAGLN, MYL9, and CNN1, the master smooth muscle transcription factor SRF, and the non-cardiac co-activator MKL1 (Supplementary Fig. 2 and Supplementary Table 3). These results further support the possibility that adult human brown adipocytes exhibit beige-like characteristics. The subcutaneous WAT–derived white preadipocyte lines also expressed some of the smooth muscle lineage–selective genes, implying that these cells may also possess ‘browning’ plasticity given appropriate external stimuli.
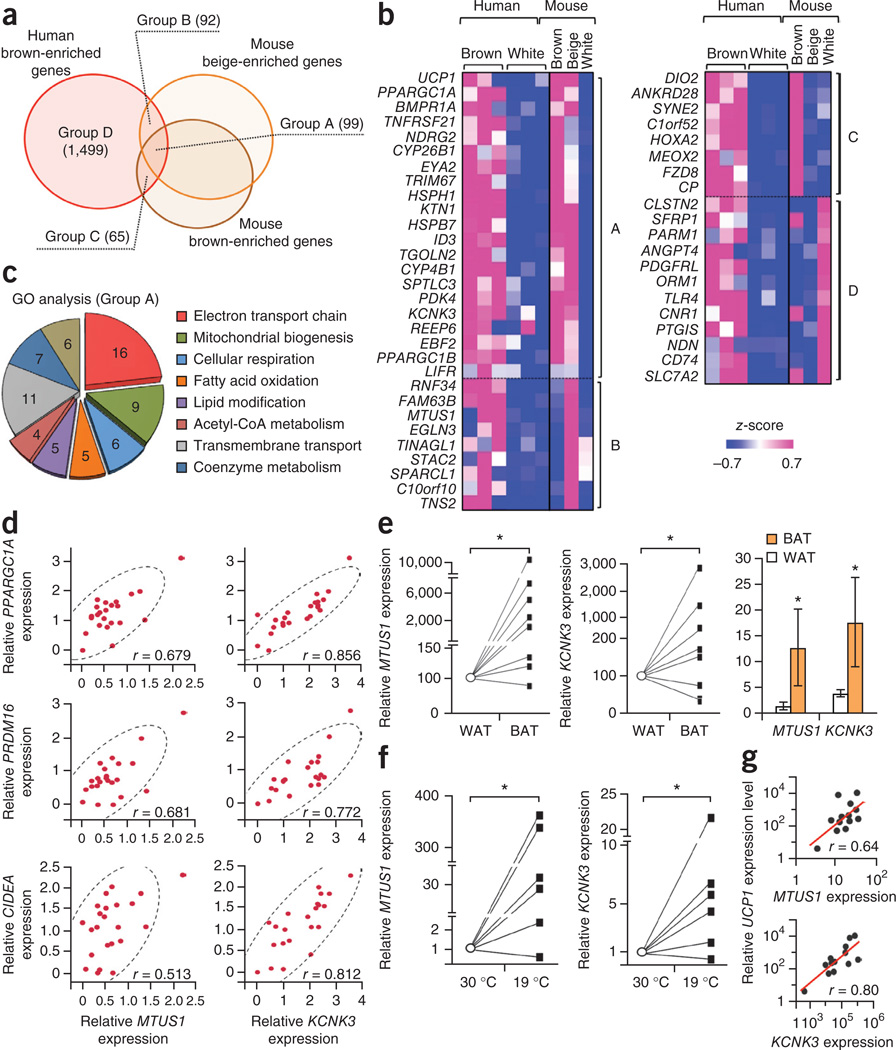
To identify human brown adipocyte–selective markers, we analyzed the overlapped gene sets between human and mouse brown adipocyte–enriched genes, which could be classified into four groups. Group A contained genes that were enriched in the differentiated clonal human brown adipocyte cultures as well as in mouse classical brown and beige adipocytes (Fig. 3a). Here we found previously defined BAT markers, such as UCP1, PPARGC1A, EBF2 (ref. 25), and BMPR1A26 (Fig. 3b and Supplementary Table 4). The other groups consisted of genes that were enriched in both human brown adipocytes and mouse beige adipocytes (group B), genes enriched in both human brown adipocytes and mouse classical brown adipocytes (group C), and genes enriched only in human brown adipocytes (group D). The subsequent gene ontology (GO) analysis found that the group A gene set was enriched with genes encoding members of the metabolic pathways involved in BAT thermogenesis, such as the electron transport chain, mitochondria biogenesis, cellular respiration, fatty acid oxidation, lipid modification, and acetyl-CoA metabolism (Fig. 3c). Next we validated the expression profiles of 36 representative human brown adipocyte–enriched genes from biopsied human BAT samples18 and integrated the correlation coefficient among these genes. The correlation analysis found that a large number (31 out of 36; 86.1%) of these genes showed significant (P < 0.05) positive correlations in expression (Supplementary Fig. 3a). We further validated the expression of human BAT marker-gene candidates in the differentiated clonal brown and white adipocyte cultures as well as in differentiated primary white adipocytes (Supplementary Fig. 3b). Based on the high correlation coefficient of the new BAT marker–gene candidates with the known BAT markers, the reproducible enrichment in all of the isolated brown adipocyte cultures as well as in human BAT depots from three independent cohorts, and the inducible expression in response to cAMP stimuli and cold temperature, we selected MTUS1 (encoding mitochondrial tumor suppressor 1) and KCNK3 (encoding potassium channel, subfamily K, member 3) as novel human BAT marker genes. MTUS1 and KCNK3 showed a highly significant correlation with previously defined BAT marker genes such as PPARGC1A, PRDM16, and CIDEA (Fig. 3d). Furthermore, MTUS1 and KCNK3 expression were significantly higher in UCP1-positive BAT depots than in UCP1-negative WAT depots isolated from the neck regions of the same individuals (eight pairs)21 (Fig. 3e). Similarly, MTUS1 and KCNK3 proteins were detected in the UCP1-positive BAT depots from the supraclavicular region (Supplementary Fig. 3c). Higher expression of KCNK3 in human BAT compared to WAT was also validated from two independent data sets27,28. Of note, MTUS1 and KCNK3 were enriched in the supraclavicular and posterior mediastinum BAT depots but not in the retroperitoneal BAT depots (Supplementary Fig. 3d), indicating that retroperitoneal BAT depots may contain a brown adipocyte population that is distinct from that in the supraclavicular and posterior mediastinum BAT depots in adult humans.
Figure 3.
Identification of human brown adipocyte markers. (a) Venn diagram of the overlapping genes enriched in human brown adipocytes, mouse classical brown adipocytes, and mouse beige adipocytes versus white adipocytes of the respective species by two-fold or more. All cells were differentiated clonal adipocytes in culture. P < 0.05 by delta method–based test. (b) Expression profiles of select genes enriched in each group. The color scale shows _z_-scored FPKM representing the mRNA level of each gene in blue (low expression)-white-red (high expression) scheme. (c) GO analysis of the gene set in Group A (GO FAT category). The area of each pie slice represents the number of genes that belong to the indicated GO terms. (d) Correlation between MTUS1 and KCNK3 mRNA expression on the _x_-axis and mRNA expression of previously defined marker genes PPARGC1A, PRDM16 and CIDEA on the _y_-axis. mRNA expression relative to the housekeeping gene TBP. n = 23 for each panel. P < 0.01 by _z_-test. (e) Gene expression of MTUS1 and KCNK3 in UCP1-positive adipose tissues (BAT) and UCP1-negative adipose tissues (WAT) from the neck region of the same individuals (eight pairs). *P = 0.017 and 0.044, respectively, by Wilcoxon signed-ranked test. The right bar graph shows the expression data without normalization to each individual. Data are expressed as means ± s.e.m. n = 8 for each group, *P < 0.05 by one-sided Welch’s _t_-test. (f) mRNA expression of MTUS1 and KCNK3 in the supraclavicular BAT isolated from six subjects under thermoneutral conditions (30 °C) and prolonged cold exposure (19 °C). Expression relative to TBP. *P = 0.035 and 0.023, respectively, by Wilcoxon signed-ranked test. (g) Correlation analysis between MTUS1 variant 3 or KCNK3 and UCP1 under prolonged cold exposure. n = 13. P < 0.01 by _z_-test.
Next we examined the regulatory pathways of MTUS1 and KCNK3 in the three differentiated clonal human brown adipocyte cultures and the supraclavicular BAT depots of adult humans. MTUS1 encodes a mitochondria-localized protein that is known to interact with the C terminus of angiotensin II type 2 receptor and regulate cell proliferation29. Among multiple alternatively spliced transcript variants, we found that variant 3 of MTUS1 was highly responsive to cAMP stimulation in human brown adipocytes (Supplementary Fig. 4a–c). Expression of KCNK3, which encodes a pH-dependent, voltage-insensitive potassium channel protein, was also higher in the brown adipocyte cultures treated by forskolin than in the vehicle-treated brown adipocyte cultures (Supplementary Fig. 5a). Importantly, expression of both MTUS1 variant 3 and KCNK3 was significantly induced in the supraclavicular BAT depots from six subjects under prolonged cold exposure (19 °C) versus thermoneutral conditions (30 °C)30 (Fig. 3f). Furthermore, MTUS1 and KCNK3 expression exhibited a significant positive correlation with UCP1 after cold stimuli (Fig. 3g). We also found that mRNA expression of Mtus1 and Kcnk3 was higher in the inguinal WAT of mice treated with the beta3-adrenergic receptor agonist CL-316,243 than in that of saline-treated mice (Supplementary Fig. 5b). Such induction was mediated by an increase in intercellular cAMP levels because MTUS1 and KCNK3 expression was significantly (P < 0.05) higher in the brown adipocyte cultures treated with either dibutyryl-cAMP or norepinephrine than in those treated with vehicle (Supplementary Fig. 5c). These results suggest that MTUS1 and KCNK3 are regulated by the sympathetic nerve–mediated cAMP pathway in mice and humans.
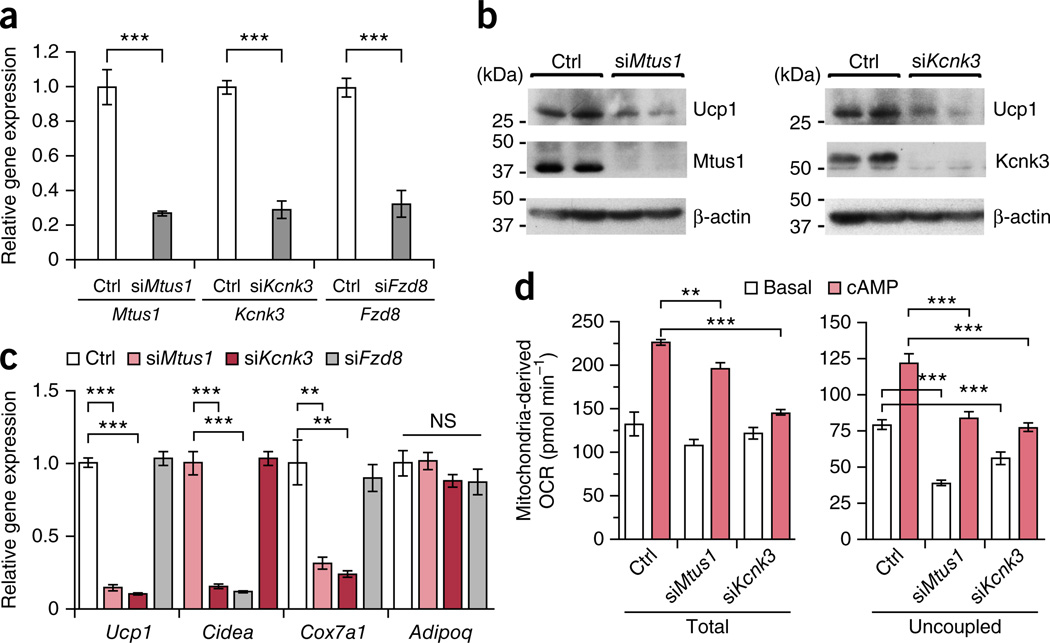
A positive correlation between the new BAT markers and UCP1 expression suggests possible functional roles of MTUS1 and KCNK3 in beige adipocytes. Because of the close relationship between adult human adipocytes and mouse beige adipocytes, we tested the functional roles of Mtus1 and Kcnk3 in beige adipocytes. To this end, differentiation potential and thermogenic function were tested in beige adipocytes from SVFs of mouse inguinal WAT17. These beige adipocytes were transfected with siRNAs targeting Kcnk3 and Mtus1 (Fig. 4a). We found that Ucp1 protein expression was substantially lower in the beige adipocytes depleted of Kcnk3 and Mtus1 than in the control siRNA-transfected cells (Fig. 4b). Expression of brown and beige adipocyte–selective molecular markers, such as Ucp1, Cidea and Cox7a, was significantly lower in the cells transfected with siRNAs targeting Mtus1 and Kcnk3 than the control siRNA-transfected cells (Fig. 4c). In contrast, knockdown of Fzd8, a mouse classical brown adipocyte–enriched gene, did not affect the brown and beige adipocyte–selective gene expression. No significant change was observed in the expression of a general adipogenic marker, adiponectin. Importantly, uncoupled cellular respiration was significantly lower in the cells depleted with Mtus1 and Kcnk3 than in the control siRNA-transfected cells upon cAMP stimulation (Fig. 4d). These results indicate that Mtus1 and Kcnk3 are required for beige adipocyte differentiation and thermogenic function.
Figure 4.
Mtus1 and Kcnk3 are required for beige adipocyte differentiation and thermogenic function. (a) Expression of Mtus1, Kcnk3, and Fzd8 in mouse inguinal WAT-derived adipocytes transfected with siRNAs targeting the indicated genes or non-targeting control (Ctrl). n = 4 for all groups. ***P < 0.001 for indicated siRNA versus control by one-sided Welch’s _t_-test. (b) Ucp1, Mtus1 and Kcnk3 protein expression from cells under the same culture condition as in a. β-actin, loading control. (c) Expression of Ucp1, Cidea, Cox7a1, and Adipoq (encoding adiponectin) from cells under the same culture condition as in a. n = 4 for all groups. **P < 0.01, ***P < 0.001 indicated siRNA versus control by one-sided Student’s _t_-test. NS, not significant. (d) Total and uncoupled cellular respiration. n = 8 for all groups. **P < 0.01, ***P < 0.001 by one-sided Welch’s _t_-test. Data are expressed as means ± s.e.m. for all bar graphs. Data are representative of three independent experiments.
It should be noted that single-cell cloning inevitably selects highly proliferative cell populations, regardless of biopsied locations and sample numbers. Possible cell-intrinsic changes that occur with obesity or aging await future investigations. While the present data do not exclude the possibility that classical brown adipocytes exist in adult humans, our unbiased global RNA-seq data analyses suggest that adult human BAT from supraclavicular regions displays molecular signatures that resemble beige adipocytes. This is in agreement with previous studies reporting that mouse beige adipocyte–enriched genes are highly expressed in adult human BAT16,18,19, and that the expression profile of tumor necrosis factor receptor superfamily, member 9 (TNFRSF9) clustered with that of UCP1 in the deeper neck adipose tissue21. This view is also supported by recent reports demonstrating that chronic cold acclimation robustly increases BAT mass in the supraclavicular region of adult humans who do not possess detectable BAT before treatment, in parallel with an increase in cold-induced energy expenditure31,32 or with an improvement in post-prandial insulin sensitivity33. The present study together with previous reports8,16,18,19,31–33 strongly indicates that adult human BAT is recruitable in response to physiological external cues such as chronic cold exposure. This research constitutes an important foundation for future human BAT studies. The isolated cells will provide new opportunities for investigating the cellular mechanisms of human BAT development and could potentially serve as a tool for cell-based disease modeling using CRISPR or TALEN technologies. These cells can also be applied to cell-based small-molecule screenings or unbiased RNAi screenings to identify novel regulators of human BAT thermogenesis. Furthermore, the newly identified genes, KCNK3 and MTUS1 variant 3, can serve as molecular markers secondary to UCP1 for assessing the number of thermogenic adipocytes within human adipose tissues.
ONLINE METHODS
General experimental approaches
No samples, mice or data points were excluded from the reported analyses. Mice were randomly assigned to two groups at the time of purchase to minimize any potential bias. No blinding was applied upon harvesting tissues after the treatment with saline or CL-316,243. RNA-seq and library constructions were performed by technical staff at the UCSF genome core who were blinded to the experimental groups. In other experiments, the investigators were not blinded to group allocation.
Isolation of clonal adipocytes from adult human BAT
Stromal vascular fractions (SVFs) were isolated from supraclavicular adipose tissues from two individuals from whom data were obtained and reported previously20 (30 and 43 years old; BMI, 23 and 26 kg/m2, respectively). Primary white preadipocytes from subcutaneous WAT were obtained from PromoCell (Heidelberg, Germany). SVFs were infected with a retrovirus expressing large T antigen (pBabe SV40 Large T antigen; Addgene) and subsequently selected using puromycin (2 µg/ml). Initially, 23 lines of sub-clonal cells were generated from the supraclavicular biopsies. Subsequently, single cell clones were obtained from the highly adipogenic sub-clonal lines by limiting dilution of cells into 96-well plates (0.2 cells/well). Wells containing cells were trypsinized and propagated in 48-well, 24-well, 12-well, 6-well, 60 mm plates, and finally 100 mm plates. A total of 65 clonal lines were originated from supraclavicular biopsies and screened for adipogenic potential. Similarly, 35 clonal lines were generated using SVFs from subcutaneous WAT with a BMI-matched individual. We obtained seven adipogenic clonal lines from supraclavicular biopsies and six adipogenic clonal lines from subcutaneous WAT. The clonally derived white adipocyte cultures after differentiation were used for Oil-red-O staining in Figure 1a; for mRNA and protein expression analyses in Figures 1b,c and 3a–c and Supplementary Figures 3b, 4b–c, and 5a, for cellular respiration assays in Fig. 1d, and for RNA-seq analyses in Figure 2a–c,e. Undifferentiated clonal white preadipocyte cell lines were used for RNA-seq analyses in Fig. 2d,e, and for cellular respiration assays in Supplementary Figure 1c. Differentiated primary non-clonal adipocytes were used in the gene expression study in Supplementary Figure 3b. As described in detail later, gene expression data of differentiated primary human adipocytes from subcutaneous WAT (human primaries A, B, and C) included in the analysis in Figure 2b were obtained from publically available microarray data sets (Accession: GSE41223).
Cell culture
Human preadipocytes were seeded in collagen-coated plates (BD Biosciences) and cultured in Advanced DMEM/F-12 medium (without phenol red) supplemented with 10% FBS, 50 IU penicillin, and 50 µg/ml streptomycin. When cells reached confluence, adipocyte differentiation was induced with medium containing 10% FBS, 5 µg/ml insulin, 1 nM T3, 0.125 mM indomethacin, 0.2 mM isobutylmethylxanthine (IBMX), and 2 µg/ml dexamethasone for 2 d. Subsequently, the cells were cultured in DMEM/F-12 medium in the presence of insulin, dexamethasone, and T3 for an additional 19 d. For rosiglitazone treatment, cells were treated with 1 µM rosiglitazone during the differentiation. For cAMP treatment, cells were incubated with 10 µM forskolin and harvested after 4 h for RNA isolation and 6 h for protein analysis. For mouse adipocyte culture, primary SVFs were isolated from the inguinal WAT of B6 male mice, according to the methods as previously reported12,17. The SVFs were cultured to confluence and adipocyte differentiation was induced with DMEM/F-12 medium (with phenol red) containing 10% FBS, 5 µg/ml insulin, 1 nM T3, 0.25 mM IBMX, 0.125 mM indomethacin, and 2 µg/ml dexamethasone. Two days after induction, cells were switched to the maintenance medium containing 10% FBS, 5 µg/ml insulin, 1 nM T3 for an additional 6 d. To induce beige adipocyte differentiation, cells were differentiated in the presence of rosiglitazone at a dose of 0.5 µM, as outlined in a previous paper17. Mycoplasma infections were not tested in the experiments.
RNAi-mediated gene knockdown
For RNAi-mediated gene knockdown, individual siRNA pools for Mtus1 (no. 065229), Kcnk3 (no. 043787) and Fzd8 (no. 045544) were obtained from GE Dharmacon. Non-targeting siRNA (no. 001810; GE Dharmacon) was used as the control. Immortalized preadipocytes derived from SVFs of mouse inguinal WAT were first reverse-transfected with siRNA (25 pmol/0.2 × 106 cells) using Lipofectamine RNAiMAX following the manufacturer’s protocol. The cells were forward-transfected 2 d after the first transfection. Cells were differentiated under an adipogenic condition that promotes beige adipocyte differentiation for 4 d (ref. 17). Knockdown of genes was validated by qRT-PCR using the primers listed in Supplementary Table 5.
RNA sequencing (RNA-seq)
RNA-seq libraries were constructed from total RNA using the Ovation RNA-seq system version 2 (NuGEN). The isolated RNA was reverse-transcribed to cDNAs using a combination of random hexameric and a poly-T chimeric primer. The cDNA libraries were subsequently amplified using the Ultralow DR library kit (NuGEN) according to the manufacturer’s instructions. Quality of the libraries was validated by Bioanalyzer (Agilent Technologies). Subsequently, high-throughput sequencing was performed using a HiSeq 2500 instrument (Illumina) at the UCSF Genomics Core Facility. Raw reads for each library were mapped using TopHat version 2.0.8 against the indexed human (hg19) or mouse (mm9) genome, downloaded from the TopHat website (http://ccb.jhu.edu/software/tophat/igenomes.shtml). The mapped reads were converted to FPKM (fragments per kilobase of exon per million fragments mapped) by running Cuffdiff 2 (http://cufflinks.cbcb.umd.edu) to determine gene expression34. The RNA-seq reads were deposited in ArrayExpress (http://www.ebi.ac.uk/arrayexpress/?) under accession numbers E-MTAB-2602 for human brown and white adipocytes and E-MTAB-2624 for mouse primary brown, beige, and white adipocytes.
Human and mouse transcriptome analyses
To compare human and mouse adipocyte transcriptomes, the gene microarray data sets from mouse beige and classical brown adipocytes were downloaded from Gene Expression Omnibus (GEO) (Accession: GSE39562) as previously reported16. Among 13 putative beige cell lines, three lines were used for our bioinformatics analysis since they were functionally validated by oxygen consumption assays, cell transplantation experiments, or UCP1 protein expression by western blotting. The following microarray data were used in the analysis shown in Figure 2: GSM971716 (designated as A), GSM971714 (B), and GSM971698 (C). Microarray data (GSM971717, GSM971718, and GSM971719) were used for mouse classical brown adipocytes and designated as A, B, and C, respectively. In addition to our human brown and white adipocyte RNA-seq data, we included a publically available microarray data set of primary human adipocytes derived from subcutaneous WAT (Accession: GSE41223). All the microarray data sets were normalized by quantile normalization35 using the dChip Software36. To combine the human and mouse data, we used ortholog information downloaded from Ensembl release 75. Clustering analysis was performed using differentially expressed genes (>2-fold) as determined by the comparison between mouse beige adipocyte lines and mouse classical brown adipocyte lines. FPKM was normalized to array signal in order to set the mean expression level of each gene equal between humans and mice. The top 800 highly expressed genes were used for the clustering analysis in Figure 2b,c. Similar clustering patterns were seen when the top 500, 1,000, 1,500, and 2,000 highly expressed genes or all the genes filtered by variance were subjected to the analyses.
Thirty-six representative human brown adipocyte–enriched genes were identified from the clustering analysis and further selected based on the additional criteria in which mRNA expression levels were high (averaged FPKM > 2.0) in differentiated human brown adipocytes and consistent among the three independent clonal cell lines (relative s.d. of FPFM < 1.0).
Multivariate statistics
Principal component (PC) was computed from the covariance matrix of the human and mouse transcriptome profiles. Hierarchical clustering (HCL) was performed with the complete linkage method using the Pearson correlation as a distance metric. The measured mRNA expressions in RNA-seq were _z_-scored and visualized as a heat-map representation in the blue-white-red scheme. All statistical analyses were performed using the JMP software version 9 (SAS Institute) or Multi Experiment Viewer37. TreeGraph 2 (ref. 38) was used to generate dendrogram trees. Gene Ontology (GO) enrichment analysis was performed in DAVID39 using the biological process terms (GO FAT category).
Quantitative PCR
Total RNA was isolated using RiboZol reagents (AMRESCO) following the manufacturer’s protocol. Quality of the RNA from all samples was validated by a NanoDrop spectrophotometer (Thermo Scientific). Reverse transcription reactions were performed using an iScript cDNA synthesis kit (Bio-Rad). Quantitative reverse transcriptase PCR (qRT–PCR) was performed with SYBR green fluorescent dye using the ViiA 7 Real-Time PCR System (Life Technologies). For the analysis of human BAT (eight pairs)21, total RNA was extracted from tissues using an RNA MiniPrep kit (Zyme Research) according to the manufacturer’s instructions. cDNA was prepared from 2 ng/µl of RNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer’s instructions. Quantitative RT-PCR assays were run in duplicates and quantified in the ABI Prism 7900 sequence-detection system. Relative mRNA expression was determined by the delta-Ct method using TATA-binding protein (TBP) as an endogenous control. The sequences of primers used in this study are provided in Supplementary Table 5.
Western blotting
Fully differentiated adipocytes were lysed in RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton-X, 10% glycerol, and cOmplete protease inhibitors (Roche)) and total protein lysates (50 µg) were boiled with 4× NuPage LDS loading buffer (Invitrogen), loaded on a 4–15% SDS-PAGE and subsequently transferred onto PVDF membranes. The blots were blocked in 5% milk in Tris-buffered saline with Tween 20 (TBS-T) and incubated overnight with rabbit anti-UCP-1 (U6382, Sigma or ab10983, Abcam) or mouse anti-β-actin (A3854, Sigma) overnight. For adipose tissues analysis, antibodies for MTUS1 (clone 1C7, Abnova) and KCNK3 (APC-024, Alomone Labs)40 were used. Light-chain-specific anti-mouse IgG (115-035-174, Jackson ImmunoResearch) was used as a secondary antibody for MTUS1.
Cellular respiration assay
Cells were plated in the gelatin-coated XF24 V7 cell culture microplates (Seahorse Bioscience) and differentiated into adipocytes. Cells were incubated in pre-warmed XF assay medium (pH 7.4) (no. 101022-100; Seahorse Bioscience) supplemented with sodium pyruvate (final concentration: 1.0 mM) and glucose (final: 25 mM) for 1 h. The oxygen consumption rate (OCR) was measured by the XFe Extracellular Flux Analyzers (Seahorse Biosciences). Cellular respiration was analyzed by using the following perturbation drugs in succession: 1 µM oligomycin, 1 µM FCCP (carbonyl cyanide-_p_-trifluoromethoxyphenylhydrazone) and 1 µM respiratory chain inhibitors (rotenone/antimycin A). The OCR purely attributed to mitochondria was determined by subtracting rotenone/antimycin A–insensitive OCR from total and uncoupled OCR. For cAMP-induced respiration, fully differentiated adipocytes were incubated with 10 µM of forskolin for 20 h. Three independent experiments were performed.
Adipose tissue samples
Human adipose tissue was collected from eight subjects (three females, five males; 49.6 ± 3.7 years) as paired sets of superficial and deep neck tissue, as described previously21. BAT samples from multiple depots were collected as described previously18. BAT samples from six subjects both under thermoneutral conditions and after prolonged cold exposure at 19 °C were collected as previously reported30. As provided in each study, written informed consent was obtained from all individuals or parents/legal guardians in the presence of a witness for each study. The study shown in Figure 3e was approved by the Human Studies Institutional Review Boards of Beth Israel Deaconess Medical Center, Joslin Diabetes Center, and Massachusetts General Hospital21. The study shown in Figure 3d and Supplementary Figure 3a,c,d was approved by the Institutional Review Board of Children’s Hospital Los Angeles18. The study shown in Figure 3f,g was approved by the University of Texas Medical Branch and Institute for Translational Sciences Institutional Review Boards30.
Animal studies
All animal experiments were performed according to procedures approved by the Institutional Animal Care and Use Committee for animal care and handling at the University of California, San Francisco. 8-week-old male C57BL/6J mice were obtained from the Jackson Laboratory. Mice were randomly assigned to different treatments (saline or CL-316,243) at the time of purchase to minimize any potential bias (n = 5 per group). Mice were intraperitoneally injected daily with 0.9% saline or beta3-adrenergic receptor-specific agonist CL-316,243 (Sigma) at a dose of 1 mg/kg for 7 d. Adipose tissues were harvested for molecular and biochemical analyses. The investigators were not blinded to sample allocation during the experiments.
Statistics
The sample sizes chosen for the experiments were calculated by power analysis (α = 0.05 and power of 0.8). The pwr package in the R statistics package, which implements power analysis as outlined by Cohen41, was used for our sample size determination. Sample size used is specified in each figure legend. Where parametric tests (_t_-test) were used, normal distribution and difference of variance were tested by Shapiro–Wilk test and _F_-test, respectively. When the null hypothesis of equal variance was rejected, Welch’s _t_-test was used. Otherwise, Student’s _t_-test was used. When normal distribution was not assumed, a nonparametric method (Wilcoxon signed-rank test) was used. Data are expressed as means ±s.e.m. The Pearson correlation and its significance (H0 = true correlation is equal to 0) were determined by the R statistics package. P < 0.05 was considered significant in all the experiments.
Supplementary Material
sup
ACKNOWLEDGMENTS
We acknowledge support from the National Institutes of Health (NIH) (DK087853 and DK097441), the UCSF Diabetes Research Center grant (DK63720), the UCSF Program for Breakthrough Biomedical Research program, the Pew Charitable Trust, and the Japan Science and Technology Agency (all to S.K.); and from the NIH (P50-GM60338) and the American Dental Association (1-14-TS-35) (both to L.S.S.). K.S. is supported by a fellowship from the Japan Society for the Promotion of Science. Y.H. is supported by the Manpei Suzuki Diabetes Foundation. I.H.N.L. is supported by the Dutch Heart Foundation.
Footnotes
Accession codes. ArrayExpress: Coordinates have been deposited with accession codes E-MTAB-2602 and E-MTAB-2624.
AUTHOR CONTRIBUTIONS
K.S. and S.K. designed the experiments. K.S., I.H.N.L., Y.H., H.H., S.B.S., M.K. and S.K. performed the cellular experiments and analyzed the data. M.C., A.M.C., L.S.S. and Y.-H.T. provided adipose tissue samples. K.S., Y.H., H.H., R.X. and S.K. analyzed adipose tissue samples. K.S. and S.K. wrote the manuscript. All authors contributed to editing the manuscript. S.K. conceived and managed the project.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Cypess AM, et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ouellet V, et al. Outdoor temperature age, sex, body mass index, and diabetic status determine the prevalence, mass, and glucose-uptake activity of 18F-FDG-detected BAT in humans. J. Clin. Endocrinol. Metab. 2011;96:192–199. doi: 10.1210/jc.2010-0989. [DOI] [PubMed] [Google Scholar]
- 3.van Marken Lichtenbelt WD, et al. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 4.Yoneshiro T, et al. Age-related decrease in cold-activated brown adipose tissue and accumulation of body fat in healthy humans. Obesity (Silver Spring) 2011;19:1755–1760. doi: 10.1038/oby.2011.125. [DOI] [PubMed] [Google Scholar]
- 5.Saito M, et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am. J. Physiol. Endocrinol. Metab. 2007;293:E444–E452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- 7.Virtanen KA, et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 8.Lidell ME, et al. Evidence for two types of brown adipose tissue in humans. Nat. Med. 2013;19:631–634. doi: 10.1038/nm.3017. [DOI] [PubMed] [Google Scholar]
- 9.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atit R, et al. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev. Biol. 2006;296:164–176. doi: 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- 11.Timmons JA, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc. Natl. Acad. Sci. USA. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature. 2013;504:163–167. doi: 10.1038/nature12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat. Med. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 14.Kajimura S, Saito M. A new era in brown adipose tissue biology: molecular control of brown fat development and energy homeostasis. Annu. Rev. Physiol. 2014;76:225–249. doi: 10.1146/annurev-physiol-021113-170252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waldén TB, Hansen IR, Timmons JA, Cannon B, Nedergaard J. Recruited vs. nonrecruited molecular signatures of brown, “brite,” and white adipose tissues. Am. J. Physiol. Endocrinol. Metab. 2012;302:E19–E31. doi: 10.1152/ajpendo.00249.2011. [DOI] [PubMed] [Google Scholar]
- 16.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohno H, Shinoda K, Spiegelman BM, Kajimura S. PPARgamma agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 2012;15:395–404. doi: 10.1016/j.cmet.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharp LZ, et al. Human BAT possesses molecular signatures that resemble beige/brite cells. PLoS ONE. 2012;7:e49452. doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee P, Werner CD, Kebebew E, Celi FS. Functional thermogenic beige adipogenesis is inducible in human neck fat. Int. J. Obes. (Lond) 2014;38:170–176. doi: 10.1038/ijo.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jespersen NZ, et al. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supraclavicular region of adult humans. Cell Metab. 2013;17:798–805. doi: 10.1016/j.cmet.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Cypess AM, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat. Med. 2013;19:635–639. doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villarroya F, Vidal-Puig A. Beyond the sympathetic tone: the new brown fat activators. Cell Metab. 2013;17:638–643. doi: 10.1016/j.cmet.2013.02.020. [DOI] [PubMed] [Google Scholar]
- 23.Tews D, et al. Comparative gene array analysis of progenitor cells from human paired deep neck and subcutaneous adipose tissue. Mol. Cell. Endocrinol. 2014;395:41–50. doi: 10.1016/j.mce.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Long JZ, et al. A smooth muscle-like origin for beige adipocytes. Cell Metab. 2014;19:810–820. doi: 10.1016/j.cmet.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajakumari S, et al. EBF2 determines and maintains brown adipocyte identity. Cell Metab. 2013;17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulz TJ, et al. Brown-fat paucity due to impaired BMP signalling induces compensatory browning of white fat. Nature. 2013;495:379–383. doi: 10.1038/nature11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svensson PA, et al. Gene expression in human brown adipose tissue. Int. J. Mol. Med. 2011;27:227–232. doi: 10.3892/ijmm.2010.566. [DOI] [PubMed] [Google Scholar]
- 28.Søndergaard E, et al. Chronic adrenergic stimulation induces brown adipose tissue differentiation in visceral adipose tissue. Diabet. Med. 2015;32:e4–e8. doi: 10.1111/dme.12595. [DOI] [PubMed] [Google Scholar]
- 29.Nouet S, et al. Trans-inactivation of receptor tyrosine kinases by novel angiotensin II AT2 receptor-interacting protein, ATIP. J. Biol. Chem. 2004;279:28989–28997. doi: 10.1074/jbc.M403880200. [DOI] [PubMed] [Google Scholar]
- 30.Chondronikola M, et al. Brown adipose tissue improves whole-body glucose homeostasis and insulin sensitivity in humans. Diabetes. 2014;63:4089–4099. doi: 10.2337/db14-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Lans AA, et al. Cold acclimation recruits human brown fat and increases nonshivering thermogenesis. J. Clin. Invest. 2013;123:3395–3403. doi: 10.1172/JCI68993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoneshiro T, et al. Recruited brown adipose tissue as an antiobesity agent in humans. J. Clin. Invest. 2013;123:3404–3408. doi: 10.1172/JCI67803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee P, et al. Temperature-acclimated brown adipose tissue modulates insulin sensitivity in humans. Diabetes. 63:3686–3698. doi: 10.2337/db14-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 36.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 38.Stöver BC, Muller KF. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics. 2010;11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 40.Inoue M, Harada K, Matsuoka H, Sata T, Warashina A. Inhibition of TASK1-like channels by muscarinic receptor stimulation in rat adrenal medullary cells. J. Neurochem. 2008;106:1804–1814. doi: 10.1111/j.1471-4159.2008.05521.x. [DOI] [PubMed] [Google Scholar]
- 41.Cohen J. Statistical Power Analysis for the Behavioral Sciences. L. Erlbaum Associates; Hillsdale NJ: 1988. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
sup