Mutations in the UGO1-like protein SLC25A46 cause an optic atrophy spectrum disorder (original) (raw)
. Author manuscript; available in PMC: 2016 Jan 31.
Published in final edited form as: Nat Genet. 2015 Jul 13;47(8):926–932. doi: 10.1038/ng.3354
Abstract
Dominant optic atrophy (DOA)1,2 and axonal peripheral neuropathy (Charcot-Marie-Tooth Type 2 or CMT2)3 are hereditary neurodegenerative disorders most commonly caused by mutations in the canonical mitochondrial fusion genes OPA1 and MFN2, respectively4. In yeast, homologs of OPA1(Mgm1) and MFN2(Fzo1) work in concert with Ugo15,6, which has no human equivalent to date7. By whole exome sequencing patients with optic atrophy and CMT2, we identified four families with recessive mutations in SLC25A46. We demonstrate that SLC25A46, like Ugo1, is a modified carrier protein that has been recruited to the outer mitochondrial membrane and interacts with the inner membrane remodeling protein, mitofilin(Fcj1). Loss-of-function in cultured cells and in zebrafish unexpectedly leads to increased mitochondrial connectivity, while severely affecting the development and maintenance of neurons in the fish. The discovery of SLC25A46 strengthens the genetic overlap between optic atrophy and CMT2, while exemplifying a novel class of modified solute transporters linked to mitochondrial dynamics.
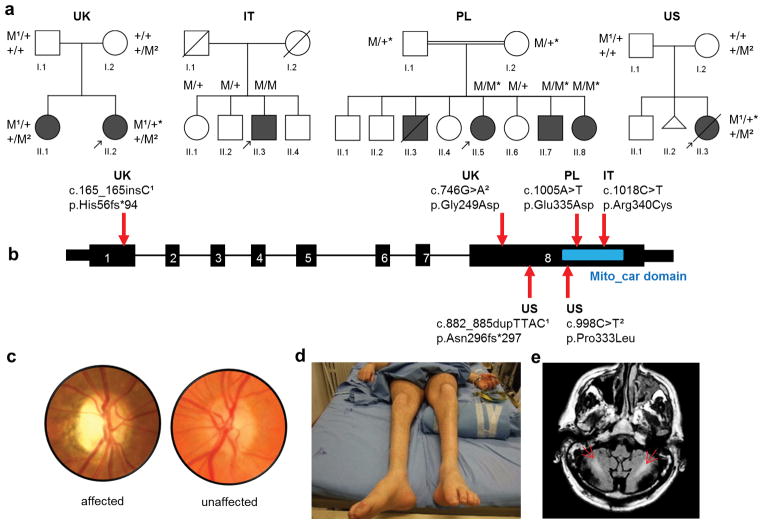
Mutations in MFN2 account for more than 20% of inherited axonal degenerative peripheral neuropathies, Charcot-Marie-Tooth type 2 (CMT2A) 3,8. A subset of these patients also develop optic nerve atrophy9. Meanwhile, mutations in OPA1 account for as much as 60–70% of dominant optic atrophy (DOA)10 and sometimes cause additional neurological symptoms referred to as optic atrophy “plus”10,11. MFN2 and OPA1 are involved in the tethering and fusion of outer and inner mitochondrial membranes4. Both CMT2 and DOA are genetically heterogeneous disorders with up to 60% of patients undiagnosed genetically 10,12. Therefore, we recruited families with both optic atrophy and axonal peripheral neuropathy to identify additional disease genes, based upon the hypothesis that causative genes would uncover new factors in common biological pathways. By applying whole exome sequencing with established methods and cut-offs for variant filtering in Mendelian inherited diseases13, we found four families with recessive variants in the nuclear-encoded mitochondrial gene SLC25A46. After excluding other candidate genes by segregation analysis, we identified the compound heterozygous mutations c.165_166insC, p.His56fs*94 and c.746G>A, p.Gly249Asp in a British family (UK), a homozygous mutation c.1005A>T, p.Glu335Asp in a Palestinian family (PL), and the compound heterozygous variants c.882_885dupTTAC, p.Asn296fs*297 and c.998C>T, p.Pro333Leu in an American family (US). We then used conventional Sanger sequencing methods to screen SLC25A46 in similar cases without a genetic diagnosis, and identified an additional family from Sardinia, Italy (IT), with the homozygous mutation c.1018C>T, p.Arg340Cys (Fig. 1, Supplementary Fig. 1, and Supplementary Table 1).
Figure 1.
Pedigrees and clinical features of optic atrophy “plus” syndromes with variants in SLC25A46. (a) Affected individuals (filled symbols); Deceased individuals (symbols with slashes); miscarriage (triangle); mutant allele (M); wild type allele (+); individuals in whom whole exomes were sequenced (*); probands (arrows). (b) Schematic gene diagram of SLC25A46, NM_138773, located on chromosome 5, cDNA: 1,257 bp, 418 aa, with positions of associated mutations indicated by arrows. The conserved mitochondrial carrier domain is indicated by a blue rectangle. (c) Image of the optic disc from patient UK II-2 shows primary temporal optic nerve pallor when compared to an unaffected individual. (d) Photograph of the legs of patient IT II-3 shows muscle wasting stereotypic of CMT2. (e) MRI shows bilateral hyperintensity of white matter (arrows) in the cerebellum, on FLAIR T2-weighted image, of patient IT II-3.
All non-truncating changes were predicted to be deleterious (Supplementary Table 1). Patients in these four families presented with similar phenotypic core features including optic atrophy, axonal CMT, as well as cerebellar atrophy (Refer to Supplementary information). Magnetic resonance spectroscopy (MRS) data in two of the patients found decreased N-Acetyl-Aspartate (NAA) and increased lactate in the central nervous system (Supplementary Fig. 2b) typical of mitochondrial disorders and suggestive of a metabolic role for SLC25A46. However, analysis of a muscle biopsy from patient IT II-3 found no ragged red fibers or cytochrome c oxidase (COX)-negative fibers (data not shown).
SLC25A46 is a member of the mitochondrial solute carrier family14 (SLC25) and is predicted to function as a transporter across the inner mitochondrial membrane15. Using the Basic Local Alignment Search tool (BLAST), we found that SLC25A46 is a reciprocal match to Ugo1 when querying between human and Schizosaccharomyces japonicus (Supplementary Table 2). Ugo1 is a modified mitochondrial solute carrier in the mitochondrial outer membrane (MOM)16,17 that operates as a mitochondrial fusion factor in Saccharomyces cerevisiae5 with no homologs identified in metazoans7. To unbiasedly determine the most similar protein to Ugo1 in metazoans, we created a bootstrapped, maximum-likelihood phylogenetic tree with all mitochondrial carrier domain-containing proteins in Homo sapiens, Caenorhabditis elegans, Saccharomyces cerevisiae, and Schizosaccharomyces pombe. This revealed SLC25A46 as the most similar to Ugo1. However, there is insufficient evidence to determine orthology (Online methods and Supplementary Fig 3) and SLC25A46 fails to complement ugo1 deletion in S. cerevisiae (data not shown).
During evolution, homologs of mitochondrial carriers, usually inner membrane proteins15, have been modified and recruited to the MOM to perform specific functions, unrelated to metabolite transport. The list includes the mammalian mitochondrial carrier homologs MTCH118 and MTCH219,20, critical players in apoptosis, and yeast Ugo15. Because human SLC25A46 is also a highly derived carrier protein as shown in supplementary Fig 4, we investigated its submitochondrial localization.
Immunocytochemistry studies in COS-7 cells demonstrate that HA-tagged SLC25A46 co-localizes more with the MOM marker (TOM20) and less with the inner membrane markers (myc-tagged mitofilin or ANT2) (Fig. 2a and supplementary Fig. 5a). We subsequently used mitochondria isolated from HEK293T cells expressing SLC25A46-HA to perform solubility and proteinase K protection assays, which demonstrated that SLC25A46 is an integral outer membrane protein (Fig. 2b and c). To gain insight into SLC25A46 function we identified interacting partners by performing an unbiased HA-immunoprecipitation assay combined with mass spectrometry analysis. Mitofilin was among the top hits in this assay (Supplementary Table. 3). Consistently, sucrose gradient sedimentation analyses found mitofilin to co-sediment with a small fraction of SLC25A46-HA, indicating that they may exist in the same high molecular mass complex (Fig. 2d). The interaction between SLC25A46 and mitofilin was further confirmed by co-immunoprecipitation assays using mitochondria isolated from cells transfected with SLC25A46-HA (Supplementary Fig. 5b) or co-transfected with SLC25A46-HA and mitofilin-myc (Fig. 2e), followed by western blot detection. These results are congruent to Ugo1, which has been shown to co-fractionate with Fcj121, the homolog of mammalian IMMT(mitofilin).
Figure 2.
SLC25A46 localizes to the outer mitochondrial membrane and interacts with mitofilin on the inner membrane. (a) High resolution confocal images of COS-7 cells co-transfected with SLC25A46-HA and Mitofilin-myc. TOM20 and Mitofilin-myc were used as markers of the outer and inner mitochondrial membranes respectively. Scale bar=1 μm. Linear profiles of the fluorescence intensity in arbitrary units along 2 μm dashed lines in merged images. (b) Mitochondria (M) isolated from SLC25A46-HA transfected HEK293T cells were submitted to brief sonication and centrifugation to fractionate soluble (S) and membrane-bound proteins. The pellet was subjected to alkaline extraction to separate soluble membrane extrinsic (CS) and membrane intrinsic (P) proteins. Equivalent volumes of each fraction were analyzed by immunoblotting with antibodies against the soluble matrix protein mHSP70, the extrinsic membrane protein SDHA, the intrinsic membrane protein COX2 and HA. (c) Mitochondria and mitoplasts prepared by hypotonic swelling of mitochondria from HEK293T cells expressing SLC25A46-HA were treated with proteinase K where indicated. After treatment samples were analyzed by immunoblotting with antibodies against the outer membrane protein TOM20, the inner membrane proteins TIM50 and COX2 and HA. (d) Co-sedimentation of SLC25A46-HA and mitoflin in a linear 7–20% sucrose gradient. Hemoglobin, a 67 kDa protein, and lactate dehydrogenase (LDH), a 130 kDa protein, were used to calibrate the gradient. M: mitochondria. Ex: total mitochondrial extract. Two exposures are shown for each protein. (e) Co-immunoprecipitation in HEK293T cells co-transfected with mitofilin-myc and SLC25A46-HA or SLC25A46-HA and myc-tagged mitochondria-targeted GFP as negative control.
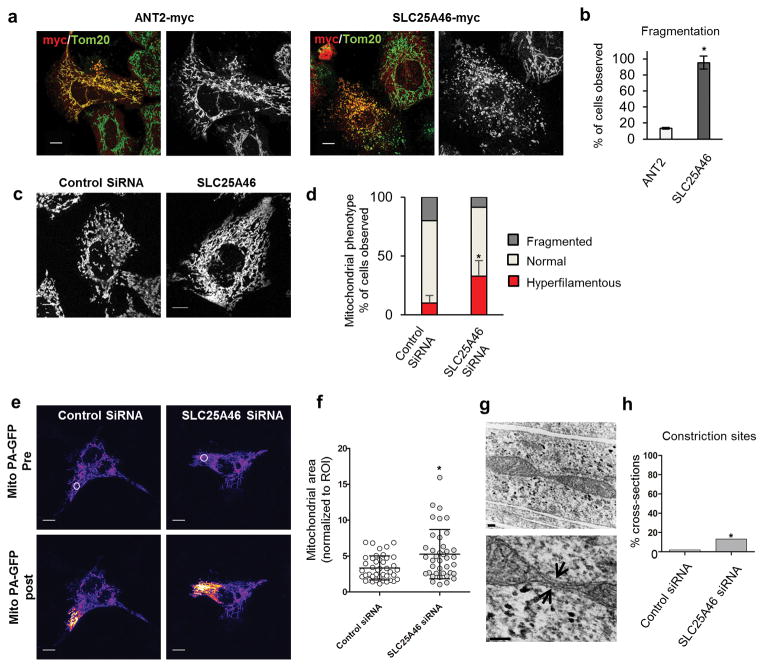
To determine if SLC25A46 plays a conserved role in mitochondrial dynamics, we compared the effects of protein overexpression to the ATP transporter ANT2 (also known as SLC25A5) and found that SLC25A46, unlike ANT2, leads to mitochondrial fragmentation in cell lines (Fig. 3a,b). Next we used siRNA to knockdown SLC25A46 and found it to cause mitochondrial hyperfusion (Fig. 3c,d). To determine the degree of mitochondrial connectivity, we observed the instantaneous diffusion of matrix targeted photoactivatable GFP, revealing that the GFP diffuses into larger areas of the knockdown cells versus controls (Fig. 3e,f).
Figure 3.
SLC25A46 levels regulate mitochondrial morphology. (a) SLC25A46 overexpression causes fragmentation of the mitochondrial network in HeLa cells in comparison to ANT2. (b) Percentage of transfected cells with fragmented mitochondria from three independent experiments. Error bars indicate standard deviation (s.d.). One-sided T-test was used to determine significance. P value = 0.00004. ANT2 n=22; SLC25A46 n=21. (c) HeLa cells treated with siRNAs and stained with Tom20. Mitochondria are more hyperfilamentous when the SLC25A46 is knocked down. (d) Quantification of mitochondrial morphology. Mean and s.d. were calculated from three independent experiments. Control siRNA n=219; SLC25A46 siRNA n=214 (e) The instantaneous diffusion of matrix-targeted, photoactivated GFP was used to assess mitochondrial connectivity. The region of interest (ROI) white circle in the pre-activation image was targeted with the laser, while the corresponding signal in the post-activation image represents the extent of the mitochondrial connectivity. (f) Quantification of the activated mitochondrial area in the post-activation image normalized to the ROI of the pre-activated image. Error bars represents s.d. and P value was calculated by one tailed T-test P = 0.0009. Control siRNA n=42; SLC25A46 siRNA n=38. (g) Representative electron micrographs of a mitochondrial cross-section with constriction sites (arrows) in SLC25A46 siRNA treated COS-7 cells. (h) Percentage of cross-sections with visible constriction sites; 2 out of 103 control versus 11 out 81 in the knockdown. Error bars represents s.d. and significance was determined by a 2-tailed Fisher’s exact test, P value 0.0028. Scale bars (a,c,e) = 10 μm; (g) = 100 nm top, 200 nm bottom.
Assuming that mitochondrial hyperfusion is controlled by the balance between fusion and fission, this phenotype could be caused by either increased fusion or loss of fission. To test the rate of mitochondrial fusion, we performed photoactivation experiments22 where we measured the extinction of activated GFP over the course of 90 minutes and found no significant difference in the fusion rates between control and knockdown cells (Supplementary Fig. 7). This in conjunction with the mitochondrial constriction sites in electron microscopy (Figure 3g,h), which appear to be fission intermediates23 suggests that delays in fission or elongation contribute more to the hyperfused morphology than an increase in fusion rate.
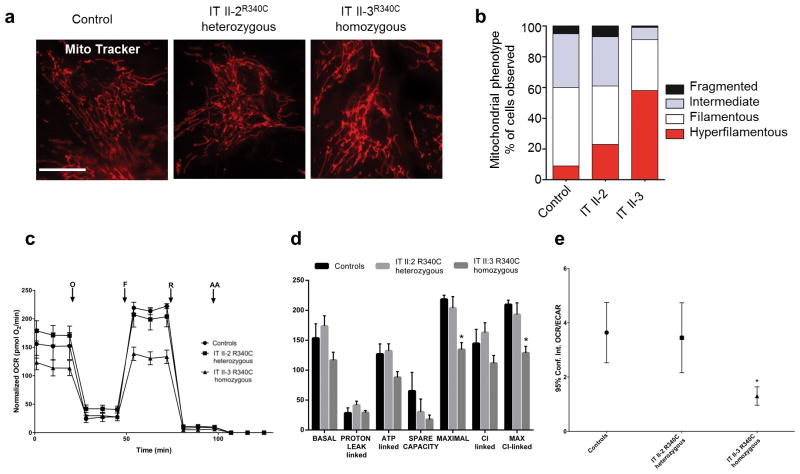
We further examined fibroblasts from proband IT II-3, which display a hyper-filamentous and interconnected network in comparison to both, the unaffected sibling IT II-2 and control, indicating the relevance of this phenomenon (Fig. 4a,b). Loss-of-function of SLC25A46 is not associated with changes in total cellular ATP, mitochondrial DNA content, or membrane potential in either siRNA-treated cell lines, or a knockdown zebrafish model (Supplementary Fig. 5b–d, or data not shown). However, patient IT II-3 fibroblasts show decreased oxygen consumption rate (OCR) and a glycolytic shift of metabolism (decreased OCR/ECAR ratio) (Fig. 4c–e) consistent with the increased lactate peak found in MRS (Supplementary Fig. 2b). A decrease in ATP synthesis driven by complex I substrates, was trending, but not significant (data not shown). These metabolic changes are congruent to previous findings in patient fibroblasts with OPA1 mutations24.
Figure 4.
Patient fibroblasts have hyper-filamentous mitochondria and are respiratory deficient. (a) Patient fibroblasts from IT II-3, homozygous for SLC25A46R340C, have a hyper-filamentous and interconnected mitochondrial network in comparison to unaffected heterozygous sibling IT II-2 and control. Scale bars = 25 μm. (b) The distribution of mitochondrial morphology into three categories by blind test in human fibroblasts: _P_=0.001, calculated with chi square test for control, IT II-2, and IT II-3. Control n=200; IT II-2 n=200; IT II-3 n = 200. (c) Oxygen consumption rate (OCR) traces in control and mutant fibroblasts, expressed as pMolesO2/min, after the injection of oligomycin (O), FCCP (F), rotenone (R) and antimycin A (AA). The homozygous IT II-3 fibroblasts show reduced OCR compared to controls and heterozygous IT II-2 fibroblasts. OCR values are normalized to protein content. Error bars indicate the standard error of mean (s.e.m.) of four independent experiments. (d) BASAL, PROTON LEAK-linked, ATP-linked, MAXIMAL, Complex I-linked, MAXIMAL Complex I-linked respiration and SPARE CAPACITY were calculated from OCR traces and reported in the graph as mean ± SEM. Dunnett’s test of control vs mutant fibroblasts was performed, * indicates p<0.01. (e) Analysis of OCR/ECAR in basal conditions. Extra-cellular acidification rate (ECAR), is an indicator of lactic acid production. The reduced OCR/ECAR ratio observed in the homozygous IT II-3 fibroblasts suggests the occurrence of a glycolitic shift. Bars indicate the confidence intervals (Conf.Int.). Analysis was performed from 5 replicate wells for each fibroblasts line. Dunnett’s test of control vs mutant fibroblasts was performed, * indicates P = 0.002.
Altogether, our data supports that unlike Ugo1, SLC25A46 acts in a pro-fission manner. Ugo1 physically interacts with both Fzo1 (MFN2) and Mgm1 (OPA1) to coordinate the simultaneous fusion of the outer and inner mitochondrial membranes7. Interestingly, mitochondrial fusion is not tightly coupled in mammalian cells, where the outer and inner membranes can fuse independently of each other25. Consistent with these findings, SLC25A46 does not interact with MFN2 or OPA1, but forms a complex with mitofilin that is independent of MFN2 (Supplementary Fig. 5b). Therefore, it is possible that SLC25A46 still functions as an adaptor protein, but the loss-of-function is more apparent on the fission mechanism where the outer and inner membranes come together at the constriction sites. Alternatively, SLC25A46 could mediate the translocation of an unknown pro-fission factor across the outer membrane, analogous to the translocation of tBID by MTCH2 during apoptosis19,26. Although the function of SLC25A46 in mitochondrial dynamics seem to deviate from the role of Ugo1 in yeast, the conservation of the interaction of both proteins with mitofilin suggest that SLC25A46 and Ugo1 might play similar roles at the cristae junctions.
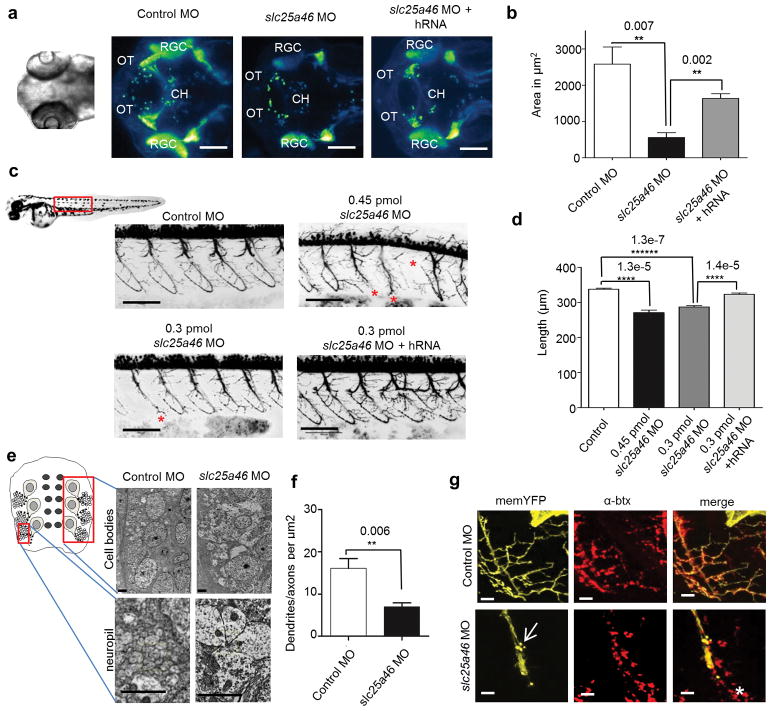
Genome wide association studies have linked SLC25A46 to atopic dermatitis27; however, the transcript has been shown to be highly expressed in the neurons affected in patients including spinal cord, cerebellum, and optic chiasm15. In order to determine the role of SLC25A46 in the vertebrate nervous system, we created a zebrafish knockdown model. Zebrafish slc25a46 is 69% identical to the human ortholog, and in agreement with previous RNA in situ hybridization data15, we found enrichment in retinal ganglion cells (RGCs) and their corresponding neuronal projections in the axons of the optic nerve and in the dendrites of the inner plexiform (Supplementary Fig. 8a–d). We then used a transient morpholino approach to knockdown the transcript and demonstrated that knockdown was effective for up to 96 hours post-fertilization (hpf) (Supplementary Fig. 5c). When injected with slc25a46 morpholino, embryos (morphants) had fewer retinal ganglion cell axons that reached the tectum by 72 hpf (Fig. 5a,b). We also saw decreased antibody staining in the inner plexiform layer suggesting that RGC dendrites are also affected (Supplementary Fig. 8e,f). Around 40% of morphant embryos injected with 0.45 picomoles (pmol) of morpholino have a severe curly-tail morphology (Supplementary Fig. 8g,h); yet, even normal-appearing larvae display alterations in swimming, indicating dysfunction of neuronal circuits (Supplementary Fig. 8i).
Figure 5.
Slc25a46 knockdown in zebrafish affects the growth and maintenance of neurons. (a) Dorsal view of 72 hpf Tg:(Islet2:egfp) embryos. Retinal ganglion cell (RGC) axons cross at the chiasm (CH) before innervating the optic tectum (OT). Scale bar=100 μm. (b) Quantification of the area of RGC-innervated tectum depicting mean and s.d. P values were calculated from a one-tailed Student’s T-test. Control MO n = 3; 0.45 pmol slc25a46 MO n = 3, 0.45 pmol slc25a46 MO + hRNA n = 3. (c) Motor neuron axonsin Tg:(Olig2:DsRed) embryos at 48 hpf. Truncated axons were commonly (*) observed from knockdown. Scale bar = 100 μm. (d) Quantification of the average common axon path length per fish with s.d. between fish. Significance was determined with a one-way ANOVA followed by a Tukey’s post hoc test (Supplementary Table 4). P values were calculated with a one-tailed Student’s T-test. Control MO n = 16; 0.45 pmol slc25a46 MO n = 14; 0.3 pmol slc25a46 MO n = 16; 0.3 pmol slc25a46 MO + hRNA n = 15. (e) Electron micrographs of zebrafish spinal cord (cell bodies top and neuropil bottom) at 48 hpf. Scale bars = 2 μm (f) Quantification of the density of neuronal processes in the neuropil. Data represents mean and s.d. P value calculated with a one-tailed Student’s T-test. Control MO n = 4; 0.45 pmol slc25a46 MO n = 4. (g) Motor neuron terminals labeled with membrane bound YFP and postsynaptic acetylcholine receptors on muscle labeled with α-btx. Axonal blebbing (arrow) and die back indicating by receptors in front of the axon (*) were commonly observed in morphant motor neurons. Scale bar = 10 μm.
We further investigated motor neurons and found significantly shorter axon tracts, many of which failed to innervate the rostral myotome at 48 hpf (Fig. 5c). The spinal cord neuropil at the level of electron microscopy showed fewer neuronal processes in slc25a46 morphants, indicating either lack of development or degeneration of dendrites (Fig. 5e,f). By stochastically labeling the motor neurons, we frequently observed axonal blebbing and dying back at the morphant motor neuron terminals specifying degeneration, which is consistent with the pathology of CMT2 (Fig. 5g). These results suggest that slc25a46 is important for both the growth and maintenance of neuronal processes.
We next investigated the mitochondria in the motor neurons in vivo. Consistent with previous findings28, we observed that in controls the majority of the mitochondria are located within the lower half of motor neuron soma, clustered at the axon initial segment. In contrast, mitochondria in degenerating neurons exhibit a reversal in cell localization favoring the apical portion of the soma and also appear to be aggregated (Fig. 6a–c). At the level of electron microscopy, we identified numerous mitochondria that appeared to be in the process of fission within the cell bodies of morphants, which was never observed in controls (Fig. 6d,e). Incomplete fission of mitochondria could inhibit transport and distribution into neuronal processes29.
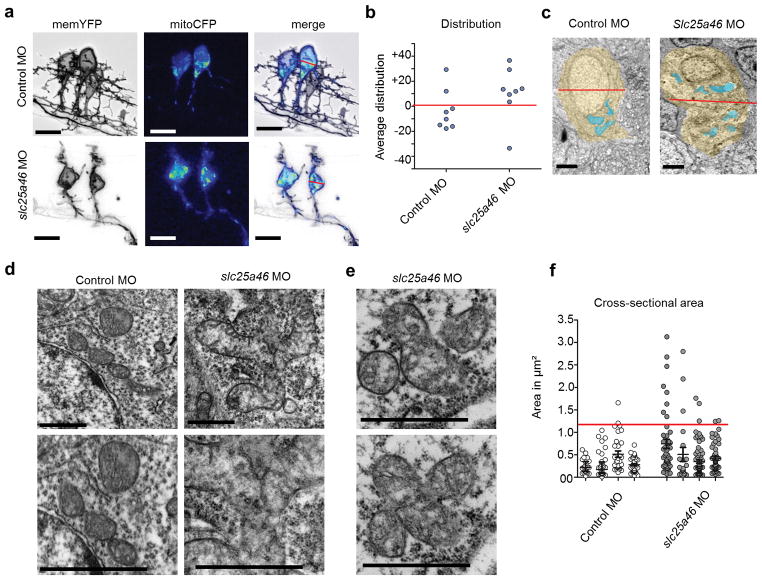
Figure 6.
Mitochondrial distribution and morphology in zebrafish motor neurons at 48 hpf. (a) Zebrafish motor neurons in vivo labeled with plasmids driving mitoCFP (blue) and memYFP (black) under the Hb9 promoter. Scale bar = 10 μm. (b) Graph depicting the distributional density of mitoCFP pixels as a percentage either above or below the midline of the soma defined as 0. In slc25a46 morphant motor neurons, mitochondria are shifted toward the top of the soma. Control MO n = 8; slc25a46 MO n = 8. P value = 0.083 by one-tailed Student’s T-test. The mitochondria in the morphants also appear to be more aggregated which is more apparent in electron microscopy images. (c) Representative electron microscopy images showing mitochondrial distribution. Soma (yellow) and mitochondria (blue). Scale bar = 1 μm. (d) Electron micrographs of mitochondria in spinal cord. Scale bar = 1 μm. Slc25a46 morphants show accumulation of large mitochondrial aggregates that appear to be in the process of fission. (e) Serial section of a clover shaped mitochondrial aggregate, suggesting that these mitochondria are fused at a narrow junction point, only visible in the right plane of section. (f) Cross-sectional areas of mitochondria with distribution of sizes show that larger aggregates are present in slc25a46 morphants. Number of fish; control MO n = 4, slc25a46 MO n = 4. Number of mitochondrial cross-sections; control MO n = 138, slc25a46 MO n = 201. Red line=3 times the s.d. of the average are found in controls. P value = 0.0006 by one-tailed Student’s T-test when comparing total mitochondrial cross-sections per group.
In conclusion, we have identified a novel mitochondrial disease gene associated with mitochondrial dynamics, optic atrophy, and CMT2. Further investigation is needed to elucidate the role of SLC25A46 in mitochondrial dynamics, but given the similar human phenotypes associated with MFN2, OPA1, and SLC25A46 mutations, these genes could be involved in common pathological mechanisms of neurodegeneration, which opens the possibility of future pathway-oriented treatments for these types of mitochondrial disorders.
Online Methods
Whole exome sequencing
Whole genomic DNA was extracted from whole blood by standard methods. Library construction was performed on dsDNA, sheared by sonication to an average size of 200 bp, in an automated fashion on an IntegenX Apollo324. After 9 cycles of PCR amplification using the Clontech Advantage II kit, 1ug of genomic library was recovered for exome enrichment using the NimbleGen EZ Exome V2 kit. Libraries were sequenced on an Illumina HiSeq2500. The Genomes Management Application tool (GEM.app) was used for variant calling using the standard pipe line13.
Sanger sequencing
Sanger sequencing was performed by standard methods on the 8 exons of the SLC25A46 gene (NM_138773.1). Primers were designed using Primer3. PCR products were amplified using 50ng DNA, with standard PCR reagents Econotaq (Lucigen, WI), on an ABI Veriti Thermocycler (Applied Biosystems, Austin, TX). PCR products were precipitated and sequencing PCR performed using BigDye Terminator Ready Reaction Mix (ABI Biosystems).
Immunofluorescence
COS-7 or HeLa cells were seeded to 75% confluence onto glass coverslips and transfected using Lipofectamine 2000 (Invitrogen) according to manufacturer’s protocol. Cells were fixed with paraformaldehyde for 20 min, permeabilized with cold methanol for 5 min and stained with anti-myc (#2276 and #2278) or anti-HA antibodies (2367 and 3724) from Cell Signaling.Tom20 (SC-11415) from Santa Cruz was used to label mitochondrial outer membrane. Corresponding Alexa-fluor-conjugated secondary antibodies (Life Technologies) were used for detection. Coverslips were mounted onto slides with DAPI mounting media (Vectashield), and imaged with a confocal microscope, Zeiss LSM710, 63X/1.4 oil objective with a pinohole of less than one Airy unit and processed with Fiji (Image J).
Mitochondrial subfractionation and proteinase K protection assay
HEK293T cells were grown in DMEM (Gibco) with 10% fetal bovine serum and incubated at 37°C. Cells were transfected with SLC25A46-HA and mitochondria were isolated 24 hours post transfection as previously reported30. To test proteins solubility, purified mitochondria were ruptured by sonication. The membrane fraction was recovered by centrifugation at 20,000 × g for 15 minutes at 4°C and was resuspended in 0.1M Na2CO3 pH 11. The sample was incubated on ice for 30 minutes and both soluble extrinsic membrane proteins and insoluble intrinsic membrane proteins were separated by centrifugation at 20,000 × g for 15 minutes at 4°C. Purified mitochondria were resuspended in 10 mM TrisHCl pH 7, 10mM KCl, 0.15mM MgSO4 buffer either containing 0.25 M sucrose or devoid of sucrose to allow mitochondrial swelling and conversion to mitoplasts31. Samples were treated with proteinase K at final concentration of 0.62 μg/ml and incubated on ice for 30 min. The reaction was inhibited with 2mM phenylmethylsulfonyl fluoride (PMSF). Mitochondria and mitoplasts were recovered by centrifugation at 20,000 × g for 15 min at 4°C and analyzed by Western blotting.
Sucrose gradient analysis
Four hundred μg of purified mitochondria were solubilized in 0.08 mL of extraction buffer (20mM TrisHCl pH 7.4, 100 mM KCl, 1 mM MgSO4, 0.5 mM PMSF) containing 1% digitonin, on ice for 15 minutes. The clarified extract was obtained by centrifugation at 20,000 × g for 15 minutes at 4°C, mixed with standard proteins (hemoglobin and lactate dehydrogenase) and applied to a linear 7–20% sucrose gradient prepared in extraction buffer containing 0.1% digitonin. Following centrifugation for 12 hours at 28,000 r.p.m. in a Beckman 55Ti rotor, the gradient was fractionated in 12 equal fractions. Each fraction was assayed for hemoglobin by absorption at 409 nm and lactate dehydrogenase activity by measuring NADH-dependent conversion of pyruvate to lactate. Subsequently, proteins contained in each fraction were concentrated by TCA precipitation and analyzed by immunoblotting.
Immunoprecipitation and mass spectrometry
HEK293T cells were transfected with SLC25A46-HA using Turbofect reagent (Thermo Scientific). After 24 hr cells were lysed with a dounce homogenizer and mitochondria were isolated with the mitochondria isolation kit for cultured cells (Pierce) following manufacture’s protocol. 800 ug of protein from mitochondria were incubated with 5 ug of anti-HA tag mouse (Cell signaling ab9110) at room temperature for 2 h. Then, the proteins/antibody mixture was incubated with protein A/G magnetic beads (Thermo Scientific) at room temperature for 1 hr on a rotator. Beads were collected using a magnetic stand and washed 3X with manufacturer’s IP Lysis/wash buffer. Proteins were eluted with a low PH elution buffer. Silver staining was performed using the Pierce Silver stain kit (Thermo scientific). Briefly, SDS-PAGE gel was washed 2X with ultra-pure water for 5 min and fixed with 30% ethanol: 10% acetic acid solution. Samples were analyzed by NuPAGE® (Invitrogen) _SDS_-PAGE Gel (4–20%) incubated with silver staining solution for 5 min followed by the developing reagent for 3 min. Reaction was stopped with 5% acetic acid for 10 min. Bands were excised from the gel using a light box and submitted for mass spectrometry analysis at Scripps Center for Metabolomics and Mass Spectrometry.
Co-immunoprecipitation
HEK293T cells were double transfected with SLC25A46-HA and mitofilin-myc or as a negative control SLC25A46 and pCMV/myc/mitoGFP (Invitrogen). After 24 hr cells were lysed with a dounce homogenizer and mitochondria were isolated with the mitochondria isolation kit for cultured cells (Pierce) following manufacture’s protocol. 800 ug of protein from mitochondria were incubated with 5 μg of either anti-HA mouse, anti-myc rabbit (Cell signaling), or control mouse and rabbit IgG at room temperature for 2 h. Then, the proteins/antibody mixture was incubated with protein A/G magnetic beads (Thermo Scientific) at room temperature for 1 hr on a rotator. Beads were collected using a magnetic stand and washed 3X with manufacturer’s IP Lysis/wash buffer. Proteins were eluted with a low PH elution buffer. Protein samples were analyzed by NuPAGE® (Invitrogen) _SDS_-PAGE Gel (4–20%) followed by western blot using appropriate antibodies. Band signal was developed with either chemiluminescent substrates West Pico or West Femto (Thermo).
Overexpression and knockdown in mammalian cells
Cells were plated to 70% confluence on glass coverslips in a 6 well plate. Plasmid transfections were carried out using Lipofectamine 2000 (Invitrogen) according to standard protocol. Immunofluorescence was carried out as previously described. Representative single plane images were acquired with a Zeiss LSM710 confocal microscope, using a 63X/1.4 oil objective processed with Fiji (Image J) and blindly assessed for fragmentation. For knockdown, cells were plated to 50% confluence onto glass coverslips in a 6 well plate. Silencer Select (Ambion) SLC25A46 (s40579) and negative control No.1 (4390843) siRNAs, were used. 25 pmol per well were transfected with Lipofectamine RNAi max (Invitrogen) according to manufactures protocol. Effective knockdown of the transcript was confirmed with RT-PCR and Q-PCR using gene specific primers (Supplementary Table 5). After 48 hours Cells were washed in PBS and fixed in 4% PFA and Immunofluorescence was carried out as previously described. Images were taken using a Zeiss LSM710, using a 63X/1.4 oil objective and processed with Fiji (Image J) LUT: grays. Representative images were assessed blindly.
Mitochondrial morphology analysis and energetic profiling of human fibroblasts
Fibroblasts were seeded onto 36 mm diameter dishes and grown in DMEM. Mitochondrial morphology was assessed by staining cells with 10nM Mitotracker Red (Life Technologies) for 30 min at 37°C. Cellular fluorescence images were acquired with an inverted Nikon Eclipse Ti-U epifluorescence microscope equipped with a back- illuminated Photometrics Cascade CCD camera (Roper Scientific). Images were collected using a 63X/1.4 oil objective. Data were acquired and analyzed using the Metamorph software (Universal Imaging Corp.). Oxygen consumption rate (OCR) and extra-cellular acidification rate (ECAR) were measured in adherent fibroblasts with a XFe24 Extracellular Flux Analyzer (Seahorse Bioscience). Each control and mutant fibroblast cell lines were seeded in 5 wells of a XF 24-well cell culture microplate (Seahorse Bioscience) at a density of 30.000 cells/well in 250 μL of DMEM and incubated for 24 hours at 37 °C in 5% CO2 atmosphere. After replacing the growth medium with 575 μL of bicarbonate-free DMEM, pH 7.3, pre-warmed, cells were incubated at 37 °C for 1 hour before starting the assay.
After baseline measurements of OCR and ECAR, OCR was measured after sequentially adding to each well 75 μL of oligomycin (O) and 75 μl of carbonyl cyanide 4-(trifluoromethoxy) phenylhy- drazone (FCCP), 75 ul of rotenone (R) and 75ul of Antimycin A (AA) to reach working concentrations of 1μM. OCR and ECAR values were normalized on protein content measured by Sulforhodamine (SRB) assay, following standard protocol. For data analysis the following parameters were evaluated32: BASAL RESPIRATION: measurement prior oligomycin injection minus non-mitochondrial respiration (measurement after antimycin A injection); PROTON LEAK-LINKED RESPIRATION: measurement after oligomycin injection minus non-mitochondrial respiration; ATP-LINKED RESPIRATION: basal respiration minus proton leak; MAXIMAL RESPIRATION: measurement after FCCP minus non-mitochondrial respiration; SPARE CAPACITY: maximal respiration minus basal respiration; CI-LINKED RESPIRATION: basal - measurement after rotenone; MAXIMAL CI-LINKED RESPIRATION: maximal respiration minus measurement after rotenone.
Photoactivation assay
The Mito-PA-GFP assay was performed as previosly described22, using a Zeiss LSM710 with a 63X/1.4 oil objective, heated stage, and carbon dioxide chamber. After acquistion of the preactivated image, a circle of aproxiamalty 5 μm was selected using the regions setup and was activated with the 405nm laser. The postactivation image was imediatley aquired using the 488 nm laser and subsquent images were taken at 30 minute increments for a total of 90 minutes. Images were processed in Fiji and dispalyed with LUT:fire. For the mitochondrial fusion assay the mean pixel intestity was calcuated for the region of interest (ROI) and normalized to the postactivation image. For the assemement of the mitochondrial conectivity, the area of the activated GFP was measured in the post activation image and normalized to the area of the activated region.
Zebrafish husbandry
Experiments were carried out using Danio rerio wild type strains: AB, TL, Tubingen and the transgenic stains, Tg(Olig2:DsRed)33 and Tg:(Islet2b:GFP)34. Adults were kept on a 14 hour light/10 hour dark cycle at 28°C. Embryos were obtained from natural crosses after removing a divider at first light and microinjections were performed into one-cell stage embryos. Embryos were reared in petri dishes of system water in a 28°C incubator with the same light dark cycle. All experiments were conducted in accordance with the University of Miami Institutional Animal Care and Use Committee guidelines.
Microinjections
The slc25a46 morpholino (Gene Tools) was targeted to the exon 3/intron 3 junction. Effective splice blocking was confirmed with diagnostic primers (Supplementary Table 5) and found to lead to the exclusion of exon 3 resulting in a frame-shift and premature stop. The standard scrambled morpholino (Gene Tools) was used for the control. 1mM stock solutions of morpholinos were diluted in 1% fast-green dye to a final concentration of 0.9 mM. Injection volumes of 333 pL or 500 pL were calibrated for a final injected amount of 0.3 picomoles and 0.45 picomoles (pmol) respectively. For the rescue experiments, human SLC25A46-HA mRNA was synthesized from the PCS2+ plasmid using mMessage Machine SP6 Kit (Ambion) and 400pg were injected.
In vivo imaging of optic nerve
Retinal ganglion cells were imaged at 72 hpf after injected slc25a46 and control morpholinos into Tg(Islet2b:eGFP)34 embyros. At 24 hpf, n-phenylthiourea (Sigma) was added to the water to suppress pigment development. Live fish were anesthetized with tricaine methanesulfonate (Sigma), embedded in 1.5% agarose, and imaged using a Leica confocal microscope 20X/air lens. 1 μm Z-stacks were processed with maximum intensity and thresholded for analysis. Calculations were made by tracing GFP positive areas. All possessing was done with Fiji (Image J). LUT: Green Fire Blue was used to make the figure.
In vivo imaging of motor neurons
Motor neuron outgrowth was assayed at 48 hpf using Tg(Olig2:Dsred)33 fish. Live fish were anesthetized with tricaine methanesulfonate (Sigma), imbedded in 1.5% agarose, and imaged using a Leica confocal microscope 20X/air lens and processed with Fiji (Image J). For analysis 1 μm Z-stacks were imaged between myotome segments 6 and 1335 and the lengths of the caudal anterior primary (CaP) axons were measured from the base of the spinal cord to the end of the rostral myotome using the Simple Neurite Tracer in Fiji (Image J). LUT: edges was used to make the figure. To stochastically label motor neurons and mitochondria, 300pL of a cocktail containing 25 ng/ul of each: HB9:Gal4-VP16, UAS-E1b:mYFP:mitoCFP 36, and transposase RNA was injected prior to the morpholinos. Embryos were presorted for YFP expression at 48 hpf before imaging. Acetylcholine receptors were labeled in 48 hpf embryos by injecting, α-bungarotoxin conjugate (Molecular Probes) into the yolk of anesthetized fish an hour before imaging with Leica confocal microscope, 63x/water lens. Motor neuron cell bodies and mitochondria were processed with LUT:edges/memYFP and 16 colors/mitoCPF. For mitochondrial distribution analysis, 1 μm Z-stacks were maximum intensity projected and auto-thresholded. The midline was measured as the half-way point between the top of the soma and the axon hillock. The area of mitochondrial pixels was calculated for the region above and below the midline, which was then converted to a percentage using Fiji (image J).
Transmission electron microscopy and analysis
COS-7 cells were cultured in 25 cm2 flasks (Corning) and treated with siRNAs as previously described. After 2 days, cells were fixed for four days in 2.5% glutaraldehyde in Millonigs phosphate buffer37, post-fixed in 1% OsO4 and uranyl acetate followed by dehydration in an ethanol series and embedded in Spurs. Zebrafish embryos were injected with 0.45 pmoles of either control or slc25a46 morpholino. The more severely affected curly morphants were selected for EM. All embryos were fixed and processed at 48 hpf as previously described, with the exception that they were embedded in LR white resin (Fisher Scientific). Ultrathin, 60 nm sections were made using a Leica microtome and were stained with cold lead citrate for 6 minutes.37 Images were acquired with a Joel JEM-1400 transmission electron microscope with Gatan camera. All images were processed and analyzed using Fiji (image J).
Plasmids
HB9:Gal4-VP16 was generated from the pmT hb9s YFP kindly provided by the Nonet Lab at Wash U. UAS-E1b:mYFP,mitoCFP was kindly provided by Bettina Schmid36 from the German Center for Neurodegenerative Diseases (DZNE). SLC25A46 and mitofilin-myc were obtained from (Origene). SLC25A46 was subcloned into pcDNA3.1 (Invitrogen) with either C-terminal HA or myc. For RNA synthesis SLC25A46-HA was also subcloned into PCS2+.
Phylogenetic analysis
The concise list of the human the mitochondrial carrier (SLC25) family was obtained from the Bioparadigms web site, while all mitochondrial carrier domain containing proteins of C.elegans, S. cerevisiae, and S. pombe were retrieved from the European Bioinformatic institute. We used Multiple Sequence Alignment by CLUSTALW to draw unrooted neighbor-joining trees. We then created a maximum-likelihood tree using PHYML by Mobyle, with 1000 bootstraps. The consensus tree was visualized and drawn using iTol.
ATP and mtDNA analysis
ATP was measured according to manufactures protocol using the ATP Determination Kit (Molecular Probes). COS-7 cells were transfected with siRNAs as previously described and homogenized in ATP assay buffer at 48 hours post transfection, or 5 whole zebrafish embryos were homogenized in ATP assay buffer at 24 hours post fertilization. Homogenates were spun at 12,000 g for 5 min to pellet tissues debris. 10 ul supernatant were added to a 97 well dish containing 90 ul of the luciferase reaction mix from the ATP Determination Kit (Molecular Probes). Luminescence was immediately read on a luminometer. ATP values were calculated by comparison to a standard curve generated from a series of ATP concentrations. Values were then normalized by the total amount of protein present in each sample. Samples were run in triplicate. mtDNA levels were quantified by quantitative PCR as described38.
RNA in situ hybridization
For RNA in situ hybridization, wild-type embryos were incubated in n-phenylthiourea (Sigma) to suppress pigment development. The slc25a46 probe sequence was PCR amplified from zebrafish cDNA (Supplementary Table 5) and the DIG RNA Labeling Mix (Roche) was used to synthesize the antisense probe. Probe hybridization and development were carried out using standard protocol39, followed by post fixation in 4% PFA and dehydration in 30% sucrose. Embryos were embedded in tissue freezing media (Triangle Biomedical Sciences), sliced with a cryostat into 30μm sections, and imaged with an Olympus BX60 compound light microscope.
Retinal ganglion cell staining
Morpholino injected larvae were fixed at 96 hpf in 4% PFA overnight and then decapitated. Zn-5 antibody (ZFIN) was used at 1:200 followed by incubation with 1:200 HRP-conjugated secondary and finally developed with 3,3′-Diaminobenzidine DAB (Vector). After DAB development fish were depigmented with H2O2 and KOH, post fixed and dehydrated in ethanol. Samples were embedded in freeing media, sliced into 30μm sections, and imaged with an Olympus BX60 compound light microscope. 8-bit grey scale numbers were calculated from TIF files of sections contained the optic nerve using Fiji image analysis such that 0=white and 256=black and was converted to percentage whereby 0=white 100=black. 5 evenly spaced pixels from the inner plexiform layer were measured to find the average intensity per eye.
Analysis of swimming
High-speed videos were taken using a Fastcam 1024PCI (Photron USA Inc., San Diego, CA, USA) with a Fujinon lens mounted in a customized behavioral chamber. Parameters were as follows: shutter speed of 1/1000 using an LED array for backlit illumination (Advance Illumination; Rochester VT; Backlight LED Illuminator), 512 × 512 resolution, a frame rate of 500 f/s to assess swimming kinematics. Three vibration-evoked behaviors, elicited with S40 stimulator using DAC timer, were recorded from a 35 mm dish containing up to 5 larvae. Videos were acquired from multiple runs containing more than 30 total larvae. Videos were analyzed using Flote40 to quantitate changes in axis curvature over time. Three representative overlapping swim traces from different fish were chosen from over 8 analyzable swim responses.
Supplementary Material
1
2
Acknowledgments
We would like to thank all families for participating in this study. We thank Prof. Antonio Torroni and Dr. Anna Olivieri for their kind help with the control population samples from Sardinia. From the University of Miami, we are thankful to Ricardo Cepeda for his excellent care and maintenance of the zebrafish facility, Jeffery Prince’s transmission electron microscopy class, Rebecca Duncan, William Browne, and Pantelis Tsouflas for advice concerning phylogenic analysis. Fabienne Poulain from the University of Utah for providing the Islet2-b fish and Bettina Schmid from the German Center for Neurodegenerative Diseases (DZNE) for donating the mitofish construct. This study was supported by NIH R01NS075764, 5R01NS072248, and U54NS065712 to S.Z. and by the CMT Association, and the Muscular Dystrophy Association. A.J.A received additional support from the Lois Pope Life Foundation development award. This study was partially supported by CCHMC Research Foundation and Division Human Genetics to T.H., R01DC012564 for Z.M.A. and by ERMION, E-RARE European consortium on optic atrophies to V.C. A.H.N. and S.M.D. were funded by the UK Oxford Partnership Comprehensive Biomedical Research Centre with funding from the Department of Health’s NIHR Biomedical Research Centers funding scheme. A.A. is supported by the National Institute of Neurological Disorders and Stroke and L.B.G. is supported by the National Institutes of Health Cellular and Molecular Biology Training Grant T32 GM007315, the National Institutes of Health Medical Scientist Training Grant T32 GM07863, and by NIH F30 NRSA NS092238. A.B. is supported by grants from the National Institutes of Health (NIH) RO1 GM071775, GM105781 and GM112179. F.F. is supported by a Development grant from the American Heart Association.
Footnotes
Accession codes
Human SLC25A46: Q96AG3
Zebrafish slc25a46: Q6DGU5
Competing financial interests
The authors declare no competing financial interests
Author contributions
A.H.N, S.Z., and J.D. initiated the project, which was subsequently developed and led jointly by J.D., S.Z., T.H., and V.C. Experiments were conceived and carried out by A.J.A, A.R., C.Z., N.P., F.V., A.B., R.B.H., L.B.G., A.M., R.S. with financial support from S.Z., T.H., A.A., and J.P. Zebrafish experiments were carried out by A.J.A., I.J.C, and S.G with financial supported from S.Z. and J.D. Patient recruitment, collection and analysis of human DNA, and genetic data was carried out by M.A.G., F.S., L.A., S.M.D, L.C., C.L.M, R.L., Z.M.A., K.L.S., X.W., L.A.K, Y.P, C.E.P, C.A.P, E.K.S., H.H.Z., O.A.A., and Y.Y. Functional MRI-MRS data was carried out by R.L. and Z.M.A. A.J.A wrote the manuscript with critical input from S.Z. and J.D.
References
- 1.Delettre C, Lenaers G, Pelloquin L, Belenguer P, Hamel CP. OPA1 (Kjer type) dominant optic atrophy: a novel mitochondrial disease. Mol Genet Metab. 2002;75:97–107. doi: 10.1006/mgme.2001.3278. [DOI] [PubMed] [Google Scholar]
- 2.Alexander C, et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. 2000;26 doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- 3.Züchner S, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–51. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 4.Chan DC. Dissecting mitochondrial fusion. Dev Cell. 2006;11:592–4. doi: 10.1016/j.devcel.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Sesaki H, Jensen RE. UGO1 encodes an outer membrane protein required for mitochondrial fusion. J Cell Biol. 2001;152:1123–34. doi: 10.1083/jcb.152.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sesaki H, Jensen RE. Ugo1p links the Fzo1p and Mgm1p GTPases for mitochondrial fusion. J Biol Chem. 2004;279:28298–303. doi: 10.1074/jbc.M401363200. [DOI] [PubMed] [Google Scholar]
- 7.Van der Bliek AM, Shen Q, Kawajiri S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a011072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braathen GJ. Genetic epidemiology of Charcot-Marie-Tooth disease. Acta Neurol Scand Suppl. 2012;126:iv–22. doi: 10.1111/ane.12013. [DOI] [PubMed] [Google Scholar]
- 9.Züchner S, et al. Axonal neuropathy with optic atrophy is caused by mutations in mitofusin 2. Ann Neurol. 2006;59:276–81. doi: 10.1002/ana.20797. [DOI] [PubMed] [Google Scholar]
- 10.Yu-Wai-Man P, Griffiths PG, Hudson G, Chinnery PF. Inherited mitochondrial optic neuropathies. J Med Genet. 2009;46:145–58. doi: 10.1136/jmg.2007.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amati-Bonneau P, et al. OPA1 mutations induce mitochondrial DNA instability and optic atrophy “plus” phenotypes. Brain. 2008;131:338–51. doi: 10.1093/brain/awm298. [DOI] [PubMed] [Google Scholar]
- 12.Züchner S, Vance JM. Emerging pathways for hereditary axonopathies. 2005:935–943. doi: 10.1007/s00109-005-0694-9. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez Ma, et al. GEnomes Management Application (GEM.app): A New Software Tool for Large-Scale Collaborative Genome Analysis. Hum Mutat. 2013;34:842–6. doi: 10.1002/humu.22305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmieri F. The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol Aspects Med. 2013;34:465–84. doi: 10.1016/j.mam.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Haitina T, Lindblom J, Renström T, Fredriksson R. Fourteen novel human members of mitochondrial solute carrier family 25 (SLC25) widely expressed in the central nervous system. Genomics. 2006;88:779–90. doi: 10.1016/j.ygeno.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 16.Hoppins S, Horner J, Song C, McCaffery JM, Nunnari J. Mitochondrial outer and inner membrane fusion requires a modified carrier protein. J Cell Biol. 2009;184:569–81. doi: 10.1083/jcb.200809099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coonrod EM, Karren MA, Shaw JM. Ugo1p is a multipass transmembrane protein with a single carrier domain required for mitochondrial fusion. Traffic. 2007;8:500–11. doi: 10.1111/j.1600-0854.2007.00550.x. [DOI] [PubMed] [Google Scholar]
- 18.Lamarca V, et al. Two isoforms of PSAP/MTCH1 share two proapoptotic domains and multiple internal signals for import into the mitochondrial outer membrane. 2007:1347–1361. doi: 10.1152/ajpcell.00431.2006. [DOI] [PubMed] [Google Scholar]
- 19.Zaltsman Y, et al. MTCH2/MIMP is a major facilitator of tBID recruitment to mitochondria. Nat Cell Biol. 2010;12:553–62. doi: 10.1038/ncb2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson AJ, Kunji ERS, Gross A. Mitochondrial carrier homolog 2 (MTCH2): the recruitment and evolution of a mitochondrial carrier protein to a critical player in apoptosis. Exp Cell Res. 2012;318:1316–23. doi: 10.1016/j.yexcr.2012.01.026. [DOI] [PubMed] [Google Scholar]
- 21.Harner M, et al. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 2011;30:4356–70. doi: 10.1038/emboj.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karbowski M, et al. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J Cell Biol. 2004;164:493–9. doi: 10.1083/jcb.200309082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polyakov VY, Soukhomlinova MY, Fais D. Fusion, fragmentation, and fission of mitochondria. Biochem Biokhimiiȃ. 2003;68:838–49. doi: 10.1023/a:1025738712958. [DOI] [PubMed] [Google Scholar]
- 24.Zanna C, et al. OPA1 mutations associated with dominant optic atrophy impair oxidative phosphorylation and mitochondrial fusion. Brain. 2008;131:352–67. doi: 10.1093/brain/awm335. [DOI] [PubMed] [Google Scholar]
- 25.Song Z, Ghochani M, Mccaffery JM, Frey TG, Chan DC. Mitofusins and OPA1 Mediate Sequential Steps in Mitochondrial Membrane Fusion. 2009;20:3525–3532. doi: 10.1091/mbc.E09-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shamas-Din A, et al. Multiple partners can kiss-and-run: Bax transfers between multiple membranes and permeabilizes those primed by tBid. Cell Death Dis. 2014;5:e1277. doi: 10.1038/cddis.2014.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirota T, et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet. 2012;44:1222–6. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- 28.Li YC, et al. Mitochondrial accumulation in the distal part of the initial segment of chicken spinal motoneurons. Brain Res. 2004;1026:235–43. doi: 10.1016/j.brainres.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 29.Narendra DP, Youle RJ. Neurodegeneration: Trouble in the cell’s powerhouse. Nature. 2012:418–419. doi: 10.1038/nature10952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tu YT, Barrientos A. The Human Mitochondrial DEAD-Box Protein DDX28 Resides in RNA Granules and Functions in Mitoribosome Assembly. Cell Rep. 2015;10:854–864. doi: 10.1016/j.celrep.2015.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fontanesi F, Clemente P, Barrientos A. Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J Biol Chem. 2011;286:555–66. doi: 10.1074/jbc.M110.188805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Divakaruni AS, Paradyse A, Ferrick Da Murphy AN, Jastroch M. Methods Enzymol. Vol. 547. Elsevier Inc; 2014. Analysis and interpretation of microplate-based oxygen consumption and pH data; pp. 309–54. [DOI] [PubMed] [Google Scholar]
- 33.Kucenas S, Snell H, Appel B. nkx2.2a promotes specification and differentiation of a myelinating subset of oligodendrocyte lineage cells in zebrafish. Neuron Glia Biol. 2010:4. doi: 10.1017/S1740925X09990123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poulain FE, Chien CB. Proteoglycan-mediated axon degeneration corrects pre-target topographic sorting errors. 2014;78:49–56. doi: 10.1016/j.neuron.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers PZ, Eisen JS, Westerfield M. Development Zebrafish and Axonal Outgrowth of Identified Motoneurons in the. 1986:6. doi: 10.1523/JNEUROSCI.06-08-02278.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plucińska G, et al. In vivo imaging of disease-related mitochondrial dynamics in a vertebrate model system. J Neurosci. 2012;32:16203–12. doi: 10.1523/JNEUROSCI.1327-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prince J, Nolen TG, Coelho L. Defensive ink pigment processing and secretion in Aplysia californica: concentration and storage of phycoerythrobilin in the ink gland. J Exp Biol. 1998;201:1595–613. doi: 10.1242/jeb.201.10.1595. [DOI] [PubMed] [Google Scholar]
- 38.Rebelo AP, Williams SL, Moraes CT. In vivo methylation of mtDNA reveals the dynamics of protein-mtDNA interactions. Nucleic Acids Res. 2009;37:6701–15. doi: 10.1093/nar/gkp727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 40.Burgess Ha, Granato M. Modulation of locomotor activity in larval zebrafish during light adaptation. J Exp Biol. 2007;210:2526–39. doi: 10.1242/jeb.003939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2