Expanding the molecular and clinical phenotype of SSR4-CDG (original) (raw)
. Author manuscript; available in PMC: 2016 Nov 1.
Published in final edited form as: Hum Mutat. 2015 Aug 27;36(11):1048–1051. doi: 10.1002/humu.22856
Abstract
Congenital disorders of glycosylation (CDG) are a group of mostly autosomal recessive disorders primarily characterized by neurological abnormalities. Recently, we described a single CDG patient with a de novo mutation in the X-linked gene, Signal Sequence Receptor 4 (SSR4). We performed whole exome sequencing to identify causal variants in several affected individuals who had either an undifferentiated neurological disorder or unsolved CDG of unknown etiology based on abnormal transferrin glycosylation. We now report, eight affected males with either de novo (4) or inherited (4) loss of function mutations in SSR4. Western blot analysis revealed that the mutations caused a complete loss of SSR4 protein. In nearly all cases, the abnormal glycosylation of serum transferrin was only slightly above the accepted normal cutoff range.
Keywords: congenital disorders of glycosylation, signal sequence receptor 4, SSR4, translocon complex, carbohydrate deficient transferrin
Congenital disorders of glycosylation (CDG) represent a growing number of mostly autosomal recessive (AR) disorders characterized by altered protein or lipid glycosylation (Freeze et al., 2014). Several examples of non-AR CDG include autosomal dominant mutations in EXT1 (MIM# 608177) and EXT2 (MIM# 608210) [Delgado et al., 2014], while mutations in TUSC3 (MIM# 601385) [Garshasbi et al., 2008; Molinari et al., 2008], SSR4 (MIM# 300934) [Losfeld et al., 2014], ALG13 (MIM# 300884) [Timal et al., 2012] and SLC35A2 (MIM# 300896) [Ng et al., 2013] cause X-linked disorders. To date over one hundred glycosylation-related disorders have been identified and > 50 involve the N-linked glycosylation pathway (Freeze et al., 2014). The majority of patients with N-linked defects are identified through a variety of biochemical methods usually involving the testing of an abundant serum glycoprotein, transferrin. Carbohydrate deficient transferrin (CDT) is a reliable and inexpensive test for identifying CDG cases; however, it cannot identify the specific gene defect. Patients can have a type I transferrin (Tf) pattern, which indicates a defect in the synthesis or transfer of the glycan from lipid-linked oligosaccharide carrier and results in the occasional absence of entire N-glycan chains. Alternatively, a type II pattern results from abnormal processing of protein bound N-glycans due to abnormal Golgi homeostasis and/or defects in trafficking complexes (Freeze, 2013). CDG patients may also be identified by genetic analysis and sometimes later confirmed by CDT. Combining both approaches is optimal.
The Translocon-Associated Protein (TRAP) complex is composed of four Signal Sequence Receptor subunits (SSR1, SSR2, SSR3 and SSR4). They form heterotetramers specifically localized to endoplasmic reticulum (ER) membrane sites where nascent secretory proteins enter the ER lumen. (Hartmann et al., 1993) These sites are referred to as translocation sites. (Hartmann et al., 1993) In addition to the TRAP complex, both the SEC61 and oligosaccharyltransferase (OST) complexes participate in the translocation process. The SEC61 complex is essential for formation of a membrane channel (Jungnickel and Rapoport, 1995), and the OST complex catalyzes co-translational transfer of N-glycans (Zimmermann et al., 2011; Pfeffer et al., 2014). While one line of evidence suggests that the TRAP complex may be required for increasing the efficiency of both the SEC61 and OST complexes, other data suggest it may play a role in how membrane proteins are inserted into the translocon (Sommer et al., 2013). Proteomic analysis has shown that these three complexes co-purify and likely work in concert for proper translocon function. (Shibatani et al., 2005)
Rare genetic disorders have been described in five subunits of the OST complex TUSC3 (MIM# 601385) [Garshasbi et al., 2008; Molinari et al., 2008], MAGT1 (MIM# 300715) [Li et al., 2011], DDOST (MIM# 602202) [Jones et al., 2012], STT3A (MIM# 601134) and STT3B (MIM# 608605) [Shrimal et al., 2013], but none in the SEC61 complex. Recently, exome sequencing identified the first and only reported case of TRAP complex deficiency, which involved a de novo mutation in SSR4 (MIM# 300934) [Losfeld et al., 2014]. And only one year later we report eight additional cases.
Once consent was provided in accordance with The Sanford – Burnham – Prebys Medical Discovery Institute Institutional Review Board, we applied exome sequencing to a cohort of undiagnosed type I CDG cases as well as individuals with unsolved neurological disorders. In some instances exome sequencing or copy number variation (CNV) analysis had already been performed and data were shared. For cDNA numbering, the use of +1 represents the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1. All variants have been deposited in the LOVD3.0 (http://databases.lovd.nl/).
Exome sequencing was performed on both affected male and female individuals by several different institutions and identified five patients (P1, P2, P5, P6, P7) suspected of SSR4-CDG. Patient 1 was part of a sequencing study involving fifty-seven confirmed CDG cases that underwent exome sequencing and analysis at the University of Washington Center for Mendelian Genomics. P2 underwent exome sequencing at Mendelics Genomic in Brazil. Exome on patients (5–7) was performed at Baylor Miraca Genetics Laboratories. Only one family (405) did not undergo exome sequencing.
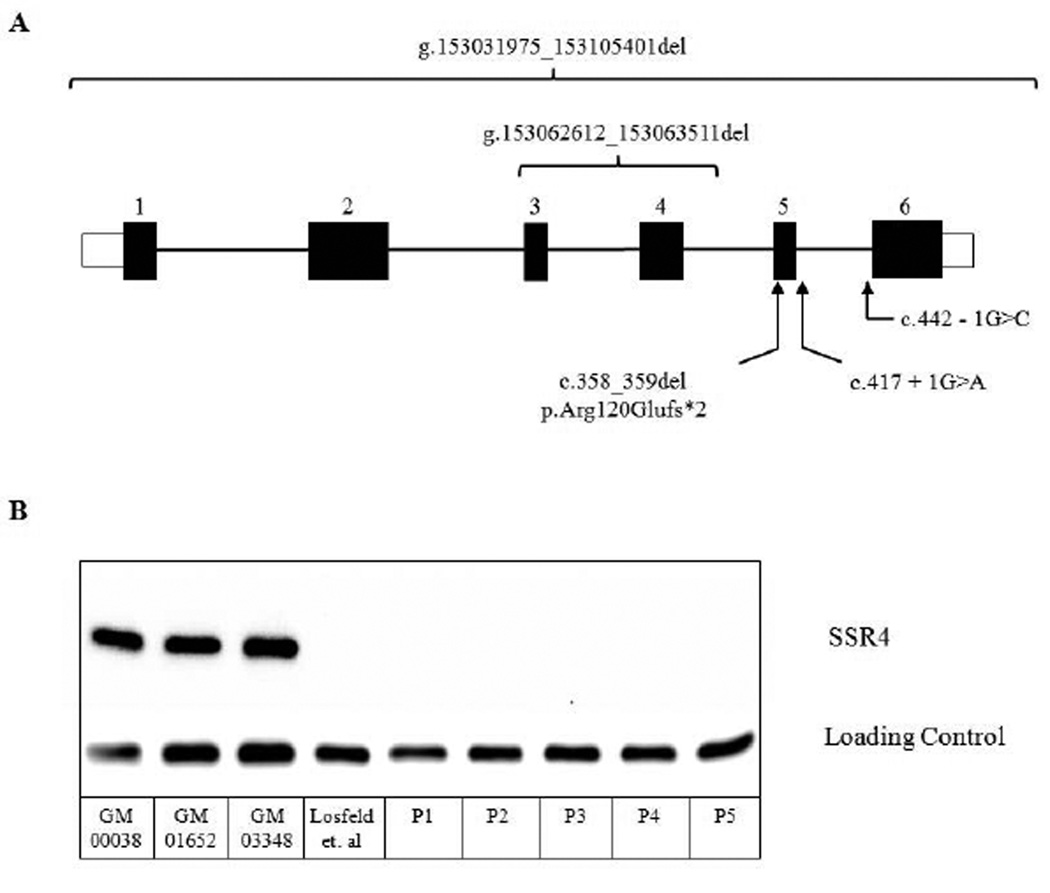
Family 323 is of Asian ancestry and exome sequencing was performed on the youngest male (P1) because he previously had a confirmed abnormal CDT test. However, exome sequencing failed to yield an obvious glycosylation-related candidate gene. Upon closer inspection of the exome results for possible exon deletions within glycosylation-related genes, we determined that this individual had a deletion within SSR4 resulting in the loss of two coding exons. Sanger sequencing of both parents confirmed the presence of a de novo deletion spanning 900bp (g.153062612_153063511del) within SSR4 (Supp. Figure S1).
Family 404 is of Brazilian origin and has two affected males (P2, P3). Exome sequencing on P2, the oldest affected sibling, identified a novel c.358_359del that results in a frameshift p.Arg120Glufs*2. Subsequent testing showed that the younger affected male (P3) was also hemizygous for the same mutation. Analysis of the parents showed that this mutation likely arose de novo in the maternal germline (Supp. Figure S1).
P5 is an affected Hispanic male from family 406 who had exome sequencing done in a CLIA certified laboratory. The initial laboratory report did not identify a clear candidate and no formal diagnosis was made. However, after the publication of the index SSR4-CDG case, an addendum was released reporting that P5 was in fact hemizygous for a novel de novo c.417+1G>A that affected a canonical donor splice site (Supp. Figure S1).
Two Hispanic males (P6, P7) from family 407, who are half-brothers, also had exome sequencing. Both P6 and P7 were found to have a novel c.442-1G>C mutation within a canonical acceptor splice site. However, unlike all the other families, the c.442-1G>C within family 407 was maternally inherited. Subsequent review of the maternal family history showed that multiple generations had both affected males and possibly mildly affected females (Supp. Figure S1). DNA from saliva samples for one cognitively impaired uncle (P8) confirmed he was hemizygous for the c.442-1G>C, as was one aunt but we could not obtain quantifiable results of her cognitive disability.
Family 405 is of European ancestry and are the only family in which exome sequencing was not performed. Instead, an Affymetrix CytoScan HD microarray was performed at Greenwood Genetics Center and showed that the affected male (P4) was hemizygous for a 73.43kb deletion (g.153031975_153105401del), which encompasses four genes PLXNB3, IDH3G, SSR4 and PDZD4. In addition to the affected hemizygous male, the mother and sister of P4 were heterozygous carriers for the deletion (Supp. Figure S1). The older sister has a milder phenotype compared to her brother and analysis of her blood samples showed skewing favoring the paternal (non-deleted) allele. The mother has a seizure disorder and blood samples showed complete skewing towards the non-deleted wild-type allele. Neither female showed an abnormal CDT suggesting that some of the shared phenotype may be due to one of the other three genes. However, it is important to note that while there was skewing towards the normal allele in blood, this may not reflect the status in the brain or other tissues. It is possible that both females could have very mild SSR4-CDG.
All the identified variants are predicted to be loss of function mutations since they are either loss of canonical splice sites, INDELs or a complete gene deletion (Fig. 1a). We used the Exome Aggregation Consortium (ExAC) database (http://exac.broadinstitute.org) [v0.3 accessed 08.06.2015] of 60,706 unrelated individuals to show none of our presented LOF variants nor any other LOF variants to be present as hetero- or hemizygotes.
Figure 1. Mutation schematic of SSR4 mutations and Western blot analysis from patient fibroblasts.
(A) Schematic representation of human SSR4 gene showing the positions of the identified mutations presented in our cohort. Coding regions are depicted as black boxes with appropriate exon number listed above. For cDNA numbering, +1 represents the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1.
(B) Western blot analysis of SSR4 protein from a total of six cases, including the index case reported by Losfeld et al. GM00038, GM01652 and GM03348 are commercially available primary fibroblast lines from apparently healthy individuals. Western blot detection of alpha-Tubulin was used as a loading control.
We predicted that all the identified mutations would result in a complete loss of SSR4 protein. To test this hypothesis we performed western blot analysis of SSR4 protein expression in primary fibroblasts from five of the eight cases (P1–5) as previously described [Losfeld et al., 2014]. SSR4 protein expression was undetectable in these samples compared to three commercially available controls (GM00038, GM01652, GM03348 – Coriell Cell Repository) (Fig. 1b). Samples were not available from P6-P8 (Family 407); however, given that they are hemizygous for a c.442-1G>C mutation, it is likely that abnormal splicing does occur which would ultimately result in either a truncated SSR4 protein or complete loss of the protein. Since only one case (P1) had CDT testing prior to exome sequencing, we performed electrospray ionizing mass spectrometry (ESI-MS) to determine the presence of the CDG biomarker in the remaining individuals. ESI-MS is a rapid and highly sensitive method for detecting transferrin and its various glyco-forms often seen in CDG patients [Lacey et al., 2001]. Specifically for a type I CDT pattern, the ratio between a fully glycosylated transferrin (di-oligo) is compared to underglycosylated forms containing only one (mono-oligo) or lacking both (a-oligo) complete glycans. Precision and accuracy of this method were reported in Lacey et al, 2001. AnalystT S/N script (AB SCIEX) was used to assess signal/noise (S/N) 500Da above or below the calculated mass of the Tf isoform. It provides a baseline and defines a peak when S/N >3. A mono-oligo / di-oligo >0.1 is considered abnormal while 0.07–0.09 has unknown clinical significance. Clearly our SSR4-CDG patients bring clinical significance to this range. Importantly, all seven cases had abnormal CDT levels only slightly above the current Mayo Medical Laboratory reference for a type I (Mono-oligo / Di-oligo > = 0.06) (P1-0.063, P2-0.09, P3-0.10, P4-0.14, P5-0.09, P6-0.09, P7-0.10). Non-glycosylated Tf was not detected in any of the cases. P8 and his affected sister were not available for testing. In those instances where the slight differences are noted, some laboratories using different methods or different cutoffs may report CLIA-approved Tf results to be “Not Significant”. We strongly advise cautious interpretation of such results because they often prematurely rule out a glycosylation abnormality as a possible cause of the clinical phenotype.
Once we confirmed SSR4-CDG both genetically and biochemically, we wanted to determine if there were consistent clinical characteristics that could be used to aid in future diagnosis.
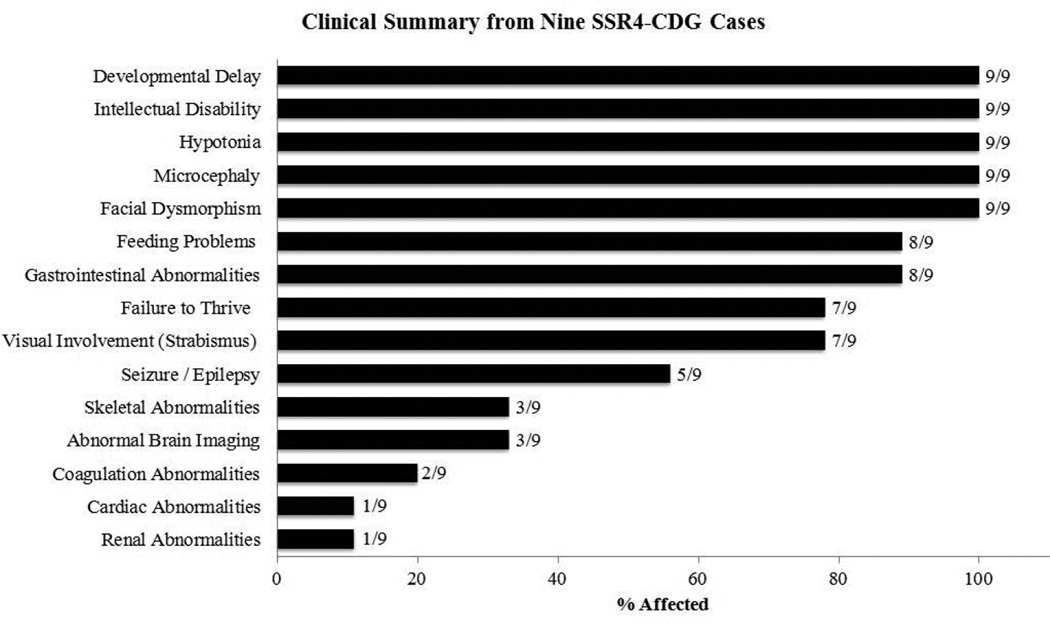
Clinical data from all the affected males presented here (n=8) and one previously reported showed many overlapping features (Fig. 2). Global developmental delays in language and social skills as well as fine and gross motor development were seen in all individuals (9/9). Other neurological deficiencies included intellectual disabilities (9/9), hypotonia (9/9), microcephaly (9/9) and seizure/epilepsy (5/9). Feeding problems (8/9) were often due to difficultly with chewing and aversions to food with certain textures. Facial dysmorphism (9/9) included deep set eyes, large mouth with widely spaced teeth, large ears and hypoplastic vermillion of upper lip. Gastrointestinal abnormalities (8/9) consisted of either reflux or vomiting. Failure to thrive (7/9) and strabismus (7/9) were also seen at a high frequency. Less commonly seen were defects associated with skeletal (3/9), hematological (2/9), cardiac (1/9) and renal system (1/9) (Fig. 2). Skeletal malformations were associated with scoliosis and joint dislocations. Hematological abnormalities consisted of mild coagulopathy. Ductus arteriosus was seen in a single patient and reported to have spontaneously resolved. The only renal abnormality was a single individual with a horseshoe kidney. Structural abnormalities of the brain included thin corpus callosum (1/9), decreased periventricular white matter (1/9) and absence of the septum pellucidum (1/9). It should be noted that while several clinical features are shared by nearly all the affected, these phenotypes are not limited to SSR4-CDG, but are commonly seen in many CDG subtypes as well as other genetic disorders. Given the vast clinical overlap with other genetic disorders, CDT testing by the most sensitive methods (ESI-MS) should be employed when these phenotypic features are encountered. In conclusion, we present an additional eight unreported cases of SSR4-CDG to expand both the genetic and clinical knowledge of this rare CDG subtype. Future studies of inherited SSR4 mutations in males should carefully assess female family members for significant, but mild intellectual disability.
Figure 2. Summary Clinical phenotype.
Clinical information was compiled on the nine affected males including the index case reported by Losfeld et al. and summarized.
Supplementary Material
Supp FigureS1
Acknowledgments
We would like to thank the families for participating in our research study. This work was supported by National Institutes of Health grant (R01DK99551) and ‘The Rocket Fund’. The University of Washington Center for Mendelian Genomics is funded by the National Human Genome Research Institute and the National Heart Lung Blood Institute (1U54HG006493) to D.N., J.S., and M.B.
References
- Delgado MA, Martinez-Domenech G, Sarrion P, Urreizti R, Zecchini L, Robledo HH, Segura F, de Kremer RD, Balcells S, Grinberg D, Asteggiano CG. A broad spectrum of genomic changes in latinamerican patients with EXT1/EXT2-CDG. Sci Rep. 2014;4:6407. doi: 10.1038/srep06407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeze HH. Understanding human glycosylation disorders: biochemistry leads the charge. J Biol Chem. 2013;288:6936–6945. doi: 10.1074/jbc.R112.429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeze HH, Chong JX, Bamshad MJ, Ng BG. Solving glycosylation disorders: fundamental approaches reveal complicated pathways. Am J Hum Genet. 2014;94:161–175. doi: 10.1016/j.ajhg.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garshasbi M, Hadavi V, Habibi H, Kahrizi K, Kariminejad R, Behjati F, Tzschach A, Najmabadi H, Ropers HH, Kuss AW. A defect in the TUSC3 gene is associated with autosomal recessive mental retardation. Am J Hum Genet. 2008;82:1158–1164. doi: 10.1016/j.ajhg.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E, Gorlich D, Kostka S, Otto A, Kraft R, Knespel S, Burger E, Rapoport TA, Prehn S. A tetrameric complex of membrane proteins in the endoplasmic reticulum. Eur J Biochem. 1993;214:375–381. doi: 10.1111/j.1432-1033.1993.tb17933.x. [DOI] [PubMed] [Google Scholar]
- Jones MA, Ng BG, Bhide S, Chin E, Rhodenizer D, He P, Losfeld ME, He M, Raymond K, Berry G, Freeze HH, Hegde MR. DDOST mutations identified by whole-exome sequencing are implicated in congenital disorders of glycosylation. Am J Hum Genet. 2012;90(2):363–368. doi: 10.1016/j.ajhg.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungnickel B, Rapoport TA. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell. 1995;82:261–270. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- Lacey JM, Bergen HR, Magera MJ, Naylor S, O'Brien JF. Rapid determination of transferrin isoforms by immunoaffinity liquid chromatography and electrospray mass spectrometry. Clin Chem. 2001;47:513–518. [PubMed] [Google Scholar]
- Li FY, Chaigne-Delalande B, Kanellopoulou C, Davis JC, Matthews HF, Douek DC, Cohen JI, Uzel G, Su HC, Lenardo MJ. Second messenger role for Mg2+ revealed by human T-cell immunodeficiency. Nature. 2011;475:471–476. doi: 10.1038/nature10246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losfeld ME, Ng BG, Kircher M, Buckingham KJ, Turner EH, Eroshkin A, Smith JD, Shendure J, Nickerson DA, Bamshad MJ University of Washington Center for Mendelian Genomics, Freeze HH. A new congenital disorder of glycosylation caused by a mutation in SSR4, the signal sequence receptor 4 protein of the TRAP complex. Hum Mol Genet. 2014;23:1602–1605. doi: 10.1093/hmg/ddt550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari F, Foulquier F, Tarpey PS, Morelle W, Boissel S, Teague J, Edkins S, Futreal PA, Stratton MR, Turner G, Matthijs G, Gecz J, et al. Oligosaccharyltransferase-subunit mutations in nonsyndromic mental retardation. Am J Hum Genet. 2008;82:1150–1157. doi: 10.1016/j.ajhg.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng BG, Buckingham KJ, Raymond K, Kircher M, Turner EH, He M, Smith JD, Eroshkin A, Szybowska M, Losfeld ME, Chong JX, Kozenko M, et al. Mosaicism of the UDP-galactose transporter SLC35A2 causes a congenital disorder of glycosylation. Am J Hum Genet. 2013;92:632–636. doi: 10.1016/j.ajhg.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Dudek J, Gogala M, Schorr S, Linxweiler J, Lang S, Becker T, Beckmann R, Zimmermann R, Forster F. Structure of the mammalian oligosaccharyl-transferase complex in the native ER protein translocon. Nat Commun. 2014;5:3072. doi: 10.1038/ncomms4072. [DOI] [PubMed] [Google Scholar]
- Shibatani T, David LL, McCormack AL, Frueh K, Skach WR. Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP, and two potential new subunits. Biochemistry. 2005;44:5982–5992. doi: 10.1021/bi047328f. [DOI] [PubMed] [Google Scholar]
- Shrimal S, Ng BG, Losfeld ME, Gilmore R, Freeze HH. Mutations in STT3A and STT3B cause two congenital disorders of glycosylation. Hum Mol Genet. 2013;22:4638–4645. doi: 10.1093/hmg/ddt312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer N, Junne T, Kalies KU, Spiess M, Hartmann E. TRAP assists membrane protein topogenesis at the mammalian ER membrane. Biochim Biophys Acta. 2013;1833:3104–3111. doi: 10.1016/j.bbamcr.2013.08.018. [DOI] [PubMed] [Google Scholar]
- Timal S, Hoischen A, Lehle L, Adamowicz M, Huijben K, Sykut-Cegielska J, Paprocka J, Jamroz E, van Spronsen FJ, Korner C, Gilissen C, Rodenburg RJ, et al. Gene identification in the congenital disorders of glycosylation type I by whole-exome sequencing. Hum Mol Genet. 2012;21:4151–4161. doi: 10.1093/hmg/dds123. [DOI] [PubMed] [Google Scholar]
- Zimmermann R, Eyrisch S, Ahmad M, Helms V. Protein translocation across the ER membrane. Biochim Biophys Acta. 2011;1808:912–924. doi: 10.1016/j.bbamem.2010.06.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp FigureS1