Targeting YAP-dependent MDSC infiltration impairs tumor progression (original) (raw)
. Author manuscript; available in PMC: 2016 Jul 1.
Abstract
The signaling mechanisms between prostate cancer cells and infiltrating immune cells may illuminate novel therapeutic approaches. Here, utilizing a prostate adenocarcinoma model driven by loss of Pten and Smad4, we identify polymorphonuclear myeloid-derived suppressor cells (MDSCs) as the major infiltrating immune cell type and depletion of MDSCs blocks progression. Employing a novel dual reporter prostate cancer model, epithelial and stromal transcriptomic profiling identified Cxcl5 as a cancer-secreted chemokine to attract Cxcr2-expressing MDSCs and, correspondingly, pharmacological inhibition of Cxcr2 impeded tumor progression. Integrated analyses identified hyperactivated Hippo-YAP signaling in driving Cxcl5 upregulation in cancer cells through YAP-TEAD complex and promoting MDSCs recruitment. Clinico-pathological studies reveal upregulation and activation of YAP1 in a subset of human prostate tumors, and the YAP1 signature is enriched in primary prostate tumor samples with stronger expression of MDSC relevant genes. Together, YAP-driven MDSC recruitment via heterotypic Cxcl5-Cxcr2 signaling reveals effective therapeutic strategy for advanced prostate cancer.
Significance
We demonstrate a critical role of MDSCs in prostate tumor progression and discover a cancer cell non-autonomous function of Hippo-YAP pathway in regulation of Cxcl5, a ligand for Cxcr2 expressing MDSCs. Pharmacologic elimination of MDSCs or blocking the heterotypic CxCl5-Cxcr2 signaling circuit elicits robust anti-tumor responses and prolongs survival.
Keywords: Cancer, prostate, genetics Yap1, MDSCs, Cxcr2, Pten, Smad4
Introduction
The tumor microenvironment (TME) is comprised of a complex mixture of tumor-associated fibroblasts, infiltrating immune cells, endothelial cells and extracellular matrix proteins and signaling molecules such as cytokines (1-3). Homotypic and heterotypic interactions between these cellular constituents play essential roles in cancer development and response to therapeutics (3, 4). Among the infiltrating immune cells, MDSCs represent a phenotypically heterogeneous population of immature myeloid cells that play a tumor-promoting role by maintaining a state of immunological anergy and tolerance (5). In particular, activated MDSCs provide a source of secreted chemokines, cytokines and enzymes which suppress local T cell activation and viability (5). In addition, MDSCs can suppress T cell activity through deprivation of nutrients, such as L-arginine and L-cysteine, and interference with T cell receptor functions via reactive oxygen species (ROS) and reactive nitrogen species.
Prostate cancer (PCa) is the most common noncutaneous malignancy in men in the United States. Similar to many other solid tumor types, PCa is characterized by a rich tumor-stroma interaction network that forms the TME (1-3). In PCa, various signaling pathways have been implicated in the crosstalk between tumor and stroma, such as androgen receptor signaling, FGF, Src, TGFβ, IGF, integrin and Hedgehog pathways (1). Interestingly, MDSC abundance in the blood correlates with circulating Prostate Specific Antigen (PSA) levels in PCa patients (6-8). MDSCs have been identified recently as a TME constituent in an indolent prostate cancer mouse model with conditional Pten deletion (9) and demonstrated to antagonize senescence during early tumorigenesis (10). However, the molecular mechanisms underlying the recruitment of MDSCs are not well understood and the extent to which MDSCs facilitate PCa progression has not been determined.
Previously, we have shown that deletion of Pten in the mouse prostate causes upregulation of Smad4 which constrains cell proliferation and invasion and, accordingly, dual deletion of Pten and Smad4 results in rapid PCa progression including metastasis (11). Comparative transcriptomic and cell profile analyses of Pten versus Pten/Smad4 deficient PCa revealed a prominent immune signature and resident MDSCs as a major TME population in Pten/Smad4 deficient tumors. Biological, molecular and pharmacological analyses established that a Yap1-mediated Cxcl5-Cxcr2 signaling axis recruits MDSCs into the TME and that MDSCs play critical roles in facilitating tumor progression. Our comprehensive analyses using PCa model coupled with clinical validation using patients samples support the view that targeting either MDSC recruitment or infiltrated MDSCs may represent a valid therapeutic opportunity in treating advanced prostate cancer.
Results
Prominent Infiltration of Immune Cells in the Ptenpc-/-Smad4pc-/- Tumor Model
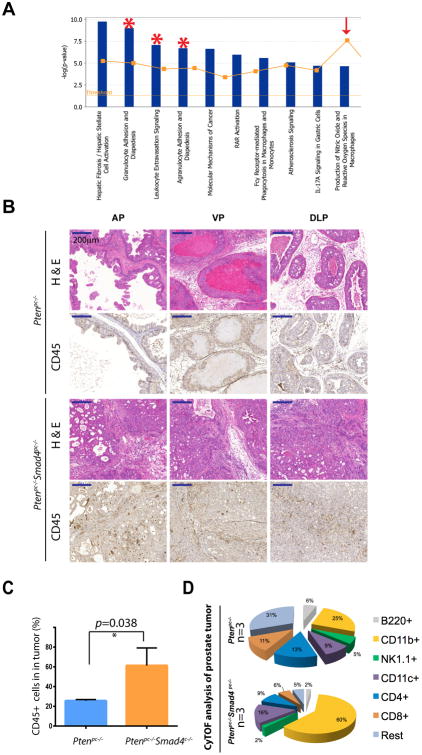
We previously reported that conditional deletion of Smad4 bypassed the senescence barrier instigated by Pten loss in the prostate epithelia, resulting in a highly proliferative and invasive prostate adenocarcinoma characterized by an exuberant stromal reaction and frequent metastasis to distant organs (11). Correspondingly, Ingenuity Pathway Analysis (IPA) revealed prominent representation of cell movement, cell proliferation, and antigen presentation as the top three categories represented in the Ptenpc-/-Smad4pc-/- tumors (11). Further analysis revealed a prominent immune signature including Granulocytes Adhesion and Diapedesis, Leukocytes Extravasation Signaling, and Agrandulocytes Adhesion and Diapedesis as 3 of the top 4 most activated pathways in Ptenpc-/-Smad4pc-/- tumors compared to those present in Ptenpc-/- tumors (Figure 1A; p value < 2.03E-7). Correspondingly, immunohistochemical staining (IHC) highlighted conspicuous infiltration of CD45+ leukocytes in Ptenpc-/-Smad4pc-/- tumors (Figure 1B). To comprehensively audit the spectrum of infiltrating immune cells in tumors, we performed mass cytometry (CyTOF) immunophenotyping (12) to catalog tumor cell type constituents from well-established tumors in 16-week old Ptenpc-/- and Ptenpc-/-Smad4pc-/- mice. Employing a 9-marker antibody panel (Supplementary Table S1), CyTOF confirmed a significant increase of CD45+ infiltrating leukocytes in Ptenpc-/-Smad4pc-/- as compared to Ptenpc-/- tumors (Figure 1C). Within the CD45+ infiltrating cells, CD11b+ myeloid cells represented a significantly increased immune population in Ptenpc-/-Smad4pc-/- as compared to Ptenpc-/- tumors (Figure 1D).
Figure 1. Prominent Infiltration of Immune Cells in the Ptenpc-/-Smad4pc-/- Tumors as Compared to Ptenpc-/- Tumors.
(A) The top 10 activated pathways in Ptenpc-/-Smad4pc-/- tumors (n=5) as compared to Ptenpc-/- tumors (n=5) identified by Ingenuity pathway analysis (IPA). (B) A significant increase in the infiltration of immune cells as shown by IHC for CD45 in Ptenpc-/-Smad4pc-/- tumors as compared to Ptenpc-/- tumors from 16 weeks old mice (n=3). AP, VP and DLP stand for anterior prostate, ventral prostate and dorsolateral prostate, respectively. Scale bar 200 μm. (C) Quantification of tumor-infiltrating CD45+ cells (AP, VP and DLP combined) in Ptenpc-/- tumors and Ptenpc-/-Smad4pc-/- from 16 weeks old mice (n=3), assessed by CyTOF. (D) Percentages of various immune cell populations within the CD45+ infiltrating immune cells in prostate tumors from 16 weeks old Ptenpc-/- and Ptenpc-/-Smad4pc-/- mice, assessed with CyTOF (9-marker) and analyzed with Flowjo. CD11b+ myeloid cells are significantly more in Ptenpc-/-Smad4pc-/- tumor as compared to Ptenpc-/- tumor (n=3, P<0.05).
CD11b+ Gr-1+ Cells are Significantly Increased in Ptenpc-/-Smad4pc-/- Tumor Model
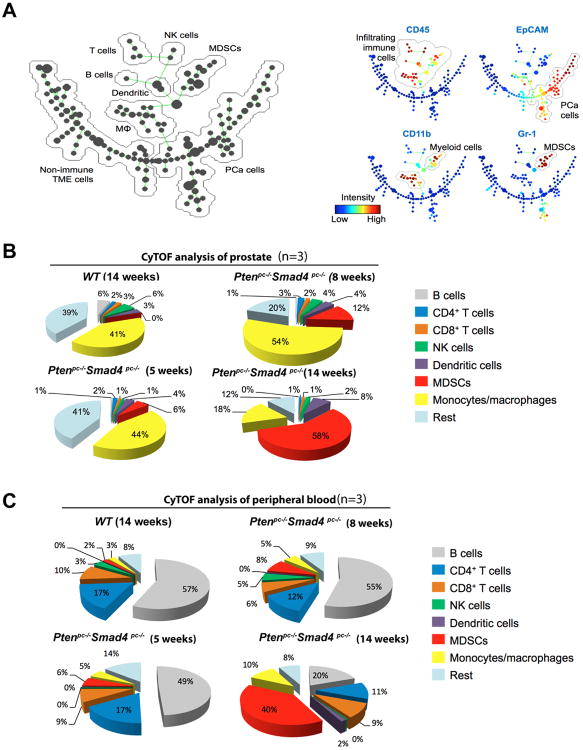
To obtain a dynamic view of peripheral and infiltrating immune cells as a function of tumor progression in the Ptenpc-/-Smad4pc-/- model, which initiates tumor development at 6-8 weeks and progresses to early invasive carcinoma by 14 weeks of age, serial CyTOF analyses using an expanded antibody panel of 17 surface markers (Supplementary Table S1) were performed on single cells from primary tumors, peripheral blood, spleen and draining lymph nodes at 5, 8, and 14 weeks of age. The detailed immunophenotyping profiles enabled construction of the SPADE derived tree (12). SPADE (spanning-tree progression analysis of density-normalized events) is a computational approach to facilitate the identification and analysis of heterogeneous cell types. SPADE of the Ptenpc-/-Smad4pc-/- model displays the complexity of the TME which is composed of epithelial tumor cells (EpCAM+ CD45-), non-immune TME cells (EpCAM- CD45-), and infiltrating immune cells (EpCAM- CD45+) that can be further grouped into various immune cell subpopulations (Figure 2A and Supplementary Figure S1A). Among the infiltrating immune cells, there was a striking age-dependent increase of CD11b+ Gr-1+ cells in tumors (Figure 2B) and peripheral blood from Ptenpc-/-Smad4pc-/- mice (Figure 2C); this trend was much less pronounced in the spleen or draining lymph nodes (Supplementary Figure S1B, for gating strategy see Supplementary Figure S1C).
Figure 2.
CD11b+ Gr-1+ Cells are Significantly Increased in Ptenpc-/-Smad4pc-/- Tumors as Compared to Ptenpc-/- Tumors. (A) SPADE tree derived from CyTOF (17-marker) analysis of whole tumor cell population from Ptenpc-/-Smad4pc-/- mice at 5-week, 8-week, and 14-week old (n=3). Live single cells were used to construct the tree. Cell populations were identified as PCa cells (EpCAM+ CD45-), non-immune TME cells (EpCAM- CD45-), T cells (CD45+ CD3+ TCRβ+), B cells (CD45+ B220+ CD19+), NK cells (CD45+ NK1.1+), dendritic cells (CD45+ CD11c+), putative MDSCs (CD45+ CD11b+ Gr-1+), and macrophages (CD45+ CD11b+ Gr-1-). On the right panels, the tree is colored by the median intensity of individual markers shown on the top to highlight infiltrating immune cells (EpCAM- CD45+), epithelial PCa cells (EpCAM+ CD45-), total myeloid cells (CD45+ CD11b+), and putative MDSCs (CD45+ CD11b+ Gr-1+). (B-C) CyTOF analysis of tumors (B) or peripheral blood (C) from 5, 8, and 14 weeks old Ptenpc-/-Smad4pc-/- mice revealed an age-dependent increase in the MDSCs infiltration. Prostate from wild type (WT) mice at 16-week old was used as control (n=3 for each genotype). See also Supplementary Figure S1.
CD11b+ Gr-1+ Cells from Ptenpc-/-Smad4pc-/- tumors are potently immunosuppressive
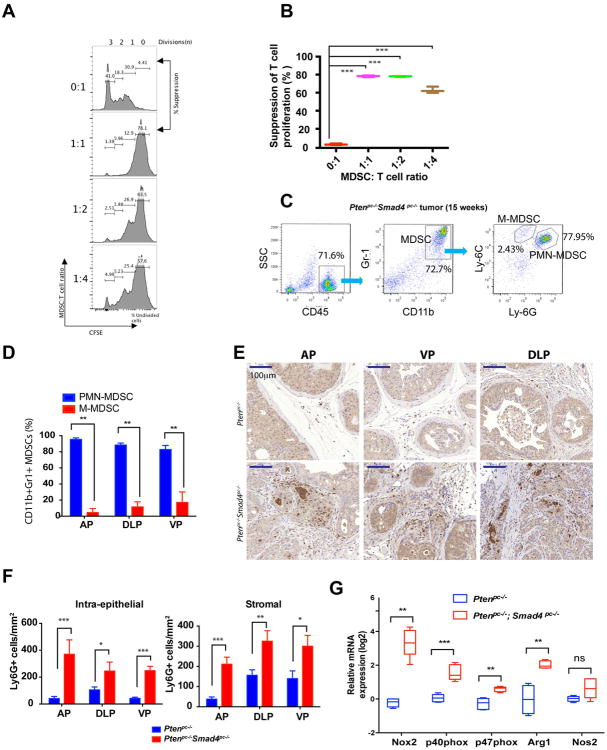
To evaluate the potential immunosuppressive activity of intratumoral CD11b+ Gr-1+ cells from Ptenpc-/-Smad4pc-/- tumors, we examined T cell proliferation using a standard cell co-culture system. These CD11b+ Gr-1+ cells strongly suppressed CD3 and CD28 antibody-induced T cell proliferation and activation (Figure 3A-B, see Supplementary Figure S2 for cell isolation strategy), establishing that CD11b+ Gr-1+ cells are indeed functional MDSCs.
Figure 3. MDSCs from Ptenpc-/-Smad4pc-/- Tumors Display Potent Immunosuppressive Activities and are dominated by PMN-MDSCs.
(A) CD11b+ Gr-1+ cells from Ptenpc-/-Smad4pc-/- tumors display potent immune-suppressive activity towards T cell activation as demonstrated by CFSE dilution assay in triplicate. (B) Summarized result from (A). (C-D) Flow cytometry analysis shows PMN-MDSCs as the major population in the infiltrated MDSCs in established Ptenpc-/-Smad4pc-/- tumors at AP, DLP and VP (n=5). (E-F) A significant increase in Ly-6G+ cells in Ptenpc-/-Smad4pc-/- tumors as compared to the Ptenpc-/- tumors as shown by IHC for Ly-6G and quantified by location of positively stained cells in the intra-epithelial or stromal compartment of the tumor at AP, DLP and VP (n=3). (G) Quantification of the mRNA expression of subunits of NADPH oxidase (Nox2, p40phox, and p47phox), Arg1 and Nos2 in the Ptenpc-/-Smad4pc-/- tumors and the Ptenpc-/- tumors (n=5). In B, D, F and G, *_P<0.05, **P<0.01, ***P<0.001._Also see Supplementary Figure S2.
MDSCs can be further classified as Ly-6G+ Ly-6CLow subset with polymorphonuclear phenotype (PMN-MDSCs) and Ly-6G- Ly-6Chigh subset with monocytic phenotype (M-MDSCs) (13). PMN-MDSCs represented the major MDSC population in Ptenpc-/-Smad4pc-/- tumors (Figure 3C-D), consistent with previously observed preferential expansion of PMN-MDSCs in tumor-bearing mice of various syngeneic models (5, 9, 13). The abundance of PMN-MDSCs was further confirmed by IHC for Ly-6G, as shown by quantification of both intra-epithelial and stromal Ly-6G+ cells in tumors from Ptenpc-/-Smad4pc-/- mice and Ptenpc-/- mice (Figure 3E-F). It has been shown previously that ROS production by PMN-MDSCs is one of the mechanisms driving immune suppression (5, 14-16). Correspondingly, IPA revealed that pathways involved in ROS and NO production are among the top pathways activated in Ptenpc-/-Smad4pc-/- tumors (Figure 1A, arrow). Consistent with the increased infiltration of PMN-MDSCs in the Ptenpc-/-Smad4pc-/- tumors, the expression of several subunits of NADPH oxidase (Nox2, p40phox, and p47phox), which are responsible for ROS production in PMN-MDSCs (5), were significantly upregulated in Ptenpc-/-Smad4pc-/- tumors relative to Ptenpc-/- tumors (Figure 3G). Moreover, Arg1, but not Nos2, was highly upregulated in the Ptenpc-/-Smad4pc-/- tumors (Figure 3G). Together, MDSCs in autochthonous Ptenpc-/-Smad4pc-/- tumors display strong T cell suppressive activity and are predominantly the PMN-MDSC subtype.
Immuno-depletion of MDSCs Impedes Tumor Progression in Ptenpc-/-Smad4pc-/- Mice
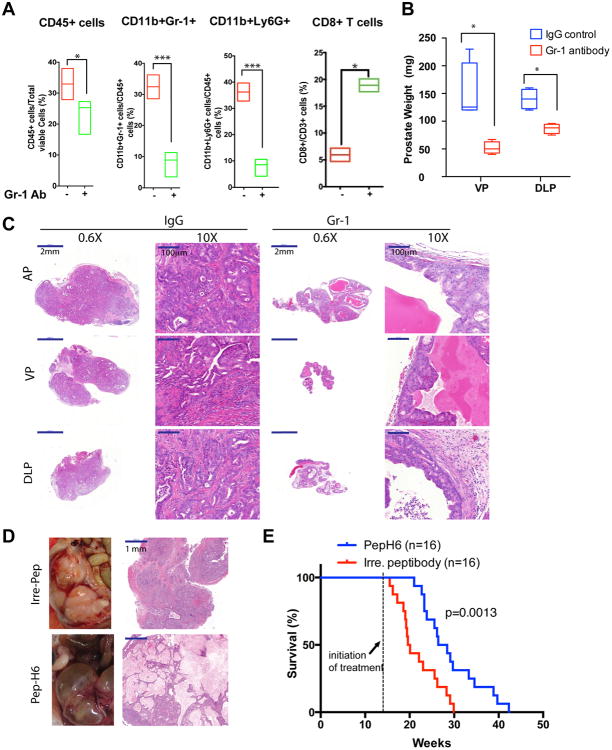
Enrichment of MDSCs in advanced Ptenpc-/-Smad4pc-/- tumors prompted us to explore the possible role of MDSCs in tumor progression. Using a well-characterized anti-Gr-1 neutralizing monoclonal antibody (clone RB6-8C5) (17), MDSCs were depleted in Ptenpc-/-Smad4pc-/- mice at 14 weeks of age, a point coincident with progression to the early invasive carcinoma stage (see Supplementary Figure S3A for treatment scheme). The potent MDSC depletion activity of anti-Gr-1 monoclonal antibody was evidenced by significantly decreased PMN-MDSCs and M-MDSCs in peripheral blood as early as day 2 post-treatment (Supplementary Figure S3B). Additionally, a systemic reduction of MDSCs in spleen, bone marrow, and prostate tumors was documented following a 30-day treatment regimen of anti-Gr-1 monoclonal antibody (Figure 4A and Supplementary Figure S3C). This MDSC depletion was accompanied by an increase of CD8+ T cells (so-called killer T cells) (Figure 4A), consistent with elimination of the T cell suppression activity of MDSCs. Importantly, in line with the CD8+ T cell expansion, we observed that the Gr-1 treated prostate displayed remarkable weight reduction in ventral and dorsolateral prostate (VP & DLP) (Figure 4B). The lack of difference in the weight of anterior prostate (AP) is likely due to the fact that AP tend to developed cysts with fluid accumulated inside the gland (18, 19), which also prevent the accurate measure of the prostate weight (Supplementary Figure S3D). Histopathological analysis revealed adenocarcinoma was the predominant pathology in mice treated with the control IgG whereas mPIN was the predominant morphological presentation in prostates from mice treated with anti-Gr-1 monoclonal antibody (Figure 4C & Supplementary Table S2). In addition, by immunohistochemical staining for CD45, Ki67, vimentin, smooth muscle actin (SMA), and Trichrome staining, we observed that tumor remnants in mice treated with anti-Gr-1 monoclonal antibody displayed markedly reduced levels of cellular proliferation, stromal reaction, and inflammation as compared to those tumors treated with control IgG antibody (Supplementary Figure S4A).
Figure 4. Targeting MDSCs with anti-Gr-1 Neutralizing Antibody or MDSC-specific Peptibody Significantly Delayed Tumor Progression in Ptenpc-/-Smad4pc-/- Mice.
(A) Administration of Gr-1 neutralizing antibody in vivo significantly reduced CD45+ infiltrating immune cells, reduced MDSCs and increased CD8+ T cells among total T cells in Ptenpc-/-Smad4pc-/- tumors (n=4), measured by flow cytometry. (B) Gr-1 antibody treatment of 14-week old mice significantly reduced the weight of VP and DLP in Ptenpc-/-Smad4pc-/- mice. (C) Gr-1 antibody remarkably altered the tumor histopathology in Ptenpc-/-Smad4pc-/- adenocarcinoma, analyzed by H&E staining of AP, VP and DLP. (D) One month of Pep-H6 peptibody treatment led to significant appearance and histology changes of the Ptenpc-/-Smad4pc-/- adenocarcinoma. (E) Kaplan-Meier survival curve showing the significant delay of mortality caused by Pep-H6 peptibody treatment of Ptenpc-/-Smad4pc-/- mice. In A and B, *_P<0.05, ***P<0.001._Also see Supplementary Figure S3 and Figure S4.
In another therapeutic trial, we also utilized the recently developed MDSC-specific peptide-Fc fusion protein (i.e., peptibodies) that have been shown to effectively eliminate MDSCs in vivo through targeting the S100A9 surface protein (20). Employing a hydrodynamic injection approach for nucleic acid delivery (21), intravenous injection of either Pep-H6 peptibody expression vector or irrelevant control peptibody vector were initiated at 14 weeks every 4 days in Ptenpc-/-Smad4pc-/- mice. Strikingly, a single injection of the Pep-H6 peptibody significantly reduced the MDSCs in the peripheral blood, whereas such effect was not observed using the irrelevant control peptibody (Supplementary Figure S4B). Pep-H6 peptibody treatment for one month led to a dramatic decrease in cancer cell content in the prostate tumors (Figure 4D) and provided significant survival benefit for tumor-bearing mice (Figure 4E). Together, our data strongly support the view that MDSC depletion blocks prostate tumor progression in the Ptenpc-/-Smad4pc-/- model.
Cxcl5-Cxcr2 signaling promotes MDSC recruitment and Cxcr2 inhibition delays tumor progression in Ptenpc-/-Smad4pc-/- mice
To elucidate the cellular origins and signaling molecules governing MDSC recruitment to prostate tumors, we incorporated mTmG dual fluorescence reporter allele into the Ptenpc-/-Smad4pc-/- model where signaling events between tumor cells and stroma can be precisely delineated. The mTmG allele (22) allows Cre-dependent GFP expression in prostate epithelial cells and ubiquitous tdTomato expression in all other non-Cre expressing cells (Figure 5A). Transcriptomic and IPA analyses of FACS-sorted GFP+ tumor cells and Tomato+ stromal cells showed distinct expression patterns by hierarchical clustering (Figure 5A) with tumor cells enriched for pathways involved in cell adhesion molecules and tight junction (consistent with their epithelial nature) and stromal cells displaying activation of more diverse pathways involved in chronic inflammation, such as cytokine/cytokine receptor interaction, and chemokine, Jak-STAT, TCR, and BCR signaling (p<0.01, data not shown). This result is consistent with the immuno- and histopathological analyses showing a massive infiltration of immune cells in the Ptenpc-/-Smad4pc-/- tumors.
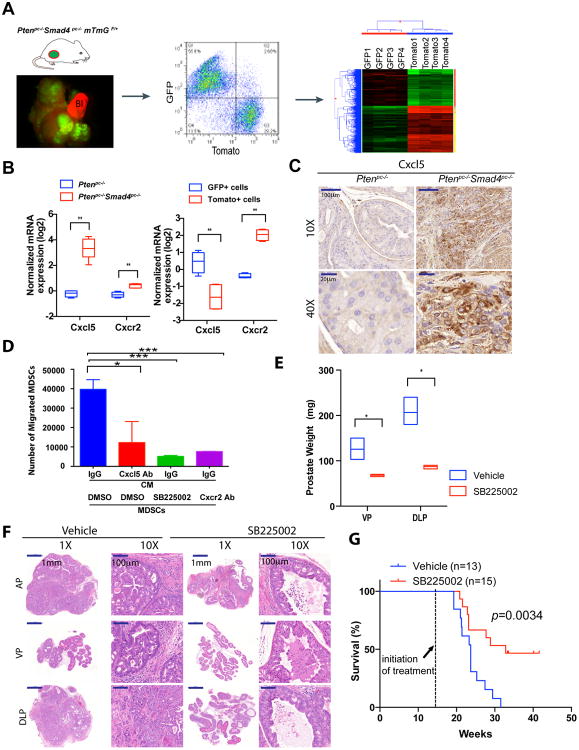
Figure 5. Cxcl5-Cxcr2 Axis Plays an Indispensable Role in Recruitment of MDSCs and Promotion of Tumor Progression.
(A) Establishment of Ptenpc-/-Smad4pc-/-mTmG+ model allows fluorescent visualization of the GFP+ tumor cells intermixed with Tomato+ stroma (Left Panel); FACS isolation of GFP+ tumor cells and Tomato+ stromal cells from the prostate adenocarcinoma (Middle Panel); microarray analysis to identify differentially expressed genes (Right Panel). In the fluorescence image, Bl denotes bladder (completely Tomato+) (n=2). (B) Quantification of mRNA expression shows that Cxcl5 and Cxcr2 were both expressed at higher levels in Ptenpc-/-Smad4pc-/- tumors than Ptenpc-/- tumors, and Cxcl5 expression was enriched in GFP+ tumor cells, whereas Cxcr2 in Tomato+ stromal cells. (n=5) (C) IHC for Cxcl5 showed significantly higher expression levels of Cxcl5 in Ptenpc-/-Smad4pc-/- tumors than Ptenpc-/- tumors (n=3). (D) Blocking the Cxcl5-Cxcr2 axis by Cxcl5 neutralizing antibody, Cxcr2 inhibitor SB225002 or Cxcr2 neutralizing antibody significantly decreased migration of MDSCs towards conditioned medium from Ptenpc-/-Smad4pc-/- tumors cells, evaluated with an in vitro transwell migration assay in triplicate. (E-F) Cxcr2 inhibitor SB225002 treatment of Ptenpc-/-Smad4pc-/- mice for 14 days (n=4) resulted in significantly reduced tumor weight of VP and DLP, and significantly delayed progression for AP prostate cancer shown by H&E staining. (G) Cxcr2 inhibitor SB225002 treatment of Ptenpc-/-Smad4pc-/- mice significantly prolonged their overall survival. In B, D and E, *_P<0.05, **P<0.01, ***P<0.001._Also see Supplementary Figure S5.
Employing this new model, we sought to identify genes that were upregulated in Ptenpc-/-Smad4pc-/- cancer cells relative to Ptenpc-/- cancer cells that might illuminate mechanisms involved in the recruitment of MDSCs by classifying the upregulated genes into either stroma- or tumor-enriched genes. To this end, our previously generated list of 242 genes with greater than 2-fold increased expression in Ptenpc-/-Smad4pc-/- relative to Ptenpc-/- tumors (11) was intersected with 486 genes preferentially expressed in Ptenpc-/-Smad4pc-/- GFP+ cancer cells relative to Tomato+ stroma cells (fold change ≥4) (Supplementary Table S3 and S4), yielding 28 genes that are markedly enriched in Ptenpc-/-Smad4pc-/- cancer cells (Supplementary Table S5). Among these 28 genes, Cxcl5, a key cytokine involved in MDSC recruitment (23, 24), is the most significantly upregulated cancer cell-specific cytokine in Ptenpc-/-Smad4pc-/- tumors as compared to Ptenpc-/- tumors (Figure 5B and Supplementary Figure S5A). Notably Cxcr2, the cognate receptor for Cxcl5, is also upregulated in Ptenpc-/-Smad4pc-/- tumors as compared to Ptenpc-/- tumors and is significantly enriched in Ptenpc-/-Smad4pc-/- Tomato+ stroma cells (Figure 5B). The upregulation of Cxcl5 expression in Ptenpc-/-Smad4pc-/- prostate tumors was further confirmed by IHC (Figure 5C). In addition, we performed FACS analysis of CD11b+ Gr-1+ cells and CD11b- Gr-1- cells from bone marrow, spleen, peripheral blood, and tumors for Cxcr2 expression. As shown in Supplementary Figure S5B, CD11b- Gr-1- cells (largely lymphocytes) are devoid of Cxcr2 expression, whereas a large fraction of CD11b+ Gr-1+ cells express Cxcr2. When Cxcr2 expression was further separated into Cxcr2high and Cxcr2+, we observed an enrichment of the Cxcr2high subpopulations in the CD11b+ Gr-1+ cells in prostate tumor compared with CD11b+ Gr-1+ cells from bone marrow, spleen or blood (Supplementary Figure S5B). This is consistent with the model of active recruitment of MDSCs by tumors through Cxcr2-mediated chemoattraction.
To validate the Cxcl5-Cxcr2 axis in the recruitment MDSCs to the TME of Ptenpc-/-Smad4pc-/- tumors, we assessed the impact of pharmacological inhibition of Cxcl5 and Cxcr2 in MDSCs using a transwell migration assay (23). First, anti-Cxcl5 neutralizing antibody pretreatment of conditioned medium (CM) derived from Ptenpc-/-Smad4pc-/- prostate cancer cell line resulted in decreased migration of MDSCs (Figure 5D); Second, Cxcr2 inhibitor SB255002 or anti-Cxcr2 neutralizing antibody pretreatment also impeded migration of MDSCs (Figure 5D); Third, in vivo blockade of the Cxcl5-Cxcr2 axis using SB255002 in 14-week Ptenpc-/-Smad4pc-/- mice over a 14-day daily dosing schedule revealed a dramatic reduction in infiltration of MDSCs in the prostate tumors (Supplementary Figure S5C-D). Notably, similar to mice treated with anti-Gr-1 neutralizing antibody, these SB225002-treated Ptenpc-/-Smad4pc-/- mice also showed significant reduction in tumor burden (VP & DLP) as compared to the vehicle-treated controls (Figure 5E & Supplementary Figure S5E). Strikingly, all SB225002-treated tumors presented with PIN pathology while control group uniformly possessed advanced adenocarcinoma (Figure 5F & Supplementary Table S2). Furthermore, SB225002 treatment significantly prolonged the overall survival of the Ptenpc-/-Smad4pc-/- mice as compared to the vehicle control (Figure 5G). Thus, we conclude that the Cxcl5-Cxcr2 axis plays a prominent role in the recruitment of MDSCs to the Ptenpc-/-Smad4pc-/- prostate TME and that inhibition of this axis profoundly impairs tumor progression.
Yap1 is activated in Ptenpc-/-Smad4pc-/- Tumors and directly regulates Cxcl5 transcription
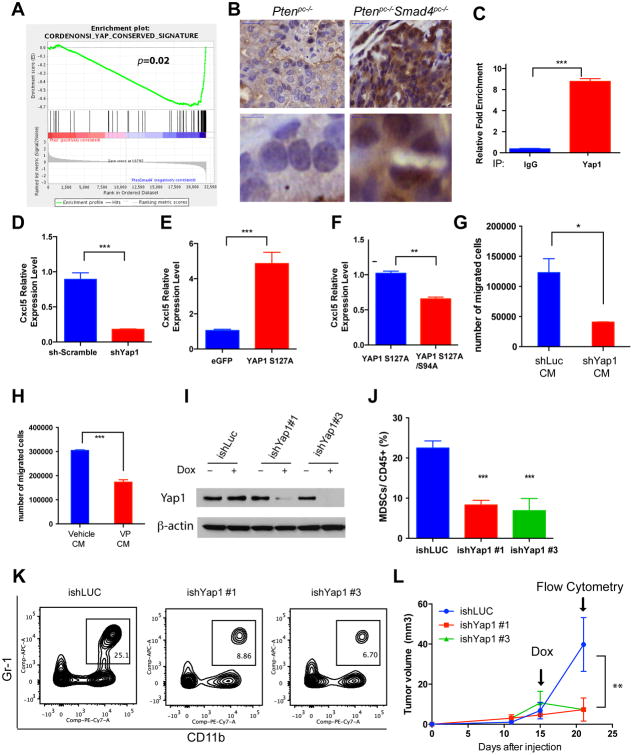
Having identified cancer cell-derived Cxcl5 as a key signaling molecule governing recruitment of MDSCs into the TME, we sought to define the molecular mechanisms underlying the strong induction of Cxcl5 expression in the Ptenpc-/-Smad4pc-/- cancer cells. As Cxcl5 expression is not significantly upregulated in the Ptenpc-/- tumors (Figure 5C), we performed unbiased Gene Set Enrichment Analysis (GSEA) to identify pathways that were activated in the Ptenpc-/-Smad4pc-/- tumors as compared to Ptenpc-/- tumors, aiming to identify potential regulators for Cxcl5 in Ptenpc-/-Smad4pc-/- tumors. The YAP oncogenic signature emerged as the second most hyperactivated pathway (Figure 6A and Supplementary Figure S6A). While it is known that the Hippo-YAP pathway plays an important role in development and cancer in organs such as liver, skin, intestine, pancreas (25-27), the role for Hippo-YAP pathway in prostate cancer biology is emerging. Specifically, Hippo pathway components LATS1/2 have been implicated in anoikis and metastasis in prostate cancer (28) and ERG-induced Yap1 activation can promote age-related prostate tumor development (29). However, beyond the cancer cell specific functions, the Hippo-Yap1 pathway has not been linked to signaling communication between cancer cells and immune cells in the TME. Consistent with the in silico analysis, IHC analysis documented a dramatic increase in the nuclear localization of Yap1 in Ptenpc-/-Smad4pc-/- cancer cells as compared to Ptenpc-/- cancer cells (Figure 6B). As Yap1, a transcriptional coactivator and the downstream mediator of Hippo signaling, is regulated posttranscriptionally by either kinase-mediated degradation or cytoplasmic sequestration (25), our findings of increased nuclear localization of Yap1 is consistent with the hypothesis that Hippo-YAP pathway is activated in the Ptenpc-/-Smad4pc-/- tumors. In addition, unbiased oPOSSUM analysis (30) indicated that TEAD1, a member of the TEAD transcription factor family that is required for Yap1 function, ranked second among the top ten transcription factors with over-represented binding sites in the 70 cancer-specific genes that were upregulated in the Ptenpc-/-Smad4pc-/- tumors as compared to the Ptenpc-/- tumors (≥1.5 fold, Z-Score=13.362, Supplementary Figure S6B-C), an observation reinforcing the relevance of the Hippo-YAP pathway. Furthermore, we identified 6 YAP/TEAD binding motifs in the promoter of Cxcl5 gene (Supplementary Figure 6D), suggesting Yap1 could be directly involved in the recruitment of MDSCs through regulating Cxcl5 expression. This hypothesis was supported by chromatin immunoprecipitation (ChIP) assay showing that Yap1 binds to Cxcl5 promoter (Figure 6C), and that shRNA-mediated knockdown of Yap1 in Ptenpc-/-Smad4pc-/- cancer cells drastically reduced the expression of Cxcl5 mRNA (Figure 6D). In addition, overexpression of a constitutively active Yap1 S127A mutant dramatically increased Cxcl5 mRNA expression in Ptenpc-/-Smad4pc-/- cell line (Figure 6E), whereas overexpression of TEAD binding defective Yap1 mutant S127A/S94A compromised its ability to activate Cxcl5 transcription (Figure 6F). To examine the effect of Yap1-dependent cytokine signaling in the regulation of MDSCs recruitment, we first prepared conditioned medium (CM) from Ptenpc-/-Smad4pc-/- cell line either infected with shRNA against Yap1 or pre-treated with Verteporfin (VP) (25), a small molecular inhibitor that disrupts Yap1-TEAD interaction. We then tested the effect of various CM on the migration of MDSCs in vitro. As shown in Figure 6G-H, we observed significantly decreased MDSCs migration in vitro when CM were from cells with either Yap1 knockdown or VP treatment.
Figure 6. Hyperactivation of Yap1 in Ptenpc-/-Smad4pc-/- Tumors Upregulates Cxcl5.
(A) GSEA analysis identified Yap1 oncogenic signature as the top activated pathways in the Ptenpc-/-Smad4pc-/- tumors compared to Ptenpc-/- tumors (n=5). (B) A significant increase in nuclear staining of Yap1 in the Ptenpc-/-Smad4pc-/- tumors compared to Ptenpc-/- tumors (n=3). (C) Chromatin immunoprecipitation shows that Yap1 can directly bind to Cxcl5 promoter using quantitative PCR in triplicates. (D) shRNA knockdown of Yap1 in Ptenpc-/-Smad4pc-/- tumor cells resulted in a dramatic reduction in Cxcl5 mRNA expression using quantitative PCR in triplicates. (E) Overexpression of a constitutively active YAP1 S127A mutant resulted in upregulation of Cxcl5 mRNA using quantitative PCR in triplicates. (F) TEAD-binding defective S127A/S94A Yap1 mutant significantly decreased Cxcl5 mRNA expression as compared to the Yap1 S127A mutant using quantitative PCR in triplicates. (G-H) Conditioned medium prepared from _Ptenpc-/-Smad4pc-/-_cells infected with Yap1 shRNA (G) or treated with Verteporfin (H), a small molecule that disrupts Yap1-TEAD interaction, induced less MDSCs migration in vitro as compared to the control conditioned medium. Transwell migration was done in triplicate for each condition. (I) Western blot analysis showed that two independent inducible shRNAs for Yap1 efficiently knockdown Yap1 expression in the _Ptenpc-/-Smad4pc-/-_cells. (J-L) Inducible Yap1 knockdown strongly suppressed the intratumoral MDSC infiltration (J-K) and tumor growth (L) of the C57BL/6-syngeneic cell line isolated from prostate tumor of Ptenpc-/-Smad4pc-/-Trp53pc-/- mice (n=5). In C, D, E and F, G, H, L **P<0.01, ***P<0.001. See also Supplementary Figure S6.
Finally, to test if targeting Yap1 in vivo can impair the infiltration of MDSCs and inhibit tumor growth, we used our recently isolated syngeneic murine prostate cancer line PPS, which is derived from the backcrossed Ptenpc-/-Smad4pc-/-Tp53pc-/- model (31) and can form subcutaneous or orthotopic tumors robustly in C57BL/6 hosts. Doxycycline-dependent shRNA knockdown of Yap1 (two independent shRNA designs #1 and #3) were established in PPS (Figure 6I), and injected subcutaneously in C57BL/6 mice. Yap1 knockdown induced by switching to doxycycline-containing drinking water resulted in a reduction of MDSCs in the intratumoral CD45+ population (Figure 6 J-K) and impaired tumor progression (Figure 6L). While the observation supports the hypothesis that targeting YAP1-dependent MDSC infiltration impairs tumor growth, we acknowledge that the tumor growth impediment by Yap1 silencing is likely due to a combined effect of both cell non-autonomous and autonomous mechanisms. Together, these findings reveal a novel function for Yap1 in the recruitment of MDSCs through direct upregulation of Cxcl5 transcription in prostate tumor cells.
YAP1 is activated in human prostate cancer and tracks with an MDSC signature
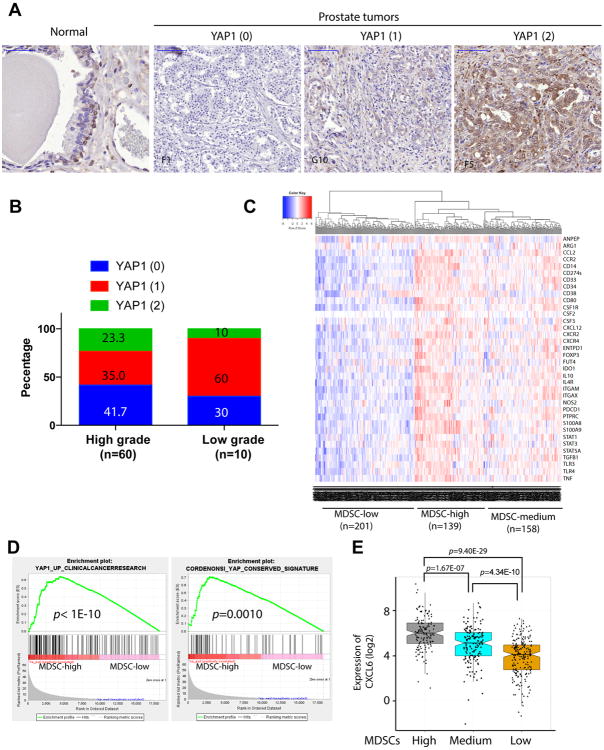
To determine whether YAP1 is overexpressed and activated in human prostate cancer, we performed immunohistochemical staining of human prostate cancer tissue microarray (TMA) for YAP1. Interestingly, YAP1 is expressed in basal cells, but not in the luminal cells of the normal human prostate (Figure 7A). In addition, we observed YAP1 is overexpressed in a subset of human prostate cancers (Figure 7A-B & Supplementary Table S6), consistent with a recent report (29). Given the lack of validated antibodies for human MDSCs for TMA analysis, we generated a list of 39 MDSC related genes curated from literature analysis (Supplementary Table S7) to generate evidence of a link between YAP1 activation and MDSC prominence in human prostate. Using the prostate TCGA RNA-seq data, unsupervised clustering with the 39-gene MDSC signature categorized 498 TCGA primary prostate tumors into three subtypes: MDSC-high (n=139), MDSC-medium (n=158), and MDSC-low (n=201) (Figure 7C), suggesting that a subset of human prostate tumors may have prominent infiltration of MDSCs. In addition, using GSEA, we found that several YAP1 signature genes are significantly overexpressed in MDSC-high samples as compared to MDSC-low samples (Figure 7D; p value < 0.005), reinforcing the link between MDSC-high prostate tumors and YAP1 transcriptional activities. Furthermore, CXCL6, the human homologue of murine Cxcl5, is expressed at a higher levels in the MDSC-high samples as compared to MDSC-low samples (Figure 7E, p=9.40E-29). Similar analysis was performed in a published dataset focused on tumor immunobiological differences in prostate cancer between African-American and European-American men (32). The 39-gene MDSC signature can cluster the 69 primary prostate tumors into MDSC-high (n=40) and MDSC-low (n=29) and Yap1 signatures were prominent in the MDSC-high groups (Supplementary Figure 7A-B). Together, these human prostate tumor findings, which parallel our murine observations, suggests that activated YAP1 is integral to MDSCs infiltration in both mouse and human prostate cancer, thus enhancing the translational value of the study.
Figure 7. YAP1 is activated in human prostate cancer and correlated with MDSCs signature and CXCL6 overexpression.
(A) IHC analysis of YAP1 expression in basal cells of normal prostate tissue and human prostate cancers. Numbers in parenthesis indicates YAP1 IHC intensity scores. (B) YAP1 IHC intensity score representation in low grade (n=10) and high grade (n=60) prostate cancer. (C) Clustering of human TCGA prostate samples into MDSC-high, MDSC-low and MDSC-medium groups using a 39-gene MDSC signature. (D) YAP1 signatures are identified in MDSC-high prostate TCGA samples. (E) CXCL6 expression is significantly higher in MDSC-high group. See also Supplementary Figure S7.
Discussion
Although a large number of studies have demonstrated a direct relationship between MDSC frequency and tumor burden (5), our understanding of the role of MDSCs in tumor progression, particularly prostate cancer, remains largely speculative. Here, using a highly invasive Pten/Smad4 deficient prostate cancer model, we established the signaling circuits involved in the recruitment of MDSCs to the TME and demonstrated a critical role of these cells in facilitating tumor progression.
Homozygous deletion of Pten in murine prostate elicited a strong senescence response that restricts tumor progression (33), thus _Pten-_deficient prostate tumors are largely indolent and progress slowly to invasive prostate adenocarcinoma without metastasis to distant organs (11, 33). Recently, it was shown that infiltrating Gr-1+ myeloid cells suppress Pten loss-induced cellular senescence through a paracrine signaling mediated by myeloid secreted Il1ra (10). We have previously reported that deletion of Smad4 leads to bypass of Pten loss-induced senescence in PCa progression, resulting in aggressive cancer cell proliferation and invasion/metastasis (11). Using the state-of-the-art CyTOF technology, we reveal that progression in the Ptenpc-/-Smad4pc-/ model is associated with abundant immune cell infiltration characterized by prominent representation of CD11b+ Gr-1+ MDSCs, which display potent immunosuppressive activities as shown by their strong antagonistic effect on T cell proliferation (Figure 3A-B).
The basis for the increased frequency of MDSCs in the TME and, specifically in Ptenpc-/-Smad4pc-/ model was not known and presumably could derive from either active chemoattraction or passive nonspecific responses to tissue stress associated with expanding tumor burden. Taking an unbiased approach to identify pathways that may recruit MDSCs, we deconvoluted cancer versus stromal cell transcriptomes by exploiting a Cre-dependent dual fluorescence lineage tracing system in the Ptenpc-/-Smad4pc-/ model. This approach identified unique immune regulatory molecules that are activated prominently in Ptenpc-/-Smad4pc-/ cancer cells, most prominently Cxcl5. We established that the Cxcl5 chemokine plays a key role in the efficient recruitment of MDSCs which enables tumor progression, as blocking Cxcl5-Cxcr2 signaling with a Cxcr2 inhibitor led to reduced MDSC infiltration with associated anti-tumor effect. It should be noted that the human homolog for murine Cxcl5 is CXCL6, and CXCL6 has been shown to be upregulated in prostate cancer as compared to normal prostate and significantly associated with high Gleason score 8-9 (34). Interestingly, it was shown that Cxcl5 promotes recruitment of MDSCs to the primary melanoma, resulting in epithelial-mesenchymal transition (EMT) and cancer cell dissemination (35). Thus, the possible role of Cxcl5/CXCL6 in prostate cancer metastasis merits further study.
Our finding of Cxcl5 as the main chemoattractant in the Ptenpc-/- Smad4pc-/- model also provided a framework to determine the cancer cell signaling pathways driving Cxcl5 upregulation. By integration of bioinformatic analysis and experimental validation, we identified that YAP1 is activated in Ptenpc-/- Smad4pc-/- prostate tumors and that Yap1 directly regulates Cxcl5 transcription and MDSCs recruitment. In addition, we showed that YAP1 is overexpressed in a subset of human prostate cancers, which is consistent with a recent publication showing a correlation of ERG and YAP1 co-expressed in a subset of human prostate cancers (29). Importantly, a 39-gene MDSCs signature clusters the prostate TCGA samples into three subtypes. By comparing the samples with high and low abundance of MDSC-related gene expression, YAP1 signatures and higher expression of CXCL6 are identified in the MDSC-high samples, which is consistent with our findings in the mouse model. Furthermore, the 39-MDSC gene signature can cluster primary prostate tumor samples from a published dataset (32) into two subtypes using MDSC-high and MDSC-low, with Yap1 signatures identified in the MDSC-high subtype. The Hippo-YAP signaling pathway is widely deregulated in human solid neoplasia and often associated with enhanced cancer cell proliferation and cancer stem cell phenotypes (25), and is implicated in the regulation of anoikis and metastasis in prostate cancer (28) and the development of age-related prostate cancers driven by ERG overexpression (29), yet how Hippo-YAP pathway regulates the TME in prostate cancer has hitherto not yet been elucidated. Our finding of a novel non-cell-autonomous function for Hippo-YAP signaling in MDSC recruitment in TME complements well the recently elucidated roles of YAP1 in promoting cell autonomous functionality of cancer cells, including enhanced tumor survival, EMT and bypass mechanism for oncogene addiction (26, 27).
Pharmacological depletion of MDSCs using Gr-1 antibody, Pep-H6 peptibody, or Cxcr2 inhibitor arrested prostate progression at the high-grade PIN stage while controls exhibited full-fledged adenocarcinoma in Ptenpc-/-Smad4pc-/ model. Given that treatment commences at 14 weeks of age (Supplementary Figure S3A), when prostate tumors have uniformly advanced to the invasive adenocarcinoma stage (11) with significant MDSC infiltration (Figure 2B), our findings support the view that anti-MDSC treatment provokes regression of advanced tumors. In addition, both Pep-H6 peptibody and Cxcr2 inhibitor treatment significantly prolonged the overall survival of the Ptenpc-/-Smad4pc-/- tumor-bearing mice. Therefore, our preclinical data suggests that pharmacological depletion of MDSCs may offer potential therapeutic benefits for advanced prostate cancer patients, particularly those deficient for PTEN and SMAD4. In line with our findings, others have demonstrated that depletion of G-MDSCs promotes the intratumoral accumulation of activated CD8 T cells and apoptosis of tumor epithelial cells in a Kras/Trp53 mouse pancreatic cancer model (36).
MDSCs are of myeloid cell lineage, and their coordinated regulation represents one of the most complex aspects of cancer-host interactions (37). The involvement of the myeloid compartment of the hematopoietic system in innate immunity, adaptive immunity as well as in regulation of TME through non-immune mechanisms, highlights the need to understand more deeply how modulating different myeloid populations, including MDSCs, can positively or negatively affect tumor growth.
Pep-H6 peptibody, targeting S100a9 expressed on MDSCs, has been shown to have minimal toxicity in treated mice (20) and potent anti-tumor activity (Figure 4D-E & Ref 18). Interestingly, Tasquinimod, a small molecular inhibitor for S100A9, has been shown to increase progression-free survival (PFS) and overall survival (OS) for metastatic castration resistant prostate cancer (mCRPC) in phase II clinical trial and has entered phase III clinical trial (38). Importantly, similar to the peptibody treatment in mice, Tasquinimod is well-tolerated and only causes minor adverse effects in human patients (38), suggesting that Tasquinimod or similar drugs that targeting S100A9 could be potentially used as chemopreventive agent for patients with high risk primary prostate cancer. The anti-proliferative mechanism may explain why targeting Cxcr2 in PCa with abundant preexisting MDSC infiltration can lead to MDSC depletion, as MDSCs have been shown to undergo active proliferation inside the prostate tumor of the Ptenpc-/- model (9). The effectiveness of targeting Cxcr2 in our model suggests targeting mechanisms that specifically regulate MDSC recruitment as well as their proliferative and survival potential in human cancers would provide therapeutic benefit for patients with PCa.
Targeting MDSCs as a cooperative approach for immunotherapy is clinically relevant, as increasing evidence indicates MDSCs represent a bona fide immunosuppressive cell population in patients with various solid tumors (39, 40). Immunosuppressive mechanisms by MDSCs in mice have been validated in humans, which include L-arginine depletion, NO and ROS production, TGFβ secretion, blocking Teff cells and inducing Treg cells, among others (39). Future studies are warranted to evaluate if combining MDSC depletion with immune checkpoint inhibitors such as anti-CTLA4, anti-PD1 and anti-PD-L1 antibodies may elicit synergistic efficacy in the preclinical models of PCa and eventually benefit patients with prostate cancer.
Methods
Mice strains
Ptenpc-/- and Ptenpc-/- Smad4pc-/- model were developed previously (11) and were backcrossed to the C57BL/6 background for more than 4 generations. B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (“mTmG”) strain was obtained from Jackson Laboratory. Mice were maintained in pathogen-free conditions at M.D. Anderson Cancer Center. All manipulations were approved under MD Anderson Cancer Center (MDACC) Institutional Animal Care and Use Committee (IACUC).
Cell lines
Ptenpc-/-Smad4pc-/- prostate cell lines have been described previously (11), which was generated in 2010. PPS, a C57BL/6-syngeneic cell line isolated from prostate tumor of _Ptenpc-/-Smad4pc-/- Tp53c-/-_mice was generated in 2013. All cell lines tested for Mycoplasma were negative within 6 months of performing the experiments. Cell line authentication was not performed.
CyTOF and Flow cytometry
Prostate tumor single cells were isolated using the Mouse Tumor Dissociation kit (Miltenyl Biotec). Single cells were isolated from spleen, lymph node, and peripheral blood using standard protocol. All isolated cells were depleted of erythrocytes by hypotonic lysis. For CyTOF analysis, cells were blocked for FcγR using CD16/CD32 antibody (clone 2.4G2, BD Biosciences) and incubated with CyTOF antibody mix for 30 minutes at room temperature. Cells were washed once and incubated with MAXPAR®Nucleic Acid Intercalator-103Rh (DVS Sciences) for 20 minutes for viability staining. Cells were fixed with 1.6% formaldehyde for 1 hour and incubated with MAXPAR®Nucleic Acid Intercalator-Ir (DVS Sciences) at 4°C overnight to stain the nuclei. The samples were analyzed with CyTOF instrument (DVS Sciences) in the Flow Cytometry and Cellular Imaging Core Facility at M.D. Anderson Cancer Center. Flow cytometry were performed using standard protocol on LSRFortessa analyzer (Becton Dickinson) and analyzed with FlowJo software (Tree Star).
T cell suppression and MDSCs migration assay
T cell suppression assay was performed as described (9) using FACS-sorted MDSCs and CFSE (Invitrogen) labeled MACS-sorted (Miltenyi) CD8+ or CD4+ T cells in anti-CD3 and anti-CD28 coated 96-well plates at an MDSC/T cell ratio of 0:1, 1:1, 1:2, 1:4, with 3.0 × 105 to 5.0 × 105 MDSCs used in each ratio. Cells were analyzed after 72 hours by Flow cytometry and the suppression of T cells is calculated as described (41). The percentage of CFSE+ cells divided in the presence of MDSCs was compared to percentage of CFSE+ divided cells in the absence of any added MDSCs. For MDSCs migration assay, equal number of FACS-sorted MDSCs, untreated or pre-treated with neutralizing antibody or inhibitor, were placed on the upper chamber of a transwell system (BD Falcon) and conditioned media (CM) from Pten/Smad4-deficient cells under various condition were added to the bottom chamber. Cells were allowed to migrate to the bottom well for 6 hours at 37 °C, 5% CO2. Migrated cells were then analyzed by flow cytometry using BD Fortessa X20. Migrated FITC positive cells were gated to count the absolute number of cells migrated through the transwell.
MSDCs depletion in vivo with Gr1 antibody, Peptibody, and Cxcr2 inhibitor SB225002
Anti-Gr-1 (clone RB6-8C5) and isotype control (clone LTF-2) were purchased from BioXcell and dosed at 200 μg/mouse (i.p.) every other day. 15 μg of endotoxin-free plasmids for irrelevant control peptibody (Irr-pep) and MDSC-specific Pep-H6 peptibody were injected into mice through tail vein using the established protocol (21) in TransIT-EE Delivery Solution (Mirus Bio LLC) every 4 days. SB225002 (Cayman Chemical) in DMSO was diluted in vehicle (0.9%NaCl, 0.3%Tween 80) for in vivo administration every other day (5 mg/kg).
Inducible Yap1 knockdown
Inducible Yap1 knockdown was constructed by cloning the two Yap1 shRNA used previously (26) from the pLKO.1 into a doxycycline inducible plasmid. Lentivirus was packaged in 293T and was used to infect PPS, a C57BL/6-syngeneic cell line isolated from prostate tumor of _Ptenpc-/-Smad4pc-/- Tp53c-/-_mice. Stable sublines were selected with puromycine (2 μg/mL) and injected subcutaneously to the flank of 5-week old male C57BL/6 mice (Jackson Laboratory). Two weeks post-injection, mice were fed with doxycycline water (2 g/L), a method used to execute doxycycline inducible expression in vivo (42). Tumors were measured and extracted 6 days later to analyze for MDSC% in infiltrating immune cells.
Computational analysis of mouse microarray data and human prostate TCGA data
RNA was isolated from FACS-sorted GFP+ and Tomato+ cells using Ptenpc-/-Smad4pc-/-mTmG+ prostate tumors, followed by microarray analysis at the MD Anderson Microarray Core facility using the Mouse Genome 430 2.0 Array (Affymetrix) to generate a Ptenpc-/-Smad4pc-/- tumor/stroma dataset GSE71319. Data set GSE25140 was downloaded from NCBI GEO database. Differentially expressed genes between two conditions (GFP+ vs. Tomato+, or Pten/Smad4 vs. Pten) were subjected to IPA, GSEA, and oPOSSUM analysis. For analysis of human prostate data, we first generated a list of 39 human MDSCs signature genes by literature mining (Supplementary Table S7). The gene expression data of 498 TCGA prostate samples was downloaded from the Broad GDAC Firehose (http://gdac.broadinstitute.org), which is the RSEM expression estimates normalized to set the upper quartile count at 1000 for gene level and then with log2 transformation. The 498 TCGA prostate samples were clustered using the 39 MDSC genes into MDSC-high, MDSC-low, and MDSC-medium (distance between pairs of samples were measured by manhattan distance, and clustering was then performed using complete-linkage hierarchical clustering). 69 samples from Wallace et al (32) were cluster into MDSC-high and MDSC-low. Differentially expressed genes between MDSC- high and MDSC-low were analyzed by GSEA. The expression of CXCL6 in MDSC-high samples is compared to MDSC-low samples using wilcoxon test.
Immunohistochemistry and Western Blot Analysis
Tissues were fixed in 10% formalin overnight and embedded in paraffin. Immunohistochemical (IHC) was performed as described earlier (11). For western blot analysis, cells were lysed on ice using RIPA buffer (Boston BioProducts) supplemented with protease and phosphatase inhibitors (Roche). YAP1 antibody was obtained from Novus Bio and Cell Signaling Technology. Cxcl5 antibodies were obtained from Bioss (Woburn, MA) and R&D Biosystems (Minneaplois, MN). Cxcr2 antibody was obtained from Bioss and R & D Biosystems. CD45 and Ly-6G antibodies were obtained from Biolegend. Prostate tissue micrcroarray was obtained from Folio Bioscience.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described (26) using YAP1 antibody from Novus. Briefly, 5ug of Rabbit IgG (Santa Cruz) or YAP1 antibody was incubate with Protein A Dynabeads magnetic beads (Invitrogen) for 4 hours, followed by extensive wash to remove unbound antibody. Antibody-Beads were then added to the chromatin and incubate for overnight. The following Primers were used for qPCR analysis: Cxcl5_S: 5′-CTCCAGTTTCCTGCCTGAAG-3′ and Cxcl5_as: 5′-GTGTGGAGATTGGGGCTCTA-3′.
Quantitative RT-PCR
RNA was isolated by RNeasy Kit (Qiagen) and reversed transcribed using Superscript III cDNA synthesis Kit (Life Technology). Quantitative PCR was performed using SYBR-GreenER Kit (Life Technology). The following primers were used: Cxcl5_Fwd: GCATTTCTGTTGCTGTTCACGCTG, Cxcl5_Rev: CCTCCTTCTGGTTTTTCAGTTTAGC; β-actin_Fwd: GAAATCGTGCGTGACATCAAAG, β-actin_Rev: TGTAGTTTCATGGATGCCACAG; Yap1_Fwd: TGAGATCCCTGATGATGTACCAC, Yap1_Rev: TGTTGTTGTCTGATCGTTGTGAT.
Statistical Analysis
Data are presented as mean ± SD unless indicated otherwise. Student's t test assuming two-tailed distributions was used to calculate statistical significance between groups. Animal survival benefit was determined by Kaplan-Meier analysis. P < 0.05 was considered statistically significant.
Supplementary Material
1
2
3
4
5
6
7
8
Acknowledgments
We thank Dr. Larry W Kwak and Dr. Hong Qin for the peptibody plasmids; Dr. Willem Overwijk and Dr. Yared Hailemichael for advise in hydrodynamic injection; Samirkumar Amin and member of the DePinho laboratory for helpful suggestions and technical support; Flow Cytometry and Cellular Imaging Core Facility (Jared Burks, Duncan Mak, and Karen Dwyer), Sequencing & Non-coding RNA Core Services (Chang-Gong Liu) at The University of Texas, MD Anderson Cancer Center (Cancer Center Support grant, CA16672).
Grant Support: The project was supported by U01CA141508 (L.C. and R.A.D.), Prostate Cancer Research Program (PCRP) W81XWH-13-1-0202 (G.W.), W81XWH-14-1-0429 (P.D.), Idea Development Award – New Investigator Option (W81XWH-14-1-0576) (X.L.) from Department of Defense, and Clayton & Modesta Williams Cancer Research Fund. Additional support was provided by Jane Coffin Childs Memorial Fund Postdoctoral Fellowship to X.L. E.J. is supported by Korean Governments (MISP) grand number 2011-0030130.
Footnotes
Conflict of Interest: The authors disclose no potential conflicts of interest.
Accession Numbers: The expression array data used in this manuscript was in GEO with accession number GSE25140 (11) and GSE71319.
Author Contributions: Conception and design: G. Wang, X. Lu, P. Dey, Y.A. Wang and R.A. DePinho.
Development of methodology: G. Wang, X. Lu, P. Dey.
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): G. Wang, X. Lu, P. Dey, E. Tapia, M.J. McArthur, P. Deng, R. Konaparthi, S. Hua, N.B. Patel, Z. Guo, P. Hou, D. Zhao, X. Shang, S. Khadka, B. Hu, Q. Chang, X. Pan, Z. Ding, Y. Shi, L. Li, M. J. McArthur, E. Tapia, P. Troncoso.
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): G. Wang, X. Lu, P. Dey, C.C. Wu, C.J. Wu, Y.A. Wang and R.A. DePinho.
Writing, review, and/or revision of the manuscript: G. Wang, X. Lu, P. Dey, Y.A. Wang and R.A. DePinho.
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): S. Jiang, Z. Fang, K. Zhao, E. Jin, W. Yao, A. Kapoor, T. N. Tieu, V. Ramamoorthy, T. Heffernan, C.C. Wu, C.J. Wu, L. Chin.
Study supervision: G. Wang, X. Lu, J. Zhang, C.J. Logothetis, Y.A. Wang, R.A. DePinho.
Other (performed pathology): M. J. McArthur, E. Tapia, P. Troncoso.
References
- 1.Karlou M, Tzelepi V, Efstathiou E. Therapeutic targeting of the prostate cancer microenvironment. Nat Rev Urol. 2010;7:494–509. doi: 10.1038/nrurol.2010.134. [DOI] [PubMed] [Google Scholar]
- 2.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–54. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Coussens L. Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. Cancer Cell. 2012;21:309–22. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg Robert A. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Talmadge JE, Gabrilovich DI. History of myeloid-derived suppressor cells. Nat Rev Cancer. 2013;13:739–52. doi: 10.1038/nrc3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vuk-Pavlović S, Bulur PA, Lin Y, Qin R, Szumlanski CL, Zhao X, et al. Immunosuppressive CD14+HLA-DRlow/- monocytes in prostate cancer. Prostate. 2010;70:443–55. doi: 10.1002/pros.21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Idorn M, Kollgaard T, Kongsted P, Sengelov L, Thor Straten P. Correlation between frequencies of blood monocytic myeloid-derived suppressor cells, regulatory T cells and negative prognostic markers in patients with castration-resistant metastatic prostate cancer. Cancer immunology, immunotherapy : CII. 2014;63:1177–87. doi: 10.1007/s00262-014-1591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brusa D, Simone M, Gontero P, Spadi R, Racca P, Micari J, et al. Circulating immunosuppressive cells of prostate cancer patients before and after radical prostatectomy: profile comparison. Int J Urol. 2013;20:971–8. doi: 10.1111/iju.12086. [DOI] [PubMed] [Google Scholar]
- 9.Garcia AJ, Ruscetti M, Arenzana TL, Tran LM, Bianci-Frias D, Sybert E, et al. Pten null prostate epithelium promotes localized myeloid-derived suppressor cell expansion and immune suppression during tumor initiation and progression. Molecular and cellular biology. 2014;34:2017–28. doi: 10.1128/MCB.00090-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Mitri D, Toso A, Chen JJ, Sarti M, Pinton S, Jost TR, et al. Tumour-infiltrating Gr-1+ myeloid cells antagonize senescence in cancer. Nature. 2014;515:134–7. doi: 10.1038/nature13638. [DOI] [PubMed] [Google Scholar]
- 11.Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–73. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjornson ZB, Nolan GP, Fantl WJ. Single-cell mass cytometry for analysis of immune system functional states. Current opinion in immunology. 2013;25:484–94. doi: 10.1016/j.coi.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. Journal of Immunology. 2008;181:5791–802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kusmartsev S, Nefedova Y, Yoder D, Gabrilovich DI. Antigen-specific inhibition of CD8+ T cell response by immature myeloid cells in cancer is mediated by reactive oxygen species. J Immunol. 2004;172:989–99. doi: 10.4049/jimmunol.172.2.989. [DOI] [PubMed] [Google Scholar]
- 15.Szuster-Ciesielska A, Hryciuk-Umer E, Stepulak A, Kupisz K, Kandefer-Szerszen M. Reactive oxygen species production by blood neutrophils of patients with laryngeal carcinoma and antioxidative enzyme activity in their blood. Acta Oncol. 2004;43:252–8. doi: 10.1080/02841860410029708. [DOI] [PubMed] [Google Scholar]
- 16.Schmielau J, Finn OJ. Activated granulocytes and granulocyte-derived hydrogen peroxide are the underlying mechanism of suppression of T-cell function in advanced cancer patients. Cancer Res. 2001;61:4756–60. [PubMed] [Google Scholar]
- 17.Pekarek LA, Starr BA, Toledano AY, Schreiber H. Inhibition of tumor growth by elimination of granulocytes. The Journal of experimental medicine. 1995;181:435–40. doi: 10.1084/jem.181.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang G, Lunardi A, Zhang J, Chen Z, Ala U, Webster KA, et al. Zbtb7a suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion. Nat Genet. 2013;45:739–46. doi: 10.1038/ng.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lunardi A, Ala U, Epping MT, Salmena L, Clohessy JG, Webster KA, et al. A co-clinical approach identifies mechanisms and potential therapies for androgen deprivation resistance in prostate cancer. Nat Genet. 2013;45:747–55. doi: 10.1038/ng.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin H, Lerman B, Sakamaki I, Wei G, Cha SC, Rao SS, et al. Generation of a new therapeutic peptide that depletes myeloid-derived suppressor cells in tumor-bearing mice. Nat Med. 2014;20:676–81. doi: 10.1038/nm.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene therapy. 1999;6:1258–66. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 22.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 23.Yang L, Huang J, Ren X, Gorska AE, Chytil A, Aakre M, et al. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008;13:23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bierie B, Stover DG, Abel TW, Chytil A, Gorska AE, Aakre M, et al. Transforming growth factor-beta regulates mammary carcinoma cell survival and interaction with the adjacent microenvironment. Cancer Res. 2008;68:1809–19. doi: 10.1158/0008-5472.CAN-07-5597. [DOI] [PubMed] [Google Scholar]
- 25.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nature reviews Drug discovery. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158:185–97. doi: 10.1016/j.cell.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–84. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes & development. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen LT, Tretiakova MS, Silvis MR, Lucas J, Klezovitch O, Coleman I, et al. ERG Activates the YAP1 Transcriptional Program and Induces the Development of Age-Related Prostate Tumors. Cancer Cell. 2015;27:797–808. doi: 10.1016/j.ccell.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon AT, Arenillas DJ, Worsley Hunt R, Wasserman WW. oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G. 2012;32:987–1002. doi: 10.1534/g3.112.003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Z, Wu CJ, Jaskelioff M, Ivanova E, Kost-Alimova M, Protopopov A, et al. Telomerase Reactivation following Telomere Dysfunction Yields Murine Prostate Tumors with Bone Metastases. Cell. 2012;148:896–907. doi: 10.1016/j.cell.2012.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallace TA, Prueitt RL, Yi M, Howe TM, Gillespie JW, Yfantis HG, et al. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 2008;68:927–36. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 33.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Q, Russell MR, Shahriari K, Jernigan DL, Lioni MI, Garcia FU, et al. Interleukin-1beta promotes skeletal colonization and progression of metastatic prostate cancer cells with neuroendocrine features. Cancer Res. 2013;73:3297–305. doi: 10.1158/0008-5472.CAN-12-3970. [DOI] [PubMed] [Google Scholar]
- 35.Toh B, Wang X, Keeble J, Sim WJ, Khoo K, Wong WC, et al. Mesenchymal transition and dissemination of cancer cells is driven by myeloid-derived suppressor cells infiltrating the primary tumor. PLoS Biol. 2011;9:e1001162. doi: 10.1371/journal.pbio.1001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stromnes IM, Brockenbrough JS, Izeradjene K, Carlson MA, Cuevas C, Simmons RM, et al. Targeted depletion of an MDSC subset unmasks pancreatic ductal adenocarcinoma to adaptive immunity. Gut. 2014;63:1769–81. doi: 10.1136/gutjnl-2013-306271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nature Reviews Immunology. 2012;12:253–68. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osanto S, van Poppel H, Burggraaf J. Tasquinimod: a novel drug in advanced prostate cancer. Future oncology. 2013;9:1271–81. doi: 10.2217/fon.13.136. [DOI] [PubMed] [Google Scholar]
- 39.Filipazzi P, Huber V, Rivoltini L. Phenotype, function and clinical implications of myeloid-derived suppressor cells in cancer patients. Cancer Immunology, Immunotherapy. 2012;61:255–63. doi: 10.1007/s00262-011-1161-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solito S, Marigo I, Pinton L, Damuzzo V, Mandruzzato S, Bronte V. Myeloid-derived suppressor cell heterogeneity in human cancers. Annals of the New York Academy of Sciences. 2014;1319:47–65. doi: 10.1111/nyas.12469. [DOI] [PubMed] [Google Scholar]
- 41.Battaglia M, Stabilini A, Roncarolo MG. Rapamycin selectively expands CD4+CD25+FoxP3+ regulatory T cells. Blood. 2005;105:4743–8. doi: 10.1182/blood-2004-10-3932. [DOI] [PubMed] [Google Scholar]
- 42.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–70. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5
6
7
8