A Screen in Mice Uncovers Repression of Lipoprotein Lipase by MicroRNA-29a as a Mechanism for Lipid Distribution Away From the Liver (original) (raw)
. Author manuscript; available in PMC: 2016 Jan 1.
Published in final edited form as: Hepatology. 2014 Nov 24;61(1):141–152. doi: 10.1002/hep.27379
Abstract
Identification of microRNAs (miRNAs) that regulate lipid metabolism is important to advance the understanding and treatment of some of the most common human diseases. In the liver, a few key miRNAs have been reported that regulate lipid metabolism, but since many genes contribute to hepatic lipid metabolism, we hypothesized that other such miRNAs exist. To identify genes repressed by miRNAs in mature hepatocytes in vivo, we injected adult mice carrying floxed Dicer1 alleles with an adenoassociated viral vector expressing Cre recombinase specifically in hepatocytes. By inactivating Dicer in adult quiescent hepatocytes we avoided the hepatocyte injury and regeneration observed in previous mouse models of global miRNA deficiency in hepatocytes. Next, we combined gene and miRNA expression profiling to identify candidate gene/miRNA interactions involved in hepatic lipid metabolism, and validated their function in vivo using antisense oligonucleotides. A candidate gene that emerged from our screen was lipoprotein lipase (Lpl), which encodes an enzyme that facilitates cellular uptake of lipids from the circulation. Unlike in energy-dependent cells like myocytes, Lpl is normally repressed in adult hepatocytes. We identified miR-29a as the miRNA responsible for repressing Lpl in hepatocytes, and found that decreasing hepatic miR-29a levels causes lipids to accumulate in mouse livers.
Conclusion
Our screen suggests several new miRNAs are regulators of hepatic lipid metabolism. We show that one of these, miR-29a, contributes to physiological lipid distribution away from the liver and protects hepatocytes from steatosis. Our results, together with miR-29a’s known anti-fibrotic effect, suggest miR-29a is a therapeutic target in fatty liver disease.
The important role that microRNAs (miRNAs) play in regulating metabolism is increasingly being recognized (1). In the liver, miRNAs appear to contribute particularly to the regulation of lipid metabolism (2). The most prominent example is miR-122, a miRNA expressed specifically in hepatocytes (3), the cells responsible for virtually all aspects of hepatic lipid metabolism. MiR-122 was shown to regulate the expression of genes involved in the synthesis of fatty acids, triglycerides (TGs), and cholesterol, and in lipid transport and storage (4–7). Another example is miR-33, which has inhibitory effects on cholesterol transport and degradation of fatty acids that make it a promising therapeutic target in cardiovascular diseases associated with dyslipidemia (8). A few other miRNAs have also been reported to be involved in hepatic lipid metabolism (2), but their contributions are incompletely understood.
Hepatic lipid metabolism entails many functions, which are encoded by many genes. Because ~1/3 of all mammalian genes are regulated by miRNAs (9), we hypothesized that other miRNAs exist that play important roles in hepatic lipid metabolism. To identify such miRNAs, we performed a hepatocyte-focused screen in mice designed to exclude biases and take miRNA expression levels into account.
Materials and Methods
Animal Studies
Dicer1fl/fl mice (10) and Dicer1fl/fl, Alb-Cre+/− mice (11) were on a mixed 129S4, C57BL/6 strain background, wildtype mice were pure C57BL/6 (Jackson Laboratory). Male 8 to 10-week-old mice were used unless specified. Liver and plasma samples were obtained after 4 hours of fasting. For high-fat diet (HFD) studies, mice were fed a Western diet (D12492, Open Source Diets) for 17 weeks before and during treatment with antisense oligonucleotide (ASO) inhibitors for 3 weeks. ASOs were dissolved in 200 µl PBS and injected intraperitoneally. 2’-fluoro/methoxyethyl-modified phosphorothioate backbone anti-miRs were generated by Regulus Therapeutics, and injected for 3 weeks at 10 µg/g body weight (twice in the first week, and once per week thereafter), or once at 20 µg/g body weight with analysis 6 days later. 2’-methoxyethyl-modified phosphorothioate backbone anti-Lpl was generated by Isis Pharmaceuticals, and injected once at 1 µg/g body weight with analysis 6 days later. All mouse experiments were approved by the Institutional Animal Care and Use Committee at the University of California San Francisco.
AAV Vector
AAV8-Ttr-Cre and AAV8-Ttr-control virus preparation, titering, and tail-vein injection were performed as described (12).
Gene Expression Profiling
Total RNA was isolated with Trizol (Qiagen). RNA concentration was measured with a Nanodrop ND-1000 and RNA quality was assessed with a Bioanalyzer 2100 (Agilent Technologies). 300 ng total RNA was amplified and hybridized to GeneChip Mouse Gene 1.0 ST microarrays (Affymetrix). Microarrays were stained and washed on a Fluidics Station 450 (Affymetrix). Probe intensities were measured using an argon laser confocal GeneArray Scanner (Hewlett-Packard). Microarray analysis was performed by the Gladstone Institutes Bioinformatics Core. Linear models were fitted for each gene using the limma package in R/Bioconductor. Moderated t statistics and the associated P values were calculated. P values were adjusted for multiple testing by controlling for false discovery rate (FDR) using the Benjamini-Hochberg method. Results were deposited in Gene Expression Omnibus (accession # GSE52526).
Quantitative PCR
Total RNA was isolated with Trizol or the miRNeasy Mini Kit (Qiagen). MRNAs were analyzed after cDNA synthesis using Superscript II (Invitrogen) or qScript cDNA SuperMix (Quanta BioSciences) in a Viia7 real time-PCR system using SYBR green (both Applied Biosystems). Gapdh was used for normalization unless stated otherwise. PCR amplification was performed at 50°C for 2 minutes and 95°C for 10 minutes, followed by 40 cycles at 95°C for 15 seconds and 60°C for 1 minute. qRT-PCR primers were designed using Primer Express software (Applied Biosystems) (Supporting Table 4). MiRNAs were analyzed using TaqMan miRNA assays (Applied Biosystems) or Exiqon miRNA qPCR SYBR Green assays. The small RNAs Sno202 and Sno429 or Snord65 were used for normalization. Relative changes in mRNA and miRNA expression were determined using the 2−ΔΔCt method.
Lipid and Lipoprotein Analysis
Peripheral blood was obtained by retro-orbital venipuncture. TGs, total cholesterol, and high-density lipoprotein (HDL) cholesterol were analyzed on an ADVIA 1800 automated clinical chemistry system (Siemens). Pooled plasma containing 1 mM EDTA was fractionated by fast protein liquid chromatography (FPLC) using a Superose 6 column (GE Healthcare) (13). FPLC cholesterol and TG levels were measured using Cholesterol E and L-Type TG M assays (Wako). Liver neutral lipids were analyzed by thin layer chromatography (TLC). For analysis of TG uptake, mice received a single injection of anti-miR-29a or anti-miR-control, followed by gavage of 200 µl olive oil containing 2 µCi of 14C-Triolein 6 days later. Gavaged mice were fasted and livers and epididymal white adipose tissues (eWATs) harvested 6 hours later. Tissues were weighed and digested in 500 µl of 1 N NaOH at 65°C for 1 hour, bleached with 50 µl 0.5 M EDTA and 100 µl 35% H202, mixed with liquid scintillation cocktail (Ultima Gold, Perkin Elmer), and analyzed using a Beckman LS 3801 liquid scintillation counter.
Statistical Analysis
Data were analyzed using 2-tailed Student t test. A P value of less than 0.05 was considered significant.
Results
Hepatocyte-Specific, Bias-Free Global MiRNA Deficiency in Adult Mice
To determine which aspects of hepatic lipid metabolism are regulated by miRNAs, we started by generating mice in which miRNA processing is disrupted in hepatocytes. For this, we used mice in which exon 24 of Dicer1 is floxed (Dicer1fl/fl mice) (10), leading to loss of enzymatic activity and global miRNA deficiency in cells that also express Cre recombinase. Because hepatocytes stop proliferating and become functionally mature in mice only after weaning (14), we analyzed the effects of global miRNA deficiency in adult mice. For this, we could not use previously reported Dicer1fl/fl mice carrying hepatocyte-specific Cre transgenes, i.e., Dicer1fl/fl mice in which Cre expression is controlled by albumin (Alb) promoter and α-fetoprotein enhancer sequences (15) or Dicer1fl/fl, Alb-Cre+/− mice (16), because they exhibit injury and death of hepatocytes by 3 or 4 weeks after birth, leading to secondary changes such as hepatocyte proliferation and liver regeneration that would have confounded our screen. Instead, we injected 8 to 10-week-old Dicer1fl/fl mice with our previously reported double-stranded adenoassociated virus serotype 8 (AAV8) vector expressing Cre from the transthyretin (Ttr) promoter (12). This vector loops out floxed sequences in all hepatocytes, but no other cells, within 48 hours after tail vein injection of 2 × 1011 viral genomes in mice. Furthermore, AAV8-Ttr-Cre does not cause liver injury or biases from genomic integration.
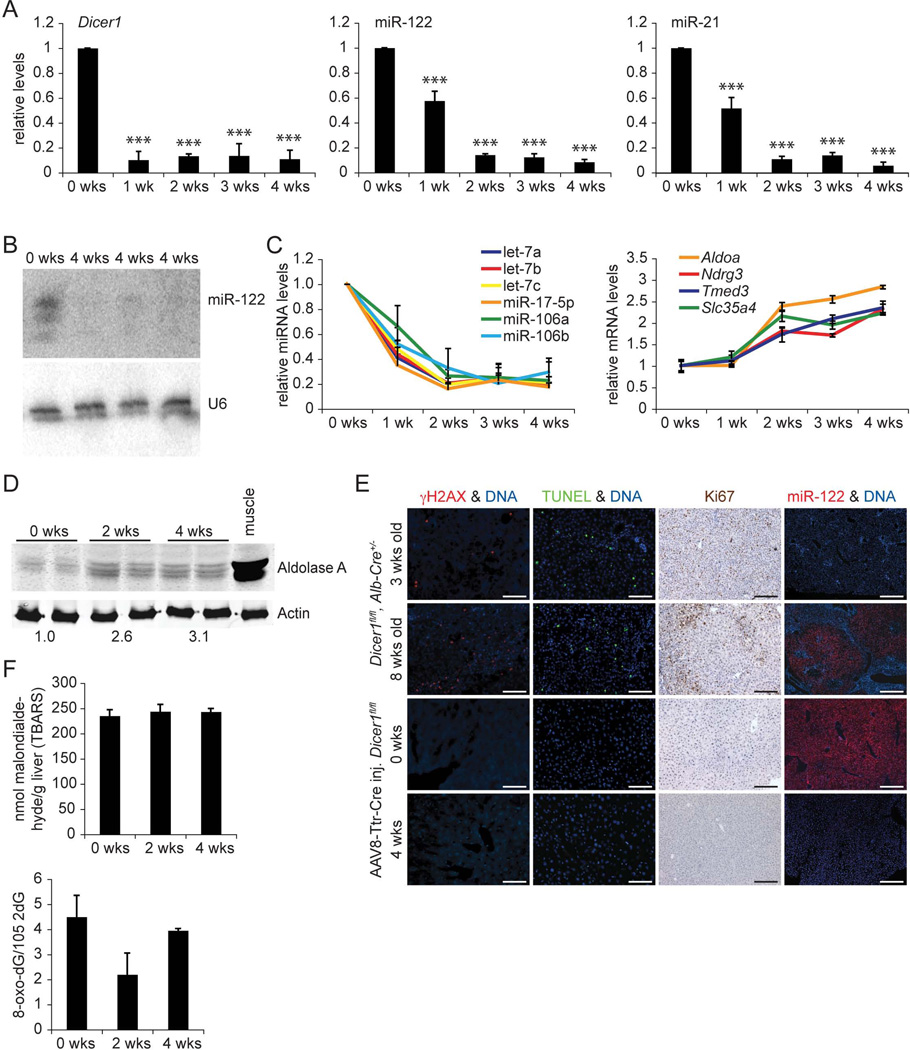
As expected, we observed rapid loss of Dicer1 mRNA in livers of Dicer1fl/fl mice after AAV8-Ttr-Cre injection (Fig. 1A). Surprisingly, miR-122 and miR-21, miRNAs predominantly expressed in hepatocytes in the liver (17), were still detectable 1 week after injection. MiR-122 and miR-21 were depleted 2 weeks after injection, suggesting high stability of these miRNAs in the quiescent hepatocytes of the adult liver. Because miR-122 is the most abundant miRNA in the liver (3), we measured its expression also by Northern blot to confirm disruption of miRNA processing in these mice (Fig. 1B). In addition, we confirmed that members of the let-7 family and miR-17-5p or miR-106 clusters, representing miRNAs expressed at high and low levels in the liver, respectively (3, 18), were also depleted (Fig. 1C). Delayed depletion of miR-122 in livers of AAV8-Ttr-Cre-injected Dicer1fl/fl mice was mirrored by slowly increasing levels of its known target genes Aldoa, Ndrg3, Tmed3, and Slc35a4 (4, 19) (Fig. 1C). Aldoa derepression translated into increased protein levels at 2 and 4 weeks after AAV8-Ttr-Cre injection, suggesting that the effects of individual miRNAs on their target genes can be discerned in this new mouse model of global miRNA deficiency in adult hepatocytes (Fig. 1D).
Fig. 1.
AAV8-Ttr-Cre-injected Dicer1fl/fl mice allow bias-free analysis of the effects of global miRNA deficiency in adult hepatocytes. (A) qRT-PCR shows loss of Dicer1 expression in livers of Dicer1fl/fl mice beginning 1 week (wk) after injection of AAV8-Ttr-Cre. qPCR shows that miR-122 and miR-21 are efficiently suppressed by 2 weeks after Dicer1 inactivation (n = 3 per time point). (B) Northern blot confirms absence of miR-122 in livers of 3 Dicer1fl/fl mice injected with AAV8-Ttr-Cre 4 weeks earlier. U6 levels show equal loading. (C) qPCR shows that miRNAs expressed in the liver at lower levels than miR-122 are also stably suppressed in Dicer1fl/fl mice injected with AAV8-Ttr-Cre (P values for all data points are at least < 0.05). qRT-PCR shows that miR-122 target genes are maximally and stably derepressed by 2 weeks after _Dicer1_ inactivation (_P_ values for all data points are at least < 0.005, except _P_ > 0.05 at 1 week after AAV8-Ttr-Cre injection, n = 3 per time point). (D) Immunoblotting shows increased levels of Aldolase A in Dicer1fl/fl mice 2 and 4 weeks after AAV8-Ttr-Cre injection. Results for 2 representative mice are shown for each time point. Gastrocnemius muscle from a wildtype mouse was used as a positive control. Fold increases relative to non-injected littermates (0 wks) are given. Actin was analyzed as a loading control. (E) Livers of 3-week-old Dicer1fl/fl, Alb-Cre+/− mice show hepatocyte DNA damage (γH2AX immunostaining) and death (TUNEL). Clonal expansion of proliferating hepatocytes (Ki67 immunostaining) capable of normal miRNA expression (miR-122 in situ hybridization) leads to high-level liver repopulation in 8-week-old Dicer1fl/fl, Alb-Cre+/− mice. Livers of adult Dicer1fl/fl mice injected with AAV8-Ttr-Cre 4 weeks earlier show no hepatocyte DNA damage or death, or repopulation with miR-122-expressing hepatocytes. For each analysis, representative stainings of at least 5 sections from 3 mice are shown. Size bars = 100 µm. (F) Thiobarbituric acid-reactive substances (TBARS) assay shows absence of lipid peroxidation in AAV8-Ttr-Cre-injected Dicer1fl/fl mice (n = 3 per time point). Levels of 8-oxo-dG are normal in livers of AAV8-Ttr-Cre-injected Dicer1fl/fl mice (n = 3 per time point). Error bars represent mean ± SD. ***P < 0.005 relative to 0 wks.
To obtain further evidence for the utility of AAV8-Ttr-Cre-injected Dicer1fl/fl mice for analyses of hepatic lipid metabolism, we excluded biases caused by hepatocyte injury. In contrast to the previously reported mouse models that used Cre transgenes to achieve global miRNA deficiency in hepatocytes (15, 16), Dicer1fl/fl mice injected with AAV8-Ttr-Cre showed no injury, DNA damage, death, or abnormal proliferation of hepatocytes, as assessed by analysis of plasma alanine aminotransferase (ALT) levels, γH2AX immunostaining, TdT-mediated dUTP nick end labeling (TUNEL), and Ki67 immunostaining, 4 weeks later (Supporting Table 1 and Fig. 1E). Furthermore, analysis of hepatic malondialdehyde and 8-oxodeoxyguanosine (8-oxo-dG) levels showed that lipid peroxidation and production of DNA adducts, common markers of hepatocyte injury in response to reactive oxygen species (ROS) (20), were not elevated in these mice (Fig. 1F). In accord with these results, liver repopulation with hepatocytes in which miRNA expression was maintained because of random Cre inactivation—as previously reported for Dicer1fl/fl, Alb-Cre+/− mice (16)—was also not observed in AAV8-Ttr-Cre-injected Dicer1fl/fl mice (Fig. 1E).
MiRNA-Regulated Aspects of Hepatic Lipid Metabolism
As a first assessment of changes in hepatic lipid metabolism due to global miRNA deficiency in hepatocytes, we analyzed the fasting plasma lipid profile of Dicer1fl/fl mice 4 weeks after AAV8-Ttr-Cre injection. Total and HDL cholesterol were decreased to levels resembling or exceeding those observed in mice in which miR-122 was inhibited with ASOs (4, 5, 19, 21), or genetically knocked out (6, 7) (Supporting Fig. 1). We also observed a moderate decrease in TG levels similar to those seen after miR-122 inhibition, but less than in miR-122 knockout mice.
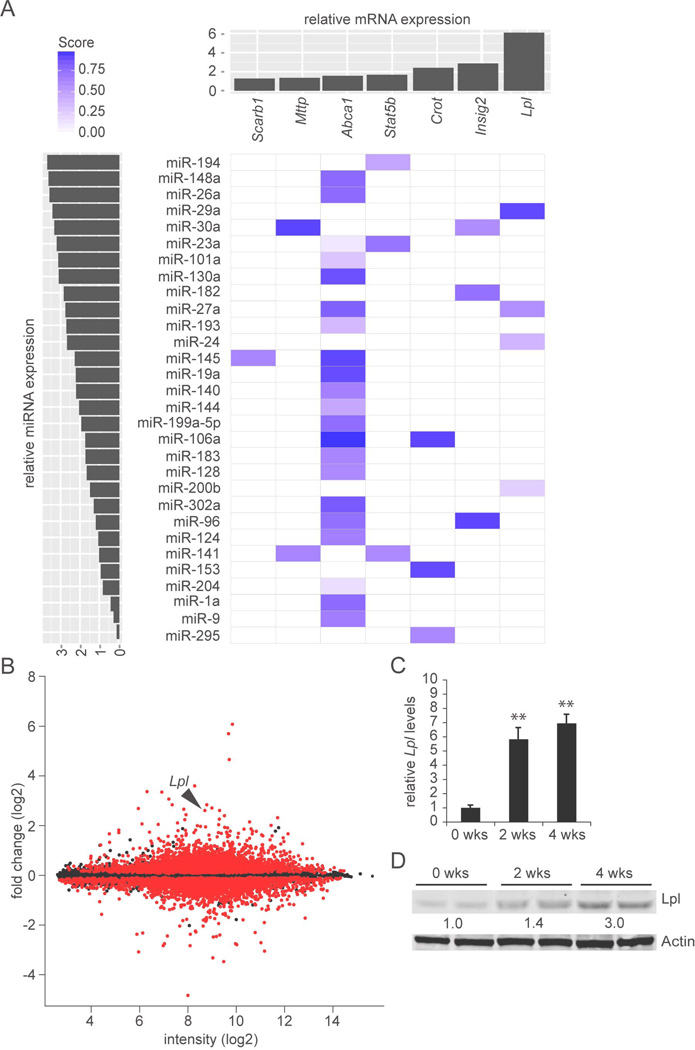
To identify miRNA-regulated genes expressed in hepatocytes responsible for the observed plasma lipid changes, we profiled global gene expression in livers of AAV8-Ttr-Cre-injected Dicer1fl/fl mice (Supporting File 1). Based on 25,931 reliably detected genes, Dicer1fl/fl mice 2 and 4 weeks after AAV8-Ttr-Cre injection clustered closely together—confirming that miRNA deficiency was stable—and separately from non-injected Dicer1fl/fl control mice (Supporting Fig. 1B and Supporting File 2). Because miRNAs typically repress their target genes (22), we focused on genes significantly upregulated in AAV8-Ttr-Cre-injected Dicer1fl/fl mice as compared to controls (> 1.25-fold upregulated and FDR P < 0.05) (Supporting File 3). Among 975 such genes, we identified 16 genes involved in lipid metabolism by Gene Ontology (GO) analysis (Supporting Table 2 and Supporting File 3). We validated these microarray results by qRT-PCR (Supporting Table 3). In addition, we investigated whether the 16 candidate genes were upregulated in an independent experiment in which control mice were injected with AAV8-Ttr-control, a sibling vector of AAV8-Ttr-Cre not expressing Cre (12) (Supporting Fig. 1C). This additional qRT-PCR analysis confirmed upregulation for 12 of the 16 candidate genes identified by microarray analysis. To ascertain both biological and clinical relevance, we focused on 7 genes that were significantly upregulated in both experiments and that contained evolutionarily conserved miRNA binding sites according to TargetScan 6.2 (Supporting File 4). To delineate the miRNAs with the highest probability of playing a significant role in the regulation of these genes, we ranked the miRNAs predicted to target them by expression level in wildtype mouse liver (23) (Fig. 2A).
Fig. 2.
MiRNAs with potential as regulators of lipid metabolism in hepatocytes. (A) Identification of miRNAs with high probability of playing a significant role in hepatic lipid metabolism. The x-axis (top) shows level of induction in livers of AAV8-Ttr-Cre-injected Dicer1fl/fl mice of lipid metabolism genes that have miRNA target sites. Results are based on global gene expression profiling of livers of Dicer1fl/fl mice 2 and 4 weeks after AAV8-Ttr-Cre injection (n = 2 per time point) as compared to non-injected Dicer1fl/fl littermate controls (n = 3). The y-axis (left) shows the miRNAs predicted to target these genes ranked by expression level in wildtype mouse liver (23). TargetScan 6.2 was used to predict probability of targeting (Score). MiRNA families consisting of several paralogs are represented by “a” paralogs. MiR-33, which is known to target Abca1 (8), was not analyzed in that study (23). (B) MA plot of global gene expression profiling of livers of Dicer1fl/fl mice after AAV8-Ttr-Cre injection. Change in gene expression is indicated on the y-axis. Intensity is shown on the x-axis. Dots representing probes with P < 0.05 comparing the 4 AAV8-Ttr-Cre-injected Dicer1fl/fl mice and 3 control mice are red, others are black. Lpl is indicated by an arrowhead. (C) qRT-PCR shows increased Lpl mRNA levels in livers of Dicer1fl/fl mice 2 and 4 weeks after AAV8-Ttr-Cre injection (n = 3 per time point). (D) Immunoblotting shows increased Lpl protein levels in livers of Dicer1fl/fl mice 2 and 4 weeks after AAV8-Ttr-Cre injection. Fold increases relative to non-injected littermates (0 wks) are given. Results for 2 representative mice are shown for each time point. Actin was analyzed as a loading control. Error bars represent mean ± SD. **P < 0.01 relative to 0 wks.
Most of the candidate genes were previously shown to play a role in hepatic lipid metabolism (24). A notable exception was Lpl, the gene that encodes lipoprotein lipase and that was most upregulated (Fig. 2B, Supporting Table 3, and Supporting Fig. 1C). Lpl catalyzes the rate-limiting step in the hydrolysis of circulating TGs, releasing fatty acids for uptake into energy-dependent tissues like heart and skeletal muscle, where they are used for β-oxidation, or adipose, where they are resynthesized into TGs and stored as energy reserves (25). In the liver, Lpl expression is gradually extinguished after birth, leading to undetectable levels in the adult (26). This tissue-specific expression pattern contributes to the physiological distribution of energy away from the liver (27, 28). Therefore, after confirming by qRT-PCR and immunoblotting that Lpl was derepressed in livers of AAV8-Ttr-Cre-injected Dicer1fl/fl mice (Fig. 2C and D), we focused on identifying the miRNAs that repress Lpl in hepatocytes.
MiR-29a Contributes to Physiological Repression of Lpl in the Adult Liver
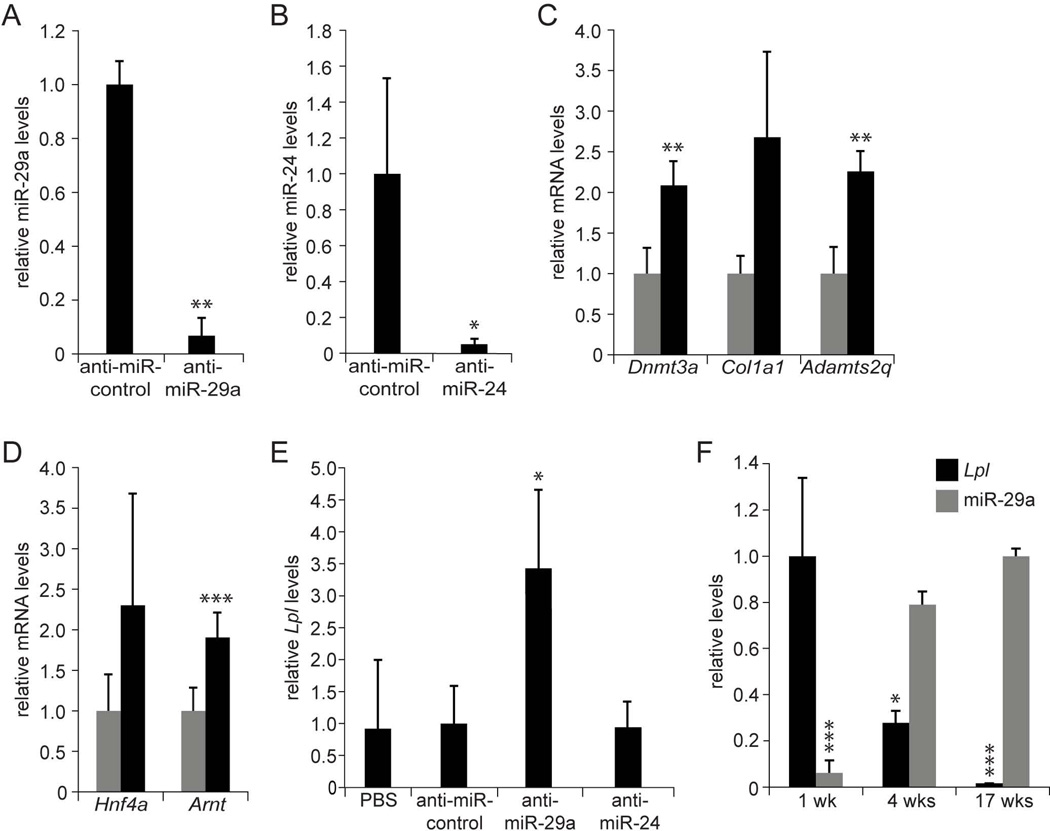
First, we determined whether the miRNAs predicted to target Lpl—miR-29a, miR-27a, and miR-24 (Fig. 2A)—were effective in doing so in vitro. Indeed, we found that transfection of mimics or inhibitors of all 3 of these miRNAs into Hepa1–6 mouse hepatoma cells efficiently decreased or increased Lpl mRNA levels, respectively (Supporting Fig. 2A and B). Next, we tested whether these miRNAs were also acting as inhibitors of Lpl in vivo. For this, we generated ASO inhibitors of these miRNAs (anti-miRs) with the same chemical properties as anti-miRs previously shown to efficiently inhibit miR-122 in the liver (29), and injected them intraperitoneally into 8 to 10-week-old C57BL/6 mice for 3 weeks. We found that anti-miR-29a and anti-miR-24 efficiently inhibited their target miRNAs in the liver (Fig. 3A and B), leading to derepression of established target genes of these miRNAs (Fig. 3C and D) (30, 31). In contrast, anti-miR-27a increased miR-27a levels and failed to derepress miR-27a target genes (data not shown). None of the 3 anti-miRs caused hepatotoxicity (Supporting Fig. 3A). Because of its paradoxical effect, we decided not to pursue miR-27a further, but focus on miR-29a and miR-24. Surprisingly, when we analyzed Lpl mRNA levels in livers of mice injected with anti-miR-29a or anti-miR-24, we found that only inhibition of miR-29a caused Lpl derepression (Fig. 3E). We ascertained by immunoblotting that increased Lpl mRNA levels translated into increased Lpl protein levels (Supporting Fig. 3C). This finding suggests a higher level of complexity of miRNA-mediated gene regulation in vivo than in vitro.
Fig. 3.
MiR-29a inhibits Lpl expression in the liver. (A) qPCR shows miR-29a inhibition in livers of mice injected with anti-miR-29a (n = 3) as compared to mice injected with anti-miR-control (n = 3) for 3 weeks. (B) qPCR shows miR-24 inhibition in livers of mice injected with anti-miR-24 (n = 3) as compared to mice injected with anti-miR-control (n = 3) for 3 weeks. (C) qRT-PCR shows derepressed miR-29a target gene levels in livers of mice injected with anti-miR-29a (black, n = 3) as compared to mice injected with anti-miR-control (grey, n = 3) for 3 weeks. (D) qRT-PCR shows derepressed miR-24 target gene levels in livers of mice injected with anti-miR-24 (black, n = 3) as compared to mice injected with anti-miR-control (grey, n = 3) for 3 weeks. (E) qRT-PCR shows increased Lpl mRNA levels in livers of mice injected with anti-miR-29a (n = 3) as compared to mice injected with anti-miR-24 (n = 3), anti-miR-control (n = 3), or PBS (n = 3) for 3 weeks. (F) qRT-PCR and qPCR show that Lpl mRNA levels decrease and miR-29a levels increase during postnatal completion of mouse liver development (1 wk, n = 6; 4 wks, n = 4; 17 wks, n = 3). Error bars represent mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.005 relative to anti-miR-control (A-E), or 1-week-old (Lpl) or 17-week-old (miR-29a) mice (F).
In addition to miR-29a, the miR-29 family consists of miR-29b and miR-29c. MiR-29a and miR-29c are located on distinct chromosomes, and exhibit differences in transcriptional regulation, but their mature sequences are identical except for 1 nucleotide (32). In accord, we found that both miR-29a and miR-29c were effectively inhibited in livers of mice injected with anti-miR-29a, whereas miR-29b, which differs from miR-29a in 5 nucleotides, appeared less affected (Supporting Fig. 3B). Because miR-29a is expressed at higher levels than miR-29c in mouse and human liver (18), we focused on miR-29a as the miR-29 family member predominantly responsible for repression of Lpl in the liver.
As evidence for a role of miR-29a in this physiological process, we found reciprocal changes in hepatic Lpl mRNA and miR-29a levels as mice progress from the postnatal phase to adulthood (Fig. 3F). We established that Lpl is a direct target of miR-29a by showing that miR-29a’s inhibitory effect on Lpl is dependent on an intact miR-29a binding site in the LPL 3’ UTR (Supporting Fig. 2C).
MiR-29a Inhibition Causes Lpl-Dependent Hepatic Lipid Uptake
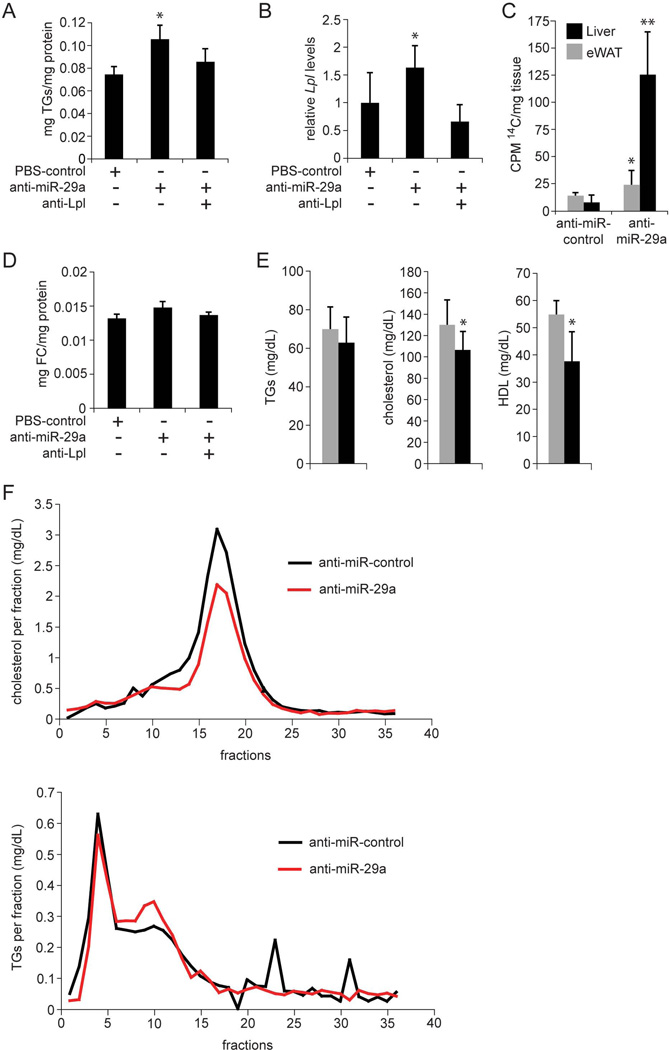
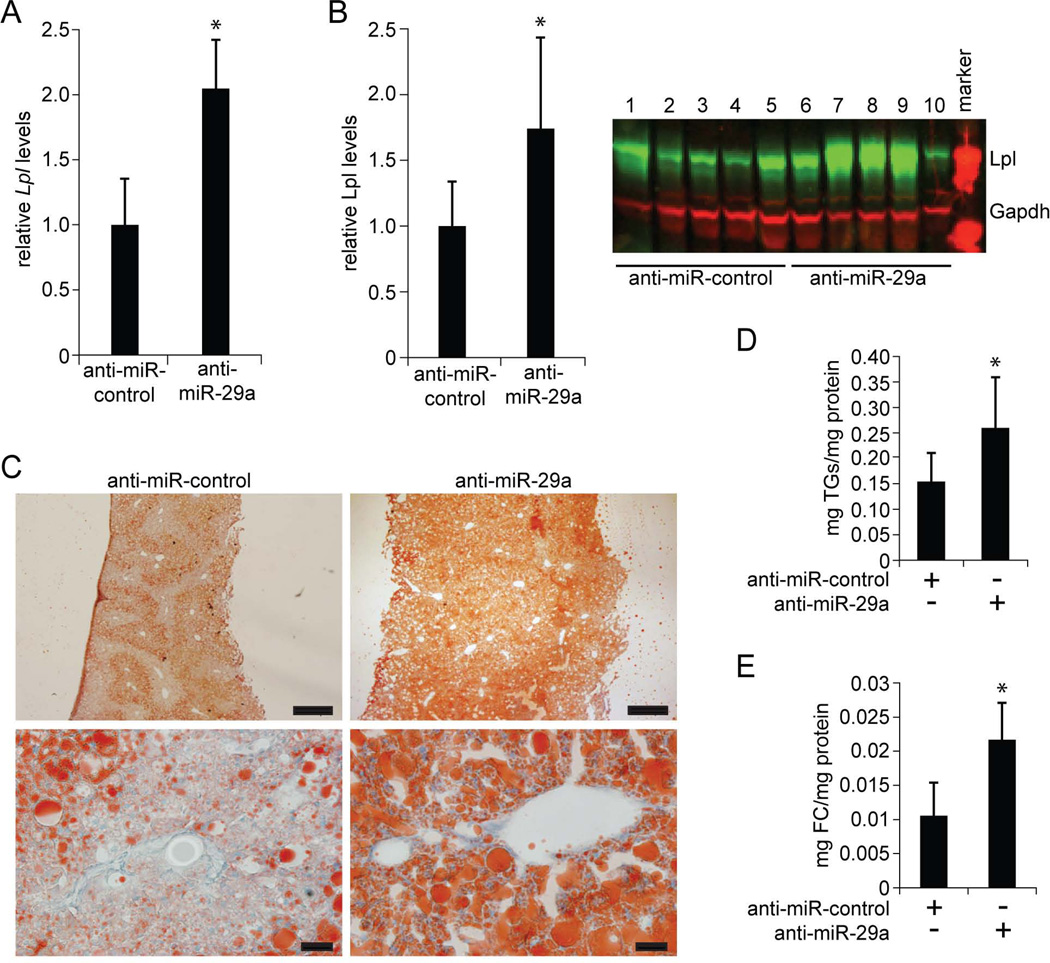
Next, we probed the functional consequences of increased Lpl expression in livers of anti-miR-29a-injected mice. Because increased Lpl expression would be expected to lead to increased uptake of fatty acids, which would be stored as TGs in the liver, we measured hepatic TG levels using TLC (33). Indeed, we found that miR-29a inhibition caused TG accumulation in the liver (Fig. 4A). Hepatic TG accumulation was prevented in mice additionally injected with ASOs that inhibit Lpl itself (anti-Lpl), showing that miR-29a inhibition increased hepatic TG content by derepressing Lpl (Fig. 4A and B and Supporting Fig. 3D). Hepatic TG accumulation can result not only from increased fatty acid uptake, but also from increased de novo synthesis of fatty acids in hepatocytes. To determine the extent to which the increased hepatic TG content in anti-miR-29a-injected mice was due to increased fatty acid uptake, we measured hepatic uptake of radiolabeled TG (14C-triolein) after intragastric gavage (34). Because dietary fatty acids absorbed by enterocytes are secreted as TGs in chylomicrons, hepatic uptake of 14C-label would require hepatic Lpl expression in this experiment. We found significantly more 14C-label in livers of anti-miR-29a-injected mice than in control mice 6 hours after gavage (Fig. 4C), which confirmed that releasing Lpl from repression by miR-29a increased hepatic uptake of TG-containing chylomicron remnants. In addition to TGs, cholesterol also increased slightly in livers of anti-miR-29a-injected mice (Fig. 4D). This finding, although not statistically significant, is consistent with previous reports showing that Lpl can contribute to uptake of HDL (35) and low-density lipoprotein (LDL) (36) cholesterol.
Fig. 4.
MiR-29a regulates liver and plasma lipids by repressing Lpl. (A) TLC shows increased TG levels in mouse livers 6 days after a single injection of anti-miR-29a (n = 3) as compared to control mice injected with PBS (n = 3). Additional injection of anti-Lpl (n = 3) prevents TG accumulation. (B) qRT-PCR shows that the increase in Lpl mRNA levels in mouse livers 6 days after injection of anti-miR-29a (n = 3) is prevented by additional injection of anti-Lpl (n = 3). Mice injected with PBS were used as controls (n = 3). (C) Counts per minute (CPM) measurement 6 hours after intragastric gavage of 14C-Triolein shows increased levels of 14C-label in livers of mice injected with anti-miR-29a (n = 4) as compared to mice injected with anti-miR-control (n = 3) 6 days earlier. 14C-Triolein uptake into eWAT is not substantially increased in anti-miR-29a-injected mice. (D) Free cholesterol (FC) levels in livers of mice injected with anti-miR-29a (n = 3) as compared to control mice injected with PBS (n = 3) 6 days earlier. Additional injection of anti-Lpl (n = 3) slightly reduces FC levels in the liver. (E) Plasma lipid profile of mice 6 days after injection of anti-miR-29a (black, n = 5) or anti-miR-control (grey, n = 5). (F) Fractionation of plasma by FPLC shows decreased LDL (fractions 10–14) and HDL (fractions 15–20) cholesterol and slightly decreased VLDL (fractions 3–6) and increased IDL and LDL (fractions 7–12) TGs in plasma of mice injected with anti-miR-29a (n = 5, pooled) as compared to mice injected with anti-miR-control (n = 5, pooled) for 3 weeks. Error bars represent mean ± SD. *P < 0.05 and **P < 0.01 relative to anti-miR-control or PBS-control.
We also analyzed the fasting plasma lipid profile of anti-miR-29a-injected mice and found changes that reflected the trends seen in AAV8-Ttr-Cre-injected Dicer1fl/fl mice, albeit to a lesser extent (Fig. 4E and Supporting Fig. 1A). These results suggest that, in addition to miR-122, miR-29a expression in hepatocytes contributes significantly to the regulation of plasma lipoprotein cholesterol levels. Indeed, plasma fractionation showed decreased levels of both HDL and LDL cholesterol in anti-miR-29a-injected mice (Fig. 4F). As in AAV8-Ttr-Cre-injected Dicer1fl/fl mice, unfractionated plasma TG levels were not significantly decreased in anti-miR-29a-injected mice (Fig. 4E). However, plasma fractionation revealed a slight decrease in very low-density lipoprotein (VLDL) TG, and an increase in intermediate-density lipoprotein (IDL) and LDL TG (Fig. 4F), which is consistent with previous reports showing that increased hepatic Lpl expression increases VLDL TG turnover (37).
MiR-29a Deficiency Promotes Hepatic Steatosis
Finally, because increased LPL expression has been observed in patients with fatty liver disease (38–40), we investigated the consequences of derepressing hepatic Lpl expression through miR-29a inhibition in a mouse model of fatty liver disease. For this, we fed 8 to 10-week-old C57BL/6 mice a HFD for 20 weeks and injected them with anti-miR-29a for the last 3 weeks. As expected, we found that Lpl mRNA and protein levels were increased in these mice (Fig. 5A and B). Furthermore, in accord with our findings in chow-fed mice (Fig. 4A–D), anti-miR-29a-injected mice showed increased hepatic lipid accumulation as illustrated by loss of periportal sparing in Oil-red-O (ORO) staining (Fig. 5C and Supporting Fig. 3E). TLC confirmed this observation and showed that the accumulating lipid contained both TGs and cholesterol (Fig. 5D and E).
Fig. 5.
MiR-29a inhibits lipid uptake into livers of mice fed HFD. (A) qRT-PCR shows increased Lpl mRNA levels in livers of HFD-fed mice injected with anti-miR-29a as compared to HFD-fed mice injected with anti-miR-control for 3 weeks (n = ≥ 4 per group). (B) Quantification of immunoblotting shows increased Lpl protein levels in livers of HFD-fed mice injected with anti-miR-29a as compared to HFD-fed mice injected with anti-miR-control for 3 weeks. Gapdh was analyzed as a loading control (n = 5 per group). (C) ORO staining shows pericentral lipid deposition in livers of HFD-fed mice injected with anti-miR-control for 3 weeks. In livers of HFD-fed mice injected with anti-miR-29a staining intensity is increased and lipid deposition is detected in both pericentral and periportal areas. For each group, representative stainings of 5 mice are shown. Size bars = 500 µm (top images) and 20 µm (bottom images). (D and E) TLC shows increased levels of TGs (D) and FC (E) in livers of HFD-fed mice injected with anti-miR-29a as compared to mice injected with anti-miR-control for 3 weeks (n = 5 per group). Error bars represent mean ± SD. *P < 0.05 relative to anti-miR-control.
Discussion
To our knowledge, we have generated the first mouse model with bias-free global miRNA deficiency in mature hepatocytes. In contrast to previously reported mouse models using hepatocyte-specific Cre transgenes to inactivate Dicer (15, 16), our AAV8-Ttr-Cre-injected Dicer1fl/fl mice showed no signs of hepatocyte injury or compensatory liver regeneration at the time of analysis. This discrepancy is most likely due to an increased sensitivity of proliferating hepatocytes to lack of miRNAs. The expression of the Alb-Cre transgene is typically initiated just before birth (11), and affords maximum recombination at 3 weeks of age in Dicer1fl/fl, Alb-Cre+/− mice (16). Dicer inactivation occurs most likely even earlier if α-fetoprotein enhancers are included as drivers of Cre expression (15). Because final liver mass is established in mice by extensive hepatocyte replication in the first 3 weeks after birth (14), hepatocytes lose Dicer and miRNA expression while proliferating in the previously reported mouse models. Proliferation has been shown to cause DNA damage and cell death in Dicer-deficient mouse embryonic fibroblasts (41), which explains findings of hepatocyte injury and compensatory liver regeneration in the previous mouse models of hepatocyte-specific global miRNA deficiency. Using AAV8-Ttr-Cre allowed us to avoid the vulnerable state of hepatocyte proliferation during postnatal liver development and disrupt miRNA processing in the quiescent hepatocytes of the adult liver.
In the absence of confounding effects from hepatocyte injury and liver regeneration, our mouse model allows screening for metabolic functions of hepatocytes that are regulated by miRNAs. Illustrating the potential of this new mouse model, the hits from our screen for miRNAs and their target genes involved in hepatic lipid metabolism include all 3 miRNAs—miR-96, miR-182, and miR-183—recently shown to play a role in Srebp2-regulated hepatic lipid synthesis (42). Srebp2 itself—and also the miR-122 target Agpat1 (7)—were not among the top hits emerging from our screen (Fig. 2A), because although they were upregulated > 1.25-fold in our microarray analysis (raw P value < 0.05) they were excluded because of high FDR. In contrast, inhibition of Mttp by miR-30, a miRNA/target gene interaction that was recently shown to limit hepatic lipoprotein assembly (43), was a top hit from our screen.
Following up on the top hits from our screen led us to identify miR-29a as a key contributor to the physiological repression of Lpl in mature hepatocytes. Factors controlling the tissue-specific expression pattern of Lpl have been extensively studied (27, 28). Our findings not only reveal that miR-29a is important for the repression of Lpl in normal adult liver, but also suggest a role for loss of hepatic miR-29a expression in the pathogenesis of fatty liver. Anti-miR-29a-injected mice developed hepatic lipid accumulation, which was aggravated by HFD feeding, a common model of fatty liver disease. Lipid accumulation in anti-miR-29a-injected mice was not limited to the pericentral area of the liver lobule as in control mice, but also affected the periportal area, which is reminiscent of observations made in adult patients that fatty liver disease begins pericentrally and that pan-lobular steatosis is indicative of advanced disease (44). As additional support for the importance of miR-29a-mediated silencing of Lpl for prevention of hepatocyte steatosis, previous studies have shown decreased miR-29a levels in livers of mouse and rat models of fatty liver disease (45–47) and increased LPL levels in livers of affected humans (38–40).
Because our ASO inhibitor failed to decrease hepatic levels of miR-27a, its contribution to regulating Lpl-mediated lipid uptake into hepatocytes remains to be determined. Although it is expressed at lower levels in the liver than miR-29a (23), miR-27a likely plays a relevant role because it is known to inhibit Lpl in vitro (48) and—by repressing secretion of the Lpl inhibitor Angptl3 from hepatocytes (49)—may additionally limit lipid uptake into the liver by promoting uptake into muscle and adipose.
Probing the transcriptional regulation of miR-29a offers a clue to the specific role miR-29a deficiency plays in fatty liver disease. Tgfβ and Nf-κb, factors that play an important role in liver inflammation, were shown to suppress miR-29a (31). Thus, suppression of miR-29a conceivably perpetuates and aggravates fatty liver disease by causing Lpl-mediated lipid uptake into hepatocytes that are already injured or that reside in liver tissue infiltrated by immune cells. The inflammatory cytokine Tnfα—known to induce Lpl expression (27, 28) and suppress miR-29b (31)—may also be involved in this pathogenic process.
In conclusion, our in vivo screen identified miR-29 as a miRNA important for the physiological distribution of lipids away from the liver and protection of hepatocytes from steatosis. MiR-29a also prevents fibrosis (31)—a frequent complication of fatty liver disease that leads to liver failure and cancer. Restoring miR-29 expression may be an effective therapy for fatty liver disease because it would target both the cause and outcome-determining complication of this disease.
Supplementary Material
Supp Material
Acknowledgment
The authors thank Pamela Derish in the Department of Surgery at UCSF for manuscript editing.
Financial Support
H. Willenbring was supported by CIRM grant RN2-00950 and NIH grant P30 DK26743. A. N. Mattis was supported by CIRM grant TG2-01153 and NIH grant K08 DK098270. R. L. Raffai was supported by NIH grant R01 HL089871 and a Merit Review grant (5I01BX000532) from the Department of Veteran Affairs. J. J. Maher was supported by NIH grants R01 DK068450 and P30 DK026743.
Abbreviations
8-oxo-dG
8-oxodeoxyguanosine
AAV
adenoassociated virus
Aldoa
aldolase A
ALT
alanine aminotransferase
Angptl3
angiopoietin-like 3
anti-Lpl
lipoprotein lipase ASO
anti-miR
microRNA ASO
ASO
antisense oligonucleotide
EDTA
ethylenediaminetetraacetic acid
FPLC
fast protein liquid chromatography
gH2AX
phosphorylated histone H2AX
GO
gene ontology
H202
hydrogen peroxide
HDL
high-density lipoprotein
HFD
high-fat diet
IDL
intermediate-density lipoprotein
Ki67
antigen Ki-67
LDL
low-density lipoprotein
let-7
lethal-7 microRNA
Lpl/LPL
lipoprotein lipase
miR-XYZ
microRNA-XYZ
miRNA
microRNA
mRNA
messenger RNA
Mttp
microsomal triglyceride transfer protein
NaOH
sodium hydroxide
Ndrg3
n-Myc downstream-regulated gene 3
Nf-κb
nuclear factor kappa b
ORO
Oil-red-O
PBS
phosphate-buffered saline
PCR
polymerase chain reaction
qPCR
quantitative PCR
qRT-PCR
quantitative reverse transcription PCR
ROS
reactive oxygen species
SD
standard deviation
Slc35a4
solute carrier family 35 member A4
Srebp2
sterol regulatory element-binding protein 2
TG
triglycerides
Tgfβ
transforming growth factor beta
TLC
thin layer chromatography
Tmed3
transmembrane emp24 domain containing 3
Tnfα
tumor necrosis factor alpha
Ttr
transthyretin
TUNEL
TdT-mediated dUTP nick end labeling
UTR
untranslated region
VLDL
very low-density lipoprotein
eWAT
epididymal white adipose tissue
Footnotes
Conflicts of Interest
Authors have nothing to disclose.
References
- 1.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sacco J, Adeli K. MicroRNAs: emerging roles in lipid and lipoprotein metabolism. Curr Opin Lipidol. 2012;23:220–225. doi: 10.1097/MOL.0b013e3283534c9f. [DOI] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 4.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 5.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 10.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 12.Malato Y, Naqvi S, Schurmann N, Ng R, Wang B, Zape J, Kay MA, et al. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J Clin Invest. 2011;121:4850–4860. doi: 10.1172/JCI59261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raffai RL, Dong LM, Farese RV, Jr, Weisgraber KH. Introduction of human apolipoprotein E4 "domain interaction" into mouse apolipoprotein E. Proc Natl Acad Sci U S A. 2001;98:11587–11591. doi: 10.1073/pnas.201279298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Apte U, Zeng G, Thompson MD, Muller P, Micsenyi A, Cieply B, Kaestner KH, et al. beta-Catenin is critical for early postnatal liver growth. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1578–G1585. doi: 10.1152/ajpgi.00359.2006. [DOI] [PubMed] [Google Scholar]
- 15.Hand NJ, Master ZR, Le Lay J, Friedman JR. Hepatic function is preserved in the absence of mature microRNAs. Hepatology. 2009;49:618–626. doi: 10.1002/hep.22656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekine S, Ogawa R, Ito R, Hiraoka N, McManus MT, Kanai Y, Hebrok M. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology. 2009;136:2304–2315. e2301–e2304. doi: 10.1053/j.gastro.2009.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song G, Sharma AD, Roll GR, Ng R, Lee AY, Blelloch RH, Frandsen NM, et al. MicroRNAs control hepatocyte proliferation during liver regeneration. Hepatology. 2010;51:1735–1743. doi: 10.1002/hep.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 20.Finkel T. Oxidant signals and oxidative stress. Curr Opin Cell Biol. 2003;15:247–254. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 21.Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su WL, Kleinhanz RR, Schadt EE. Characterizing the role of miRNAs within gene regulatory networks using integrative genomics techniques. Mol Syst Biol. 2011;7:490. doi: 10.1038/msb.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jump DB. Fatty acid regulation of hepatic lipid metabolism. Curr Opin Clin Nutr Metab Care. 2011;14:115–120. doi: 10.1097/MCO.0b013e328342991c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies BS, Beigneux AP, Fong LG, Young SG. New wrinkles in lipoprotein lipase biology. Curr Opin Lipidol. 2012;23:35–42. doi: 10.1097/MOL.0b013e32834d0b33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peinado-Onsurbe J, Staels B, Deeb S, Ramirez I, Llobera M, Auwerx J. Neonatal extinction of liver lipoprotein lipase expression. Biochim Biophys Acta. 1992;1131:281–286. doi: 10.1016/0167-4781(92)90026-v. [DOI] [PubMed] [Google Scholar]
- 27.Preiss-Landl K, Zimmermann R, Hammerle G, Zechner R. Lipoprotein lipase: the regulation of tissue specific expression and its role in lipid and energy metabolism. Curr Opin Lipidol. 2002;13:471–481. doi: 10.1097/00041433-200210000-00002. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Eckel RH. Lipoprotein lipase: from gene to obesity. Am J Physiol Endocrinol Metab. 2009;297:E271–E288. doi: 10.1152/ajpendo.90920.2008. [DOI] [PubMed] [Google Scholar]
- 29.Davis S, Propp S, Freier SM, Jones LE, Serra MJ, Kinberger G, Bhat B, et al. Potent inhibition of microRNA in vivo without degradation. Nucleic Acids Res. 2009;37:70–77. doi: 10.1093/nar/gkn904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oda Y, Nakajima M, Mohri T, Takamiya M, Aoki Y, Fukami T, Yokoi T. Aryl hydrocarbon receptor nuclear translocator in human liver is regulated by miR-24. Toxicol Appl Pharmacol. 2012;260:222–231. doi: 10.1016/j.taap.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 31.Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 32.Kriegel AJ, Liu Y, Fang Y, Ding X, Liang M. The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol Genomics. 2012;44:237–244. doi: 10.1152/physiolgenomics.00141.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koliwad SK, Streeper RS, Monetti M, Cornelissen I, Chan L, Terayama K, Naylor S, et al. DGAT1-dependent triacylglycerol storage by macrophages protects mice from diet-induced insulin resistance and inflammation. J Clin Invest. 2010;120:756–767. doi: 10.1172/JCI36066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yen CL, Cheong ML, Grueter C, Zhou P, Moriwaki J, Wong JS, Hubbard B, et al. Deficiency of the intestinal enzyme acyl CoA:monoacylglycerol acyltransferase-2 protects mice from metabolic disorders induced by high-fat feeding. Nat Med. 2009;15:442–446. doi: 10.1038/nm.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rinninger F, Kaiser T, Mann WA, Meyer N, Greten H, Beisiegel U. Lipoprotein lipase mediates an increase in the selective uptake of high density lipoprotein-associated cholesteryl esters by hepatic cells in culture. J Lipid Res. 1998;39:1335–1348. [PubMed] [Google Scholar]
- 36.Loeffler B, Heeren J, Blaeser M, Radner H, Kayser D, Aydin B, Merkel M. Lipoprotein lipase-facilitated uptake of LDL is mediated by the LDL receptor. J Lipid Res. 2007;48:288–298. doi: 10.1194/jlr.M600292-JLR200. [DOI] [PubMed] [Google Scholar]
- 37.Merkel M, Weinstock PH, Chajek-Shaul T, Radner H, Yin B, Breslow JL, Goldberg IJ. Lipoprotein lipase expression exclusively in liver. A mouse model for metabolism in the neonatal period and during cachexia. J Clin Invest. 1998;102:893–901. doi: 10.1172/JCI2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sookoian S, Gianotti TF, Rosselli MS, Burgueno AL, Castano GO, Pirola CJ. Liver transcriptional profile of atherosclerosis-related genes in human nonalcoholic fatty liver disease. Atherosclerosis. 2011;218:378–385. doi: 10.1016/j.atherosclerosis.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 39.Westerbacka J, Kolak M, Kiviluoto T, Arkkila P, Siren J, Hamsten A, Fisher RM, et al. Genes involved in fatty acid partitioning and binding, lipolysis, monocyte/macrophage recruitment, and inflammation are overexpressed in the human fatty liver of insulin-resistant subjects. Diabetes. 2007;56:2759–2765. doi: 10.2337/db07-0156. [DOI] [PubMed] [Google Scholar]
- 40.Pardina E, Baena-Fustegueras JA, Llamas R, Catalan R, Galard R, Lecube A, Fort JM, et al. Lipoprotein lipase expression in livers of morbidly obese patients could be responsible for liver steatosis. Obes Surg. 2009;19:608–616. doi: 10.1007/s11695-009-9827-5. [DOI] [PubMed] [Google Scholar]
- 41.Mudhasani R, Zhu Z, Hutvagner G, Eischen CM, Lyle S, Hall LL, Lawrence JB, et al. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J Cell Biol. 2008;181:1055–1063. doi: 10.1083/jcb.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeon TI, Esquejo RM, Roqueta-Rivera M, Phelan PE, Moon YA, Govindarajan SS, Esau CC, et al. An SREBP-responsive microRNA operon contributes to a regulatory loop for intracellular lipid homeostasis. Cell Metab. 2013;18:51–61. doi: 10.1016/j.cmet.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soh J, Iqbal J, Queiroz J, Fernandez-Hernando C, Hussain MM. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat Med. 2013;19:892–900. doi: 10.1038/nm.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chalasani N, Wilson L, Kleiner DE, Cummings OW, Brunt EM, Unalp A, Network NCR. Relationship of steatosis grade and zonal location to histological features of steatohepatitis in adult patients with non-alcoholic fatty liver disease. J Hepatol. 2008;48:829–834. doi: 10.1016/j.jhep.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin X, Ye YF, Chen SH, Yu CH, Liu J, Li YM. MicroRNA expression pattern in different stages of nonalcoholic fatty liver disease. Dig Liver Dis. 2009;41:289–297. doi: 10.1016/j.dld.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 46.Li S, Chen X, Zhang H, Liang X, Xiang Y, Yu C, Zen K, et al. Differential expression of microRNAs in mouse liver under aberrant energy metabolic status. J Lipid Res. 2009;50:1756–1765. doi: 10.1194/jlr.M800509-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pogribny IP, Starlard-Davenport A, Tryndyak VP, Han T, Ross SA, Rusyn I, Beland FA. Difference in expression of hepatic microRNAs miR-29c, miR-34a, miR-155, and miR-200b is associated with strain-specific susceptibility to dietary nonalcoholic steatohepatitis in mice. Lab Invest. 2010;90:1437–1446. doi: 10.1038/labinvest.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang M, Wu JF, Chen WJ, Tang SL, Mo ZC, Tang YY, Li Y, et al. MicroRNA-27a/b regulates cellular cholesterol efflux, influx and esterification/hydrolysis in THP-1 macrophages. Atherosclerosis. 2014;234:54–64. doi: 10.1016/j.atherosclerosis.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 49.Vickers KC, Shoucri BM, Levin MG, Wu H, Pearson DS, Osei-Hwedieh D, Collins FS, et al. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2013;57:533–542. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp Material