Impermanence of bacterial clones (original) (raw)
Abstract
Bacteria reproduce asexually and pass on a single genome copied from the parent, a reproductive mode that assures the clonal descent of progeny; however, a truly clonal bacterial species is extremely rare. The signal of clonality can be interrupted by gene uptake and exchange, initiating homologous recombination that results in the unique sequence of one clone being incorporated into another. Because recombination occurs sporadically and on local scales, these events are often difficult to recognize, even when considering large samples of completely sequenced genomes. Moreover, several processes can produce the appearance of clonality in populations that undergo frequent recombination. The rates and consequences of recombination have been studied in Escherichia coli for over 40 y, and, during this time, there have been several shifting views of its clonal status, population structure, and rates of gene exchange. We reexamine the studies and retrace the evolution of the methods that have assessed the extent of DNA flux, largely focusing on its impact on the E. coli genome.
Keywords: clonality, homologous exchange, recombination, genome evolution, E. coli
Reproduction by binary fission virtually guarantees the clonality of a bacterial lineage. Apart from mutations and other rare events that might modify chromosome integrity during replication, the primary sequence of DNA in all daughter and descendent cells remains identical, generation after generation after generation. Unlike animals, in which parthenogenetic forms are ecologically constrained and relatively short-lived over evolutionary timescales (1–3), asexually reproducing bacteria have persisted since the origin of cellular life and represent the most diverse and widespread organisms on the planet. Naturally, the vast diversity present in bacteria could have arisen solely by asexual means—there has certainly been sufficient time and large enough population sizes to allow for enormous numbers of mutations (and combinations of mutations) to be experienced. Moreover, it seems as though some of the most extraordinary innovations in the history of life have occurred without intervention of the sexual process (4).
Bacteria as Clonal Organisms
Despite their obligatory asexual mode of reproduction, the clonality of bacterial lineages can be disrupted by sex, or at least by what we refer to as sex. In bacteria, sex is the inheritance of genetic material from any source aside from their one parent cell and can occur by any of several processes. Foreign DNA can be introduced by cell-to-cell contact, transmitted to the cell by an infectious agent, or acquired directly from the environment; and, therefore, genes can be obtained from organisms representing any domain of life, and even from entities (i.e., viruses and phages) that are not classified to any domain of life. Moreover, events of sex in bacteria occur without known regularity and usually constitute a very small portion of the genome. In fact, sexually acquired DNA need not involve recombination at all but can persist as a heritable extrachromosomal element—yielding a situation where the genome has changed but clonality is preserved. Taken together, sex in bacteria shares few features with those normally associated with sex in eukaryotes: it is simply the uptake of any genetic material that might eventually be vertically or horizontally transmitted (5–7).
Discovering Clonality in Natural Populations
By the 1950s, the numerous mechanisms by which bacteria could obtain new DNA sequences—conjugation (8), transformation (9), and transduction (10)—had been characterized, but the incidence of these processes and the extent of their effects on the diversification of bacterial clones remained unknown. Multilocus enzyme electrophoresis (MLEE), applied to bacteria by the 1970s (11), was able to supply quantifiable information about the forces that shape the allelic and genotypic variation in natural populations. The first large-scale population genetic survey of Escherichia coli concluded that the strain variation within hosts was generated by the “regular” occurrence of recombination and that the species as a whole was in linkage equilibrium (12). This supposition led the author (12) to construe that selective forces caused the preponderance of certain alleles and deviations from the random assortment of alleles over loci.
A more refined analysis incorporating several additional loci revealed that E. coli was, in fact, essentially clonal, with recombination rates perhaps on the order of that of mutation rates (13). In that study, evidence indicating that E. coli had a clonal population structure came from several sources. First, despite extensive allelic diversity at each of the 20 loci assayed, only a small number of genotypes were recovered, reflecting the infrequent reassortment of alleles. Second, strains of the same (or very similar) multilocus genotype were present in unrelated and geographically distant hosts (and, in one case, an infant from Massachusetts harbored a strain identical to the laboratory type specimen E. coli K-12, originally isolated in California in 1922), attesting to the long-term stability and wide geographic distribution of individual clones. Additionally, single locus variants (SLVs; strains identical at all but one locus) usually differed by the presence of a unique allele, suggesting that these polymorphisms arose by mutation rather than by recombination. Expanded studies on E. coli from diverse sets of hosts reported almost the same results (14, 15), and, around the same time, Ørskov and Ørskov devised the “clone concept” for E. coli pathogens to explain their findings that certain serotypic combinations were recovered repeatedly from temporally and geographically unassociated hosts (16). Although serological classification is based on cell surface factors whose variation could result from selection caused by the interactions between bacteria and hosts, the concordance between the population structures defined by serotyping and by MLEE provided strong support for the view that E. coli is predominantly clonal (17).
Just How Clonal Are Bacteria?
Broad application of MLEE illuminated the clonal nature of the bacterial populations (18, 19). Discovering that the majority of species displayed a nonrandom association of alleles among loci (linkage disequilibrium) led to the view that rates of recombination are typically low in bacteria isolated from their natural habitats—but, unfortunately, there is trouble with this interpretation. Whereas the occurrence of linkage equilibrium can be attributed to recombination, the converse need not be true: i.e., linkage disequilibrium is not always indicative of a lack of recombination. Numerous factors, quite apart from the lack of recombination, can cause linkage disequilibrium, and the possibility that the clonality observed in most bacterial populations derives from sources other than the lack of recombination was brought to the forefront in a PNAS paper by Maynard Smith et al. (20) that asked (and was entitled, as is this subsection): “How clonal are bacteria?”.
Some bacterial species may be truly clonal: i.e., they experience no recombination. However, several circumstances will give the appearance of clonality, even in species that undergo regular bouts of recombination. Scenarios particularly relevant to bacteria in which this circumstance is evident are when recombination proceeds within genetically or geographically isolated subpopulations and when there has been epidemic expansion (or periodic selection) of a particular strain. In these cases, sampling a mixture of strains from multiple subpopulations, or only the progeny of the ephemeral epidemic strain, will both yield evidence of strong disequilibrium, and the challenge is to distinguish such cases from true clonality. Maynard Smith et al. (20) tried to differentiate these cases both by partitioning the samples into subgroups to determine how the observed extent of recombination changed and by confining analyses to individual genotypes (as opposed to the entire sample, which may contain multiple isolates of the epidemic clone). Despite its vagaries, reanalysis of MLEE data using their approach (20) yielded species that were completely clonal (e.g., Salmonella enterica) and others that were panmictic (e.g., Neisseria gonorrheae), as well as some with intermediate population structures. Surprisingly, they did not apply their methods to E. coli, the bacterial species for which the most comprehensive MLEE data were then available (∼5,000 isolates); however, its population structure was thought to most closely resemble that of its sister group Salmonella.
Entering the Sequencing Era
MLEE, by assaying allelic variants in a handful of loci scattered around the genome, is limited to the detection of rather large-scale events of recombination, typically those involving regions much larger than a bacterial gene, leaving events occurring on a much smaller scale unnoticed. The advent of sequence-based analyses remedied this situation by offering resolution of allelic variation at the level of the individual nucleotide. Once it became possible to generate nucleotide sequences for homologs in multiple isolates, the question became a matter of how best to detect, and to assess the amounts and effects, of recombination. The methods fell into two general groups: tree-based approaches, which examined incongruencies in the phylogenies inferred from different genes; and alignment-based approaches, which examined the distribution of polymorphic sites.
E. coli was the focus of several studies, which found, with few exceptions, that the topology and strain groupings based on gene sequences matched those originally designated by MLEE, as would be expected in a predominantly clonal species. Attempts to quantify the relative impact of recombination vs. mutation on the genetic diversity of E. coli began by reconstructing the likely origin of each variable site from alignments of homologous sequences. It is obvious that ascribing a polymorphism to either a recombinational or a mutational event is based on several assumptions and is often problematic. For example, clustered substitutions in the gene sequence of a strain are more-than-likely introduced in a single recombinational event whereas a sporadic variant is typically viewed as arising from mutation, although such single-site variants could also originate through recombination from near-identical relatives. Another useful signal of recombination is that of homoplasies—characters inferred to be shared by two or more strains but not present in their common ancestor—which can arise in multiple strains by gene transfer but also by independent mutations.
Looking for tracts of clustered substitutions in five strains of E. coli, Milkman and Bridges (21) enumerated 10 recombinational replacement events in regions estimated to have incurred 50 nucleotide substitutions, yielding a rate of retained replacements to retained substitutions of 0.2 (and again implying that E. coli is largely clonal and that its genetic diversity was generated chiefly by mutations). Despite these results, it was argued that simply enumerating events of recombination and mutation does not convey their actual contributions because recombination can change multiple nucleotides—so, in order for the processes to be compared directly, both need to be expressed on a per-nucleotide-site basis. Examining the branching orders for four genes in 12 strains of E. coli, Guttman and Dykhuizen (22) detected inconsistencies attributable to three recombination events within a group of strains that had no polymorphisms attributable to mutations, resulting in a rate of nucleotide change by recombination that was an estimated 50 times that of mutation. Note, however, that they found that the phylogenies for all genes were largely congruent, such that the few recombinational events, although affecting a disproportionate number of nucleotides, did not disrupt the clonal nature of E. coli populations.
Moving from MLEE to MLST
Nucleotide sequencing methods added new dimensions to analysis of bacterial populations and led to the widespread use of a multilocus sequence typing (MLST) approach, in which six or seven gene fragments (of lengths suitable for Sanger sequencing) were PCR-amplified and sequenced for each bacterial strain (23–25). MLST is, in many ways, an extension of MLEE, in that it indexes the allelic variation at multiple housekeeping genes in each strain. Naturally, MLST had advantages over MLEE, the most prominent of which was its high level of resolution, its reproducibility, and its portability, allowing any researchers to generate data that could be easily processed and compared across laboratories.
Similar to MLEE, most applications of MLST assign a unique number to each allelic variant (regardless of its number of nucleotide differences from a nonidentical allele), and each strain is designated by its multilocus genotype: i.e., its allelic profile across loci. However, the sequence information generated for MLST proved extremely useful for examining the role of mutation and recombination in the divergence of bacterial lineages (26–28). Focusing on SLVs (i.e., allelic profiles that differed at only one locus), Feil et al. (29) tabulated those in which the allelic variants differed at single sites, indicating an SLV generated by mutation, or at multiple sites, taken as evidence of an SLV generated by recombination. (Actually, their complementary analysis based on homoplasy revealed that perhaps half of allelic variants differing at a single site also arose through recombination.) Their calculations of r/m (the ratio of substitutions introduced by recombination relative to mutation) for Streptococcus pneumoniae and Neisseria meningitidis ranged from 50 to 100, on the order of what Guttman and Dykhuizen (22) estimated in E. coli.
Current practice is to use r and m to denote per-site rates of recombination and mutation, and ρ and θ to denote events of recombination and mutation, respectively; however, these notations have been applied somewhat indiscriminately and their values derived by disparate methods, often hindering comparisons across studies. Vos and Didelot (30) revisited the MLST datasets for scores of bacterial taxa and recalculated r and m in a single framework, thereby allowing direct comparisons of the extent of recombination in generating the clonal divergence within species. The r/m values ranged over three orders of magnitude, and there was no clear association between recombination rates and bacterial lifestyle or phylogenetic division. Additionally, there were several cases where the values that they obtained were clearly at odds with previous studies: for example, they found _S. enterica_—the most clonal species based on MLEE—to have among the highest r/m ratios, even higher than that of Helicobacter pylori, which is essentially panmictic. Contrarily, r/m of E. coli was only 0.7, substantially lower than some previous estimates. Such discrepancies are likely due to the methods used to identify recombinant sites, the specific datasets that were analyzed, and the effects of sampling on recognition of recombination.
The population structure of E. coli was viewed as largely clonal because recombination was either limited to particular genes and to particular groups of strains. A broad MLST survey involving 100s of E. coli strains looked at the incidence of recombination within the well-established subgroups (clades) that were originally defined by MLEE (31). Although the mutation rates were similar for all seven genes across all subgroups, recombination rates differed substantially. Moreover, that study found a link between recombination and virulence, such that subgroups comprising pathogenic strains of E. coli displayed increased rates of recombination.
Clonality in the Genomic Era
Even if recombination occurs infrequently and affects small regions of the chromosome, the clonal status of the lineage will erode, making it difficult to establish the degree of clonality without sequences of entire genomes. Complete genome sequences now offer the opportunity to decipher the impact of recombination on bacterial evolution; but, admittedly, comparing sets of whole genomes is much more computationally challenging than analyzing the sequences from a few MLST loci and still suffers from many of the same biases. Although many of the same analytical problems arise when examining any set of sequences, the advantages of using full genome sequences are that they show the full scale of recombination events occurring through the genome, that they are better for defining recombination breakpoints, and that they can reveal how recombination might be related to certain functional features of genes or structural features of genomes.
The first comprehensive analysis of recombinational events occurring throughout the E. coli genome, conducted by Mau et al. (32), considered the complete sequences of six strains and used phylogenetic and clustering methods to identify recombinant segments within regions that were conserved in all strains. (32). Although they inferred one long (∼100-kb) stretch of the chromosome that underwent a recombination event in these strains, they reported that the typical length of recombinant segments was only about 1 kb in length, which was much shorter than that reported in studies based in more limited portions of the genome; and furthermore, they estimated that the extent of recombination was higher than previous estimates. The short size of recombinant fragments suggested that recombination occurred primarily by events of gene conversion rather than crossing-over, as is typical in eukaryotes, and by transduction and conjugation, which usually involve much larger pieces of DNA. Shorter segments of DNA could result from the partial degradation of longer sequences or could directly enter the cell through transformation, but E. coli is not naturally transformable, and its occurrence has been reported only under specific conditions (33, 34).
A second study on E. coli (35) focused on a diverse set of 20 complete genomes and used population-genetics approaches (36, 37) to detect recombinant fragments. In this analysis, the length of recombinant segments was much shorter than previous estimates (only 50 bp) although the relative impact of recombination and mutation on the introduction of nucleotide polymorphism was very close to that estimated with MLST data (r/m ≈ 0.9) (30). The study (35) also asked how the effects of recombination differed along the chromosome and identified several (and confirmed some) recombination hotspots, most notably, two centering on the rfb and the fim operons (38, 39). These two loci are involved in O-antigen synthesis (rfb) and adhesion to host cells (fim), and, because these two cellular features are exposed to phages, protists, or the host immune system, they are thought to evolve quickly by diversifying selection (40).
Aside from these hotspots, smoother fluctuations of the recombination rate are apparent over broader scales. Chromosome scanning revealed a decrease in the recombination rate in the ∼1-Mb region surrounding the replication terminus (35). Several hypotheses have been proposed to account for this transition in recombination rate along the chromosome, including: (i) a replication-associated dosage effect, which leads to a higher copy number and increased recombination rate (due to this increased availability of homologous strands) proximate to the replication origin; (ii) a higher mutation rate nearer to the terminus, resulting in an effectively lower value r/m ratio (41); and (iii) the macrodomain structure of the E. coli chromosome, in which the broad region spanning the replication terminus is the most tightly packed and has a reduced ability to recombine due to physical constraints (42). (An alternate hypothesis, combining features of i and ii posits that the homogenizing effect of recombination serves to reduce the rate of evolution of conserved housekeeping genes, which are disproportionately located near the replication origin.) In fact, each of the hypotheses that attempt to account for the variation in r/m values along the chromosome remain blurred by the tight association of mutation, selection, and recombination; therefore, caution is needed when interpreting this metric.
A more recent study involving 27 complete E. coli genomes used a Bayesian approach, implemented in ClonalFrame (43), to detect recombination events (44). Again, the r/m ratio was near unity; however, recombination tracts were estimated to be an order of magnitude longer than the previous based on many of the same genomes (542 bp vs. 50 bp), but still shorter than original estimates of the size of recombinant regions. That study (44) defined a third hotspot around the aroC gene, which could be involved in host interactions and virulence.
These analyses, all based on full genome sequences, estimated similar recombination rates for E. coli, confirming previous observations that, on average, recombination introduces as many nucleotide substitutions as mutations. Despite rather frequent recombination, this amount of DNA flux does not blur the signal of vertical descent for genes conserved among all strains (i.e., the “core genome”) (35). Unfortunately, the delineation of recombination breakpoints is still imprecise and highly dependent on the particular method and the dataset used to recognize recombination events. In all cases, similar sets of genes were overly affected by recombination, particularly fast-evolving loci that encoded proteins that were exposed to the environment, involved in stress response, or considered virulence factors. And, in fact, the recombination hotspots in Pseudomonas aeruginosa have been recently shown to include similar gene categories (45).
How to Best Assess the Impact of Recombination on E. coli Evolution
The large variety of methods that have been developed to detect recombination (46) reflects the fact that there are numerous technical and conceptual difficulties associated with the identification of the specific tracts of DNA that have been involved in gene exchange. As might be expected, the power and accuracy of these algorithms are maximized when a donor sequence is included (imparting the source of homology between unrelated lineages) and when the recombinant sequence introduces many polymorphic nucleotides (43, 46). Therefore, homoplasies—characters that are inferred to be shared by, but not present in, the common ancestor of lineages—represent robust signals of recombination and provide a very fine (i.e., per nucleotide site) resolution of recombination maps, as have been performed recently for sequenced strains of Staphylococcus aureus (47). Homoplasic sites allow detection of internal recombination events (i.e., recombinant polymorphic sites that are included in the dataset) but ignore polymorphic sites that were introduced by external, unsampled sources. Unsampled polymorphism can be introduced by closely related lineages (that acquired new mutations and would go undetected because they mimic vertical inheritance) or by divergent unsampled lineages. Although approaches based on homoplasies could miss the latter cases of recombination—virtually all approaches overlook the former—the increasing number of sequenced genomes and the long history of MLEE and MLST analyses suggest that current sampling of E. coli genomes is ample. However, it remains possible that several new major lineages have yet to be discovered (48, 49).
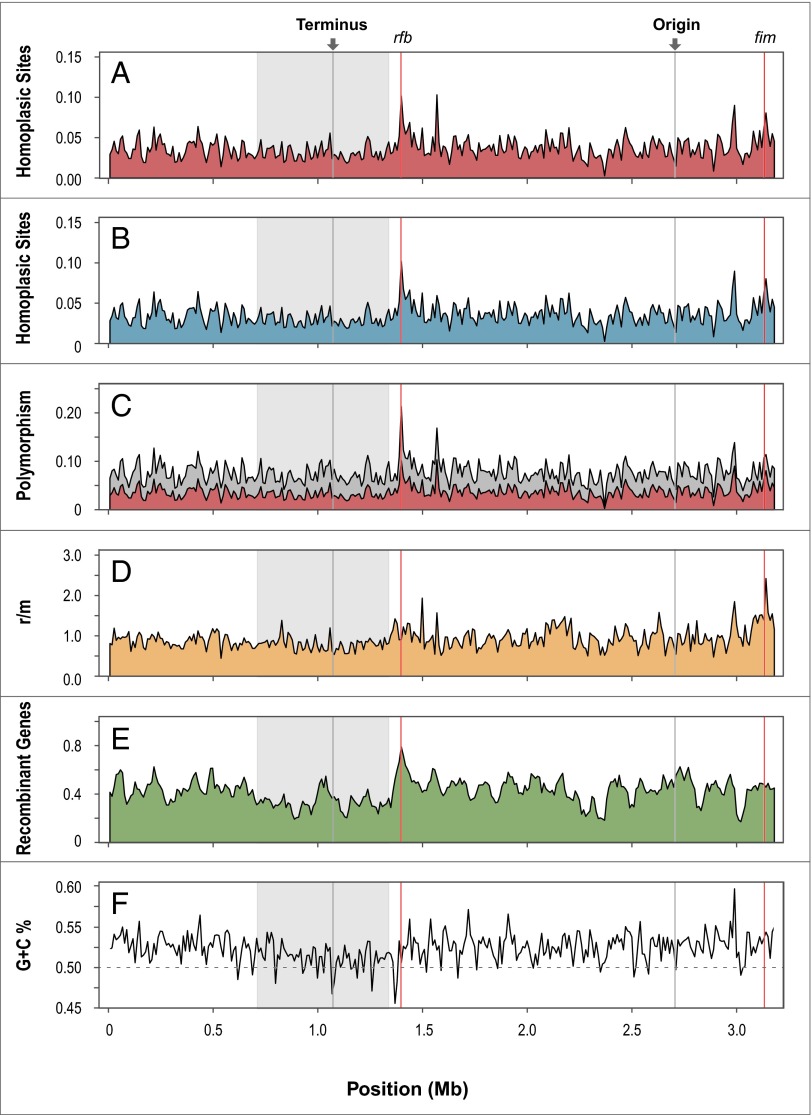
To obtain the recombinational landscape of the E. coli genome (Fig. 1), we focused our analysis on the detection of homoplasies among the genes common to a subset of the complete genomes [www.ncbi.nlm.nih.gov/genome (June 2014 version)]. The complete set of orthologous genes was defined by best bidirectional hit with USEARCH (global) with 80% identity (50) among the annotated genes present in the 65 sequenced strains. Adjacent core genes were merged into 1,131 core fragments that conserved gene order and intergenic regions, aligned with MAFFT v7 (51), and concatenated into a single large contig. The maximum likelihood distances D between the core gene sets were computed with TREE-PUZZLE (52), allowing us to select a subset of 19 genomes in a manner that maximized the nucleotide diversity of their core genomes and to retain only a single representative of nearly identical strains.
Fig. 1.
Recombinational landscape of E. coli. Genomic features and recombination parameters were calculated for the entire core genome of concatenated orthologs shared by 19 fully sequenced strains of E. coli. (A) Proportion of homoplasic sites, inferred by a distance metric that uses fixed branch lengths (see How to Best Assess the Impact of Recombination on E. coli Evolution) in nonoverlapping 10-kb windows spanning the core genome. (B) Proportion of homoplasic sites, inferred by a phylogenetic topology method (implemented in recHMM) (54), in nonoverlapping 10-kb windows spanning the core genome. (C) Proportion of the total variation (polymorphism) present in nonoverlapping 10-kb windows (gray) that is attributable to homoplasic sites (red). (D) Relative impact of recombination (π r) and mutation (π m) on the polymorphism in nonoverlapping 10-kb windows spanning the core genome based on the r/m (π r/π m) statistic. (Polymorphisms identified with the distance metric, as in A.) (E) Detecting recombination within core fragments (orthologous gene contigs) spanning the core genome with PHI (10,000 permutations; P < 0.001) (57). Because this method detects recombination on a gene-by-gene (not site-by-site) scale, results are presented for 50-kb sliding windows with 10-kb steps. (F) G+C contents in nonoverlapping 10-kb windows spanning the core genome.
Homoplasies appearing in the concatenation of core fragments (∼3.2 Mb, representing 3,022 genes) were detected by two methods: (i) Distance method: Based on the distance D between genomes, a polymorphic site composed of two alleles N0 and N1 was considered homoplasic when max (D N1N1) > min (D N0N1), with N0 and N1 the major and minor alleles, respectively. (Triallelic sites were subjected to the same pairwise comparison, but tetraallelic sites (n = 357) were not considered.); and (ii) Topology method: A maximum likelihood tree was inferred from the concatenation of core fragments for all 19 genomes using RAxML v7.2 (53), and polymorphic sites were considered homoplasic when incongruent with the topology of the core genome tree using recHMM (54, 55). Both the distance and the topology methods yielded similar results (Fig. 1 A and B) and detected 116,288 and 112,400 homoplasies, respectively, of which 110,143 were common to both methods. We confined subsequent analyses to homoplasic sites identified by the distance method because it is faster and less sensitive to unsolved internal branches.
Homoplasies arise from recombination but can also result from mutations that occur independently in the lineages in question. Fortunately, the two processes can often be distinguished because a single recombination event is likely to introduce multiple homoplasies that display the same incongruent pattern (i.e., clusters of polymorphic sites that have the same distribution among lineages). To establish whether homoplasies arose from recombination or from convergent mutations, we looked for the signatures of congruent homoplasies in 1-kb windows across the entire concatenation. Almost half (46%) of the homoplasic sites have a nearby (within 500-bp) homoplasic site displaying the same distribution among strains, suggesting that they were introduced in the same recombination event, not by convergent mutations. By simulating the accumulation of the current polymorphism in the E. coli genome, and assuming that it was introduced exclusively by random mutations, we estimate that only 2.4% of polymorphic sites would be homoplasic due to independent mutations, indicating that convergent mutations have a negligible contribution relative to recombination in the introduction of homoplasies.
Using homoplasic sites, we mapped the incidence of recombination across the entire E. coli genome. This method could possibly underestimate the frequency of recombination events by failing to recognize recombinant segments from unsampled lineages; however, it clearly illustrates the extent to which recombination varies in its effects across the chromosome. These results confirm the existence of the recombination hotspot near the fim gene and suggest the presence of several additional recombination hotspots. The previously identified rfb hotspot is situated in a region of high polymorphism (Fig. 1_C_); however, the relative frequencies of homoplasic to nonhomoplasic polymorphisms is not particularly high (Fig. 1_D_), which suggests that this hotspot might in fact be attributable to diversifying selection as opposed to recombination. Our analysis, based solely on homoplasic sites, also recognized the region surrounding the replication terminus as having a reduced rate of recombination. Overall, the relative frequency of homoplasic and nonhomoplasic polymorphism suggests a near equal contribution of recombination and mutation in the introduction of polymorphic sites (r/m = 0.92, SD ± 0.26), which is consistent with previous analyses based on full genome data (Table 1).
Table 1.
A selective history of E. coli clonality
| Year of study | Extent of recombination reported | Method used to assess variation | No. of strains examined | No. of loci examined | Source |
|---|---|---|---|---|---|
| 1973 | Common (population assumed to be panmictic) | MLEE | 829 | 5 isozymes | (12) |
| 1980 | Rare (on the order of the mutation rate) | MLEE | 109 | 20 isozymes | (13) |
| 1990 | ρ/θ = 0.2 | Nucleotide sequencing & restriction mapping | 9 | One 15-kb region | (21) |
| 1991 | “Tremendous amount” | Nucleotide sequencing | 73 | 15 regions | (82) |
| 1993 | Rate inestimable | Nucleotide sequencing | 37 | One 4.4-kb region | (83) |
| 1993 | r/m = 1–10 | Nucleotide sequencing | 4–13 | 5 genes | (84) |
| 1994 | r/m = 50 | Nucleotide sequencing | 12 | 4 genes | (22) |
| 2000 | Common | MLST | 14 | 7 genes | (85) |
| 2006 | ρ/θ = 0 | MLST | 21–83 | 16 genes | (86) |
| 2006 | “Higher than previously thought” | Whole genome sequencing | 6 | All shared genes | (34) |
| 2006 | “Low” | MLST | 44 | 12 genes | (6) |
| 2006 | r/m = 0.32–2.14 | MLST | 432 | 7 genes | (31) |
| 2008 | r/m = 0.70 | MLST | 44 | 7 genes | (30) |
| 2009 | r/m = 0.90 | Whole genome sequencing | 20 | All shared genes | (35) |
| 2012 | r/m = 1.02 | Whole genome sequencing | 27 | All shared genes | (44) |
| 2015 | r/m = 0.92 | Whole genome sequencing | 19 | All shared genes | This study |
Aside from contributing to the variation of individual genes, recombination also seems to affect how the chromosome itself evolves. At the terminus of replication, the lower recombination rate coincides with a reduction in the G+C content (35), as is observed in other species (56) (Fig. 1_F_). This effect becomes even more noticeable when detecting recombination at larger scales, as with the computational method PHI (pairwise homoplasy index) (Fig. 1_E_) (57). In that mutations are universally biased toward A and T (58, 59) and recombination influences the effectiveness of selection (60), these two effects, in combination, could result in a reduced ability of low-recombining loci to purge slightly deleterious (and A+T-biased) mutations. This background selection model is supported by the decrease of polymorphism and indications of purifying selection on nonsynonymous sites near the terminus (35). Moreover, there is additional evidence that selection serves to elevate genomic G+C contents in bacteria (61, 62). Alternatively, a lower recombination rate near the replication terminus could lower the G+C content of the region by minimizing the G+C-biased repair of recombination-induced mismatches by biased gene conversion (63).
Beyond the Core Genome
Most genome-wide analyses of recombination have been limited to the regions constituting the core genome, but this approach ignores the accessory genes—those that are not ubiquitous among strains—and their neighboring intergenic regions. Such regions are just as prone to recombination events; however, their sporadic distributions make their identification and analysis somewhat more difficult. There are many classes of accessory genes, such as mobile elements (e.g., prophages, transposons), which are known to be associated with elevated rates of recombination. In both E. coli and S. aureus, it was recently shown that core genes in the vicinity of accessory genes or mobile elements experience higher recombination rates (44, 47). Chromosome loci with the highest homologous recombination rates (recombination hotspots) have also been associated with nonmobilizable genomic islands in E. coli (e.g., the fim locus). These heightened rates of recombination could be due to selection—elements can encode adaptive traits that confer an advantage to their acquisition (64)—and the absence of site-specific integrases or transposases within many of these elements suggests that many rely on recombination to propagate in the population. Additionally, many recombination hotspots in E. coli seem to be evolving under diversifying selection, supporting a general role of homologous exchange in spreading both beneficial alleles and beneficial accessory genes (35).
The ability of recombination to spread beneficial alleles (and purge deleterious alleles) has been known for some time (65); however, its effect on the dynamics of bacterial genes and genomes remains obscure. Studies on Vibrio cyclitrophicus and Burkholderia pseudomallei both suggest than genes, rather than genomes, reach fixation into the population (66, 67), but these species undergo much higher recombination rates than E. coli (30). The population structure of E. coli, in which certain genotypes dominate the population, would indicate that periodic selection (selective sweeps) lead to occasional epidemic structures in E. coli and other species that experience local or low rates of recombination.
Genomic Determinants of Bacterial Clonality
What determines whether a bacterial population is clonal or panmictic? Several genomic features have been linked to the ability of bacteria to modulate the amount of DNA uptake and exchange within and between populations.
Firstly, recombination efficiency is connected to the extent of sequence identity. mutS mutants of E. coli demonstrate low levels of sexual isolation, suggesting that mismatch repair plays a central role in the frequency of recombination (68). Recombination initiation requires minimal substrate lengths of 23–27 identical nucleotides, termed “minimal efficient processing segments” (MEPS) (69). The frequency of MEPS decreases exponentially with sequence divergence, suggesting that the clonal or panmictic status of a species depends on its level of polymorphism and its population structure. Moreover, this requirement would imply that more divergent strains display lower frequencies of DNA exchange, compatible with clonal evolution, whereas closely related strains recombine more frequently. As highlighted previously (in Just How Clonal Are Bacteria?), frequent recombination, when confined to close relatives, would produce populations that possess all of the hallmarks of clonality, making it difficult to determine the actual clonal status of the species.
Secondly, several additional barriers to DNA acquisition and exchange occur in bacteria (70); and among them, restriction-modification (R-M) systems vary substantially among species and strains (71). By selectively degrading incoming DNA according to their sequence and methylation patterns, these systems can influence the range and extent of DNA exchange between cells and populations, and a recent study highlighted the role of R-M systems in regulating sequence exchange within B. pseudomallei (67).
Third, the mobile element repertoires, which can be highly variable among strains (72, 73), are likely to determine the capacity for DNA transfer by mediating transduction and conjugation, and by providing templates for homologous exchange. Additionally, mobile elements integrated into the E. coli genome sometimes encode enzymes catalyzing homologous exchange (74, 75): For example, the defective prophage rac encodes the RecT recombinase, which can supplement recombination functions in RecBCD mutants (76), and is typically more promiscuous than the RecBCD pathway (77, 78). Moreover, there is wide variation among E. coli strains in the repertoires of complete or partially degraded prophages, implying that strains can rapidly acquire and lose recombination genes depending on their particular set of mobile elements. This dynamic reservoir of ready-to-use recombination enzymes might serve to promote fluctuations in recombination rates within and among lineages.
Finally, there can be counterselection against recombination in some genomes arising from the epistatic interactions among alleles at different loci (79, 80). In this scenario, genes whose products are involved in multiprotein complexes or depend on specific protein–protein interactions would sustain fewer nonsynonymous substitutions introduced by recombination (analogous to barriers to gene exchange proposed in the “complexity hypothesis” (81), in which highly interacting proteins are not susceptible to horizontal acquisition).
Conclusions
The perception of the clonal status of E. coli has changed substantially over the past four decades, mostly due to the maturation of allele-typing methods, to increased awareness of effects of strain and locus sampling, and to the development of new recombination-detection algorithms. Genome sequencing should overcome many of the remaining obstacles to evaluating recombination occurring in bacterial populations, and recombination-detection methods have been improved to provide better resolution of sequence history and population structures. Given the abundance of lineages that have been sampled in E. coli and other species, the use of homoplasies as signals of recombination should help to increase the application and accuracy of these methods. When considering E. coli, several studies have converged on the view that recombination contributes almost as much as mutations to the introduction of nucleotide polymorphism. However, despite its occurrence, the rate and scale of recombination does not destroy the clonal status of E. coli. Estimations of the size of recombinant segments in E. coli are imprecise but mostly range from 50–500 bp, indicating that DNA exchange acts primarily by gene conversion. Moreover, recombination rates are highly variable among genes and along the chromosome, and it remains difficult to tease apart the relative contributions of mutations, recombination, and selection to the observed variation.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution IX: Clonal Reproduction: Alternatives to Sex,” held January 9–10, 2015, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/ILE_IX_Clonal_Reproduction.
This article is a PNAS Direct Submission.
References
- 1.Maynard Smith J. The Evolution of Sex. Cambridge Univ Press; Cambridge, UK: 1978. [Google Scholar]
- 2.Barton NH, Charlesworth B. Why sex and recombination? Science. 1998;281(5385):1986–1990. [PubMed] [Google Scholar]
- 3.Lynch M, Bürger R, Butcher D, Gabriel W. The mutational meltdown in asexual populations. J Hered. 1993;84(5):339–344. doi: 10.1093/oxfordjournals.jhered.a111354. [DOI] [PubMed] [Google Scholar]
- 4.Abby SS, Rocha EPC. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 2012;8(9):e1002983. doi: 10.1371/journal.pgen.1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redfield RJ. Do bacteria have sex? Nat Rev Genet. 2001;2(8):634–639. doi: 10.1038/35084593. [DOI] [PubMed] [Google Scholar]
- 6.Narra HP, Ochman H. Of what use is sex to bacteria? Curr Biol. 2006;16(17):R705–R710. doi: 10.1016/j.cub.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 7.Levin BR, Cornejo OE. The population and evolutionary dynamics of homologous gene recombination in bacterial populations. PLoS Genet. 2009;5(8):e1000601. doi: 10.1371/journal.pgen.1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lederberg J, Tatum EL. Gene recombination in Escherichia coli. Nature. 1946;158(4016):558. doi: 10.1038/158558a0. [DOI] [PubMed] [Google Scholar]
- 9.Avery OT, Macleod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumonococcal types: Induction of transformation by a deoxyribonucleic acid fraction isolated from pnuemococcus type III. J Exp Med. 1944;79(2):137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zinder ND, Lederberg J. Genetic exchange in Salmonella. J Bacteriol. 1952;64(5):679–699. doi: 10.1128/jb.64.5.679-699.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baptist JN, Shaw CR, Mandel M. Comparative zone electrophoresis of enzymes of Pseudomonas solanacearum and Pseudomonas cepacia. J Bacteriol. 1971;108(2):799–803. doi: 10.1128/jb.108.2.799-803.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milkman R. Electrophoretic variation in Escherichia coli from natural sources. Science. 1973;182(4116):1024–1026. doi: 10.1126/science.182.4116.1024. [DOI] [PubMed] [Google Scholar]
- 13.Selander RK, Levin BR. Genetic diversity and structure in Escherichia coli populations. Science. 1980;210(4469):545–547. doi: 10.1126/science.6999623. [DOI] [PubMed] [Google Scholar]
- 14.Whittam TS, Ochman H, Selander RK. Multilocus genetic structure in natural populations of Escherichia coli. Proc Natl Acad Sci USA. 1983;80(6):1751–1755. doi: 10.1073/pnas.80.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caugant DA, Levin BR, Selander RK. Distribution of multilocus genotypes of Escherichia coli within and between host families. J Hyg (Lond) 1984;92(3):377–384. doi: 10.1017/s0022172400064597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ørskov F, Ørskov I. From the National Institutes of Health: Summary of a workshop on the clone concept in the epidemiology, taxonomy, and evolution of the enterobacteriaceae and other bacteria. J Infect Dis. 1983;148(2):346–357. doi: 10.1093/infdis/148.2.346. [DOI] [PubMed] [Google Scholar]
- 17.Achtman M, et al. Clonal analysis of Escherichia coli O2:K1 isolated from diseased humans and animals. Infect Immun. 1986;51(1):268–276. doi: 10.1128/iai.51.1.268-276.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selander RK, et al. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51(5):873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selander RK, Musser JM, Caugant DA, Gilmour MN, Whittam TS. Population genetics of pathogenic bacteria. Microb Pathog. 1987;3(1):1–7. doi: 10.1016/0882-4010(87)90032-5. [DOI] [PubMed] [Google Scholar]
- 20.Smith JM, Smith NH, O’Rourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90(10):4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milkman R, Bridges MM. Molecular evolution of the Escherichia coli chromosome. III. Clonal frames. Genetics. 1990;126(3):505–517. doi: 10.1093/genetics/126.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guttman DS, Dykhuizen DE. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science. 1994;266(5189):1380–1383. doi: 10.1126/science.7973728. [DOI] [PubMed] [Google Scholar]
- 23.Maiden MC, et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95(6):3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feil EJ, Spratt BG. Recombination and the population structures of bacterial pathogens. Annu Rev Microbiol. 2001;55:561–590. doi: 10.1146/annurev.micro.55.1.561. [DOI] [PubMed] [Google Scholar]
- 25.Maiden MC, et al. MLST revisited: The gene-by-gene approach to bacterial genomics. Nat Rev Microbiol. 2013;11(10):728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spratt BG, Hanage WP, Feil EJ. The relative contributions of recombination and point mutation to the diversification of bacterial clones. Curr Opin Microbiol. 2001;4(5):602–606. doi: 10.1016/s1369-5274(00)00257-5. [DOI] [PubMed] [Google Scholar]
- 27.Hanage WP, Fraser C, Spratt BG. Sequences, sequence clusters and bacterial species. Philos Trans R Soc Lond B Biol Sci. 2006;361(1475):1917–1927. doi: 10.1098/rstb.2006.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Didelot X, Maiden MC. Impact of recombination on bacterial evolution. Trends Microbiol. 2010;18(7):315–322. doi: 10.1016/j.tim.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feil EJ, Smith JM, Enright MC, Spratt BG. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics. 2000;154(4):1439–1450. doi: 10.1093/genetics/154.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2009;3(2):199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- 31.Wirth T, et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol Microbiol. 2006;60(5):1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mau B, Glasner JD, Darling AE, Perna NT. Genome-wide detection and analysis of homologous recombination among sequenced strains of Escherichia coli. Genome Biol. 2006;7(5):R44. doi: 10.1186/gb-2006-7-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baur B, Hanselmann K, Schlimme W, Jenni B. Genetic transformation in freshwater: Escherichia coli is able to develop natural competence. Appl Environ Microbiol. 1996;62(10):3673–3678. doi: 10.1128/aem.62.10.3673-3678.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsen SD, et al. Natural plasmid transformation in Escherichia coli. J Biomed Sci. 2002;9(3):246–252. doi: 10.1007/BF02256071. [DOI] [PubMed] [Google Scholar]
- 35.Touchon M, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5(1):e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Padhukasahasram B, Wall JD, Marjoram P, Nordborg M. Estimating recombination rates from single-nucleotide polymorphisms using summary statistics. Genetics. 2006;174(3):1517–1528. doi: 10.1534/genetics.106.060723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beaumont MA, Zhang W, Balding DJ. Approximate Bayesian computation in population genetics. Genetics. 2002;162(4):2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milkman R, Jaeger E, McBride RD. Molecular evolution of the Escherichia coli chromosome. VI. Two regions of high effective recombination. Genetics. 2003;163(2):475–483. doi: 10.1093/genetics/163.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weissman SJ, et al. Clonal analysis reveals high rate of structural mutations in fimbrial adhesins of extraintestinal pathogenic Escherichia coli. Mol Microbiol. 2006;59(3):975–988. doi: 10.1111/j.1365-2958.2005.04985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seed KD, et al. Evolutionary consequences of intra-patient phage predation on microbial populations. eLife. 2014;3:e03497. doi: 10.7554/eLife.03497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mira A, Ochman H. Gene location and bacterial sequence divergence. Mol Biol Evol. 2002;19(8):1350–1358. doi: 10.1093/oxfordjournals.molbev.a004196. [DOI] [PubMed] [Google Scholar]
- 42.Valens M, Penaud S, Rossignol M, Cornet F, Boccard F. Macrodomain organization of the Escherichia coli chromosome. EMBO J. 2004;23(21):4330–4341. doi: 10.1038/sj.emboj.7600434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175(3):1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Didelot X, Méric G, Falush D, Darling AE. Impact of homologous and non-homologous recombination in the genomic evolution of Escherichia coli. BMC Genomics. 2012;13:256. doi: 10.1186/1471-2164-13-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dettman JR, Rodrigue N, Kassen R. Genome-wide patterns of recombination in the opportunistic human pathogen Pseudomonas aeruginosa. Genome Biol Evol. 2015;7(1):18–34. doi: 10.1093/gbe/evu260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin DP, Lemey P, Posada D. Analysing recombination in nucleotide sequences. Mol Ecol Resour. 2011;11(6):943–955. doi: 10.1111/j.1755-0998.2011.03026.x. [DOI] [PubMed] [Google Scholar]
- 47.Everitt RG, et al. Mobile elements drive recombination hotspots in the core genome of Staphylococcus aureus. Nat Commun. 2014;5:3956. doi: 10.1038/ncomms4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walk ST, et al. Cryptic lineages of the genus Escherichia. Appl Environ Microbiol. 2009;75(20):6534–6544. doi: 10.1128/AEM.01262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo C, et al. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc Natl Acad Sci USA. 2011;108(17):7200–7205. doi: 10.1073/pnas.1015622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 51.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18(3):502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 53.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 54.Zhou Z, et al. Transient Darwinian selection in Salmonella enterica serovar Paratyphi A during 450 years of global spread of enteric fever. Proc Natl Acad Sci USA. 2014;111(33):12199–12204. doi: 10.1073/pnas.1411012111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 56.Daubin V, Perrière G. G+C3 structuring along the genome: A common feature in prokaryotes. Mol Biol Evol. 2003;20(4):471–483. doi: 10.1093/molbev/msg022. [DOI] [PubMed] [Google Scholar]
- 57.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172(4):2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hershberg R, Petrov DA. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 2010;6(9):e1001115. doi: 10.1371/journal.pgen.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hildebrand F, Meyer A, Eyre-Walker A. Evidence of selection upon genomic GC-content in bacteria. PLoS Genet. 2010;6(9):e1001107. doi: 10.1371/journal.pgen.1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8(3):269–294. [PubMed] [Google Scholar]
- 61.Balbi KJ, Rocha EP, Feil EJ. The temporal dynamics of slightly deleterious mutations in Escherichia coli and Shigella spp. Mol Biol Evol. 2009;26(2):345–355. doi: 10.1093/molbev/msn252. [DOI] [PubMed] [Google Scholar]
- 62.Raghavan R, Kelkar YD, Ochman H. A selective force favoring increased G+C content in bacterial genes. Proc Natl Acad Sci USA. 2012;109(36):14504–14507. doi: 10.1073/pnas.1205683109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Galtier N, Piganeau G, Mouchiroud D, Duret L. GC-content evolution in mammalian genomes: The biased gene conversion hypothesis. Genetics. 2001;159(2):907–911. doi: 10.1093/genetics/159.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Middendorf B, et al. Instability of pathogenicity islands in uropathogenic Escherichia coli 536. J Bacteriol. 2004;186(10):3086–3096. doi: 10.1128/JB.186.10.3086-3096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fisher RA. The Genetical Theory of Natural Selection. Clarendon; Oxford: 1930. [Google Scholar]
- 66.Shapiro BJ, Alm EJ. Comparing patterns of natural selection across species using selective signatures. PLoS Genet. 2008;4(2):e23. doi: 10.1371/journal.pgen.0040023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nandi T, et al. Burkholderia pseudomallei sequencing identifies genomic clades with distinct recombination, accessory, and epigenetic profiles. Genome Res. 2015;25(1):129–141. doi: 10.1101/gr.177543.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matic I, Rayssiguier C, Radman M. Interspecies gene exchange in bacteria: The role of SOS and mismatch repair systems in evolution of species. Cell. 1995;80(3):507–515. doi: 10.1016/0092-8674(95)90501-4. [DOI] [PubMed] [Google Scholar]
- 69.Shen P, Huang HV. Homologous recombination in Escherichia coli: Dependence on substrate length and homology. Genetics. 1986;112(3):441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baltrus DA. Exploring the costs of horizontal gene transfer. Trends Ecol Evol. 2013;28(8):489–495. doi: 10.1016/j.tree.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 71.Oliveira PH, Touchon M, Rocha EP. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 2014;42(16):10618–10631. doi: 10.1093/nar/gku734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bobay LM, Rocha EP, Touchon M. The adaptation of temperate bacteriophages to their host genomes. Mol Biol Evol. 2013;30(4):737–751. doi: 10.1093/molbev/mss279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guglielmini J, Quintais L, Garcillán-Barcia MP, de la Cruz F, Rocha EP. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011;7(8):e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bobay LM, Touchon M, Rocha EP. Manipulating or superseding host recombination functions: A dilemma that shapes phage evolvability. PLoS Genet. 2013;9(9):e1003825. doi: 10.1371/journal.pgen.1003825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Garriss G, Waldor MK, Burrus V. Mobile antibiotic resistance encoding elements promote their own diversity. PLoS Genet. 2009;5(12):e1000775. doi: 10.1371/journal.pgen.1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kolodner R, Hall SD, Luisi-DeLuca C. Homologous pairing proteins encoded by the Escherichia coli recE and recT genes. Mol Microbiol. 1994;11(1):23–30. doi: 10.1111/j.1365-2958.1994.tb00286.x. [DOI] [PubMed] [Google Scholar]
- 77.Noirot P, Gupta RC, Radding CM, Kolodner RD. Hallmarks of homology recognition by RecA-like recombinases are exhibited by the unrelated Escherichia coli RecT protein. EMBO J. 2003;22(2):324–334. doi: 10.1093/emboj/cdg027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martinsohn JT, Radman M, Petit MA. 2008. The lambda Red proteins promote efficient recombination between diverged sequences: Implications for bacteriophage genome mosaicism. PLoS Genet 4:e1000065(5) [DOI] [PMC free article] [PubMed]
- 79.Kondrashov FA, Kondrashov AS. Multidimensional epistasis and the disadvantage of sex. Proc Natl Acad Sci USA. 2001;98(21):12089–12092. doi: 10.1073/pnas.211214298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Friedman J, Alm EJ, Shapiro BJ. Sympatric speciation: When is it possible in bacteria? PLoS ONE. 2013;8(1):e53539. doi: 10.1371/journal.pone.0053539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: The complexity hypothesis. Proc Natl Acad Sci USA. 1999;96(7):3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dykhuizen DE, Green L. Recombination in Escherichia coli and the definition of biological species. J Bacteriol. 1991;173(22):7257–7268. doi: 10.1128/jb.173.22.7257-7268.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Milkman R, Bridges MM. Molecular evolution of the Escherichia coli chromosome. IV. Sequence comparisons. Genetics. 1993;133(3):455–468. doi: 10.1093/genetics/133.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Whittam TS, Ake SE. Genetic polymorphisms and recombination in natural populations of Escherichia coli. In: Takahata N, Clark AG, editors. Mechanisms of Molecular Evolution. Sinauer; Sunderland, MA: 1993. pp. 223–246. [Google Scholar]
- 85.Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS. Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000;406(6791):64–67. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- 86.Pérez-Losada M, et al. Population genetics of microbial pathogens estimated from multilocus sequence typing (MLST) data. Infect Genet Evol. 2006;6(2):97–112. doi: 10.1016/j.meegid.2005.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]