Genomic evidence for the Pleistocene and recent population history of Native Americans (original) (raw)
. Author manuscript; available in PMC: 2016 Jan 31.
Published in final edited form as: Science. 2015 Jul 21;349(6250):aab3884. doi: 10.1126/science.aab3884
Abstract
How and when the Americas were populated remains contentious. Using ancient and modern genome-wide data, we find that the ancestors of all present-day Native Americans, including Athabascans and Amerindians, entered the Americas as a single migration wave from Siberia no earlier than 23 thousand years ago (KYA), and after no more than 8,000-year isolation period in Beringia. Following their arrival to the Americas, ancestral Native Americans diversified into two basal genetic branches around 13 KYA, one that is now dispersed across North and South America and the other is restricted to North America. Subsequent gene flow resulted in some Native Americans sharing ancestry with present-day East Asians (including Siberians) and, more distantly, Australo-Melanesians. Putative ‘Paleoamerican’ relict populations, including the historical Mexican Pericúes and South American Fuego-Patagonians, are not directly related to modern Australo-Melanesians as suggested by the Paleoamerican Model.
It is generally agreed that ancestral Native Americans are descendants of Siberian peoples who traversed the Bering Land Bridge (Beringia) from northeast Asia in Late Pleistocene times, and though consensus has yet to be reached, it is mostly conceded that the Clovis archaeological complex, dating to ca. 13 KYA, does not represent the first migration as long supposed (1–7). Archaeological evidence accumulated over the last two decades indicates that people were south of the North American continental ice sheets more than a millennium earlier and had reached as far south as southern South America by at least ca. 14.6 KYA (1–3). Interpretations differ, however, regarding the precise spatio-temporal dynamics of the peopling process, owing to archaeological claims for a significantly earlier human presence pre-dating the Last Glacial Maximum (LGM; ca. 20 KYA) (8–10), and conflicting interpretations of the number and timing of migrations from Beringia based on anatomical and genetic evidence (11–16). Much of the genetic evidence is from studies of mitochondrial DNA (mtDNA) and Y-chromosome, which as single, uniparentally inherited loci are particularly subject to genetic drift and sex-biased demographic and cultural practices.
Among the principal issues still to be resolved regarding the Pleistocene and recent population history of Native Americans are: (i) the timing of their divergence from their Eurasian ancestors; (ii) whether the peopling was in a single wave or multiple waves, and, consequently, if the genetic differences seen between major subgroups of Native Americans (e.g., Amerindian and Athabascan) result from different migrations or in situ diversification in the Americas (5, 6, 17, 18); (iii) if the migration involved ca. 15,000 years of isolation in the Bering Strait region, as proposed by the Beringian Incubation Model to explain the high frequency of unique and widespread American mitogenomes and private genetic variants (19–22); and, finally, (iv) if there was post-divergence gene flow from Eurasia and possibly even population replacement in the Americas, the latter suggested by the apparent differences in skull morphology between some early (‘Paleoamerican’) remains and those of more recent Native Americans (23–27). We address these issues using genomic data derived from modern populations, supplemented by ancient specimens that provide chronologically controlled snapshots of the genetics of the peopling process as it unfolded.
We sequenced 31 genomes from present-day individuals from the Americas, Siberia and Oceania to an average depth of ca. 20X: Siberians – Altai (n = 2), Buryat (n = 2), Ket (n=2), Koryak (n = 2), Sakha (n = 2), Siberian Yupik (n = 2); North American Native Americans – Tsimshian (n =); southern North American and Central and South American Natives – Pima (n = 1), Huichol (n = 1), Aymara (n = 1), Yukpa (n = 1); and, Oceanians – Papuan (n = 14) (28) (Table S1). All the genome-sequenced present-day individuals were previously genotyped using single nucleotide polymorphism (SNP) chips (4, 29–35) except for the Aymara individual that was SNP chip genotyped in this study (tables S3 and S4). They were selected on the basis of their ancestry profiles obtained with ADMIXTURE (36) to best represent their respective populations, and to minimize recent genetic admixture from populations of western Eurasian origin (28). For populations represented by more than one individual, we also verified from the genotype data that the sequenced individuals did not represent close relatives (28). We additionally sequenced 23 genomes from ancient individuals dating between ca. 0.2-6 KYA from North and South America, with an average depth ranging between 0.003X and 1.7X, including specimens affiliated to putative relict Paleoamerican groups such as the Pericúes from Mexico and Fuego-Patagonians from the southernmost tip of South America (23, 26–28) (table S5). Finally, we generated SNP chip genotype data from 79 present-day individuals belonging to 28 populations from the Americas and Siberia (28) (table S4). All the aforementioned datasets were analyzed together with previously published genomes and SNP chip genotype data (Tables S1, S3, and S4), masking the data for recent European admixture in some present-day Native American populations (28).
The structure of Native American populations and the timing of their initial divergence
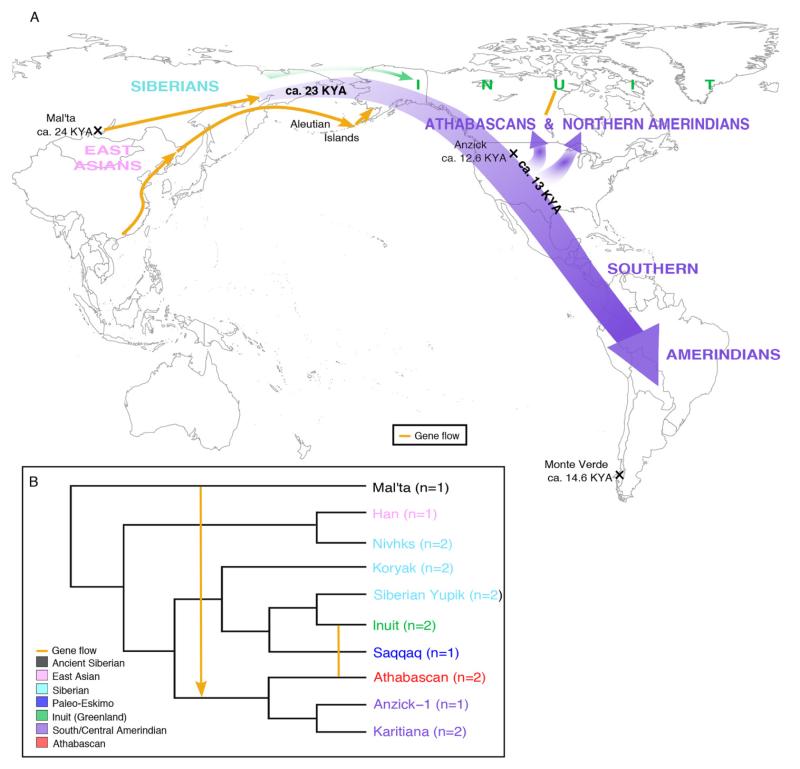
We explored the genetic structure of Native American populations in the context of worldwide populations using ADMIXTURE (36), employing a reference panel consisting of 3,053 individuals from 169 populations (table S3) (28). The panel included SNP chip genotype data from present-day individuals generated in this study and previously published studies, as well as the 4,000 year-old Saqqaq individual from Greenland (29) and the 12,600 year-old Anzick-1 (Clovis culture) individual from Montana (5) (table S3). When assuming four ancestral populations (_K_=4), we found a Native American-specific genetic component, indicating a shared genetic ancestry for all Native Americans including Amerindians and Athabascans (fig. S4). Assuming _K_=15, there is structure within the Native Americans. Athabascans and northern Amerindians (primarily from Canada) differ from the rest of the Native Americans in sharing their own genetic component (fig. S4). As reported previously, Anzick-1 falls within the genetic variation of southern Native Americans (5), while the Saqqaq individual shares genetic components with Siberian populations (fig. S4) (29).
To ascertain the population history of present-day American populations in relation to worldwide populations, we generated admixture graphs with TreeMix (28, 37). All the modern Siberian and Native American genomes sequenced in this study, except for the North American Tsimshian genome that showed evidence of recent western Eurasian admixture (28), were used for this analysis, together with previously published genomes from Africa (Yoruba) (38), Europe (Sardinian, French) (38), East Asia (Dai, Han) (38), Siberia (Nivkh) (39) and the Americas (Karitiana, Athabascan, Greenlandic Inuit) (5, 38, 39) (table S1). The ancient individuals included in the analysis were Saqqaq, Anzick-1 and the 24,000 year-old Mal’ta child from south-central Siberia (4). TreeMix affirms that all Native Americans form a monophyletic group across all ten migration parameter values, with further diversification into two branches, one representing Amerindians (represented in this analysis by Amerindians from southern North America and Central and South America) and the other Athabascans (Fig. 1B and fig. S5). Paleo-Eskimos and Inuit were supported as a separate clade relative to the Native Americans, as reported previously (Fig. 1B and fig. S5) (29, 39). Our results show that the Siberian Yupik and Koryak are the closest Eurasian populations to the Americas, with the Yupik likely representing back-migration of the Inuit into Siberia (Fig. 1B and fig. S5).
Fig. 1. Origins and population history of Native Americans.
(A) Our results show that the ancestors of all present-day Native Americans, including Amerindians and Athabascans, derived from a single migration wave into the Americas (purple), separate from the Inuit (green). This migration from East Asia occurred no later than 23 KYA and is in agreement with archaeological evidence from sites such as Monte Verde (50). A split between the northern and southern branches of Native Americans occurred ca. 13 KYA, with the former comprising Athabascans and northern Amerindians and the latter consisting of Amerindians in northern North America and Central and South America including the Anzick-1 individual (5). There is an admixture signal between Inuit and Athabascans and some northern Amerindians (yellow line); however, the gene flow direction is unresolved due to the complexity of the admixture events (28). Additionally, we see a weak signal related to Australo-Melanesians in some Native Americans, which may have been mediated through East Asians and Aleutian Islanders (yellow arrows). Also shown is the Mal’ta gene flow into Native American ancestors some 23 KYA (yellow arrow) (4). It is currently not possible for us to ascertain the exact geographical locations of the depicted events; hence, the positioning of the arrows should not be considered a reflection of these. B. Admixture plot created on the basis of TreeMix results (fig. S5) shows that all Native Americans form a clade, separate from the Inuit, with gene flow between some Native Americans and the North American Arctic. The number of genome-sequenced individuals included in the analysis is shown in brackets.
To assess the pattern of the earliest human dispersal into the Americas, we estimated the timing of the divergence of ancestral Native Americans from East Asians (hereafter, including Siberians) using multiple methods. There is still some debate regarding mutation rates in the human genome (40), and this uncertainty could affect our estimates and results.
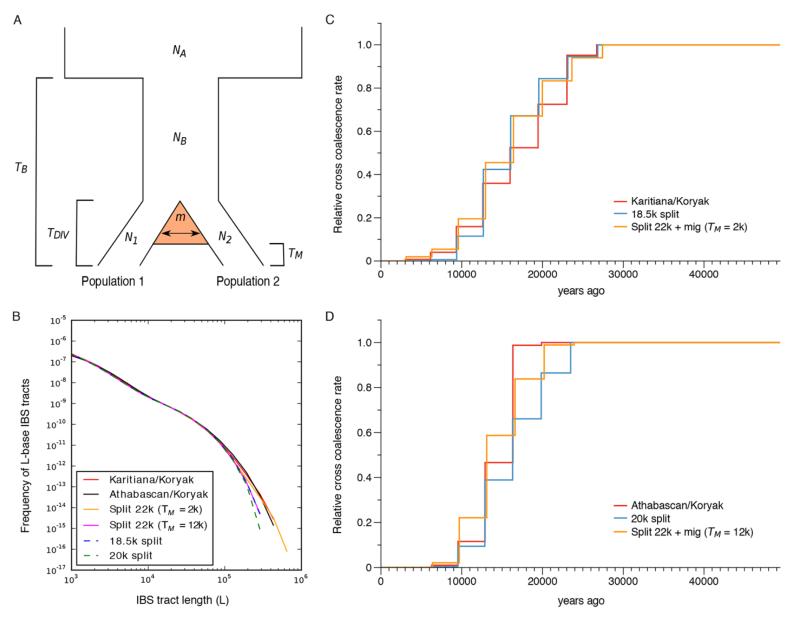
We applied diCal2.0 (28) (Method 1), a new version of diCal (41) extended to handle complex demographic models involving multiple populations with migration (42), and an identity-by-state (IBS) tract method (43) (Method 2) to the modern genome dataset (28). With these, we first estimated divergence times between Native Americans and the Koryak of Siberia, one of the genetically closest sampled East Asian populations to Native Americans (fig. S5), using demographic models that reflect a clean split between the populations (28). With both diCal2.0 and IBS tract method, the split of Native Americans (including Amerindians and Athabascans) from the Koryak dates to ca. 20 KYA (28) (tables S11A and S12 and fig. S15).
We further applied diCal2.0 to models with gene flow post-dating the split between Native Americans and Koryak (Fig. 2A) and found that they provided a better fit to the data than the models without gene flow (28). Overall, simulated databased on the models inferred using diCal2.0 and real data show very similar IBS tract length distributions (Fig. 2B) and relative cross coalescence rates (CCR) between pairs of individuals estimated using the Multiple Sequentially Markovian Coalescent (MSMC) method (Method 3) (28, 44) (Figs. 2, C and D). This serves as a confirmation for the model estimates from diCal2.0. We evaluated all the three methods using simulations under complex demographic models, and additionally investigated the effects of switch-errors in haplotype phasing on the estimates (28).
Fig. 2. Divergence estimates between Native Americans and Siberian Koryak.
(A) The demographic model used allows for continuous gene flow between populations 1 and 2, starting from the time TDIV of divergence and ending at TM. The backward probability of migration per individual per generation is denoted by m. The bottleneck at TB captures the out-of-Africa event. (B) The red and black solid curves depict empirical distributions of IBS tracts shared between Karitiana-Koryak and Athabascan-Koryak, respectively. The orange, pink, dashed blue and dashed green curves depict IBS tracts shared between the two population pairs, simulated under two demographic models based on results from diCal2.0. Overall, for Karitiana-Koryak and Athabascan-Koryak, the migration scenarios (orange and pink, respectively) match the empirical curves (red and black, respectively) better than the clean split scenarios (dashed blue and dashed green, respectively), with more long IBS tracts showing evidence of recent common ancestry between Koryaks and Native Americans. (C and D) Relative cross coalescence rates (CCR) for the Karitiana-Koryak and Athabascan-Koryak divergence (red), respectively, including data simulated under the two demographic models in panel B. In both cases, the model with gene flow (orange) fits the data (red) better than the clean split model (blue). The migration model explains a broader CCR tail in the case of Karitiana-Koryak and the relatively late onset of the CCR decay for Athabascan-Koryak.
We then applied the diCal2.0 model that allows for gene flow between populations after their split to estimate divergence times for Native Americans from more geographically and genetically distant East Asian groups, including the Siberian Nivkh and Han Chinese. As before, the divergence estimates for Amerindians and Athabascans were very similar to one another, ca. 23 KYA (table S11B and figs. S18 and S21).
Hence, our results suggest that Amerindians and Athabascans were, by three different methods, consistently equidistant in time to populations that were sampled from different regions of East Asia, including some proximate to Beringia, and with varied population histories. This suggests that these two major Native American sub-groups are descendants of the same source population that split off from ancestral East Asians during the LGM. It is conceivable that harsh climatic conditions during the LGM may have contributed to the isolation of ancestral Native Americans, ultimately leading to their genetic divergence from their East Asian ancestors.
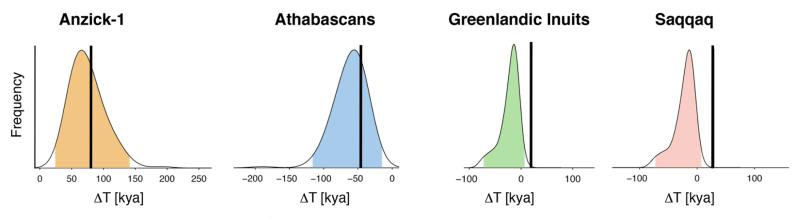
We also modeled the peopling of the Americas using a climate-informed spatial genetic model (CISGeM), in which the genetic history and local demography is informed by paleoclimatic and paleovegetation reconstructions (28, 45), and found the results to be in accordance with the conclusion of a single migration source for all Native Americans. Using present-day and ancient high coverage genomes, we found that Athabascans and Anzick-1, but not Greenlandic Inuit and Saqqaq (29, 39), belong to the same initial migration wave that also gave rise to present-day Amerindians from southern North America and Central and South America (Fig. 3), and that this migration likely followed a coastal route, given our current understanding of the glacial geological and paleoenvironmental parameters of the Late Pleistocene (fig. S31).
Fig. 3. Testing migrations into the Americas using a climate-informed model.
Estimates of difference in genetic divergence between Amerindians (from southern North America and Central and South America) or Koryak versus Athabascan and Greenlandic Inuit and the ancient Saqqaq and Anzick-1 genomes (black vertical lines), compared to posterior probability distribution predicted from a climate-informed spatial genetic model reconstructing a single wave into the Americas (curves, the colored part represents the 95% credibility interval). ΔT for population X is defined as T(X,Koryak)-T(X,Central and South Amerindians) (28). Both Anzick-1 and the Athabascans were part of the same wave into the Americas to which other Amerindian populations from southern North America and Central and South America belonged, while the Inuit and Saqqaq are the descendants of different waves (observed values outside the 95% credibility interval).
In all cases, the best fit of the demographic models to the IBS tract distribution and relative CCR by MSMC required gene flow between Siberian and Native American populations after their initial split (Figs. 2, B to D). We also found strong evidence for gene flow between Athabascans and the Inuit (table S11B) supported by results from ADMIXTURE (fig. S4), TreeMix (fig. S5), _D_-statistics employing both whole genome and SNP chip genotype data (28, 46, 47) (figs. S6 and S8A), and outgroup f3 statistics using whole genome data (28, 47) (Fig. S12). We attempted to estimate the divergence times between Inuit and Siberians as well as Inuit and Native Americans (table S11 and figs. S19 and S25 to S27), but our analyses were complicated by gene flow between Inuit and Athabascans as well as complex admixture patterns among Arctic groups (fig. S5).
We tested the duration and magnitude of post-split gene flow between Native Americans and Siberians using diCal2.0 by introducing stopping time of gene flow as a free parameter (28). We still obtained the highest likelihood for a divergence time of 22 KYA between Amerindians and Siberians as well as Athabascans and Siberians, although estimates for gene flow rate and end of the gene flow differ (table S11C and fig. S22). Significant gene flow between Athabascans and Siberians seems to have stopped ca. 12 KYA (Table S11C), suggesting a link to the breaching of the Beringian Land Bridge by rising sea levels (48).
Overall, our results support a common Siberian origin for all Native Americans, contradicting claims for an early migration to the Americas from Europe (49), with their initial isolation and entrance into the Americas occurring no earlier than 23 KYA, but with subsequent admixture with East Asian populations. This additionally suggests that the Mal’ta-related admixture into the early Americans (4), representing ancestors of both Amerindians and Athabascans (Fig. 1 and fig. S5), occurred sometime after 23 KYA, following the Native American split from East Asians.
Subsequent in situ diversification of Native American groups
That Amerindian and Athabascan groups were part of the same migration implies that present-day genetic differences observed between them must have arisen later, after ca. 23 KYA. Using the clean-split model in diCal2.0 on the modern genomes dataset, we estimated that Athabascans and Karitiana diverged ca. 13 KYA (95% confidence interval of ca. 11.5-14.5 KYA, estimated from parametric bootstrap results) (table S11A, fig. S16), which is consistent with results from MSMC (fig. S27) (28).
Where the divergence between Karitiana and Athabascans occurred is not known. However, several independent lines of evidence suggest that it is more likely to have occurred in lower latitude North America instead of eastern Beringia (Alaska). These include the equidistant split times of Amerindians and Athabascans to Asian populations, the relatively brief interval between their estimated divergence date range and the age of Anzick-1 (12.6 KYA) (5), and lastly, the geographic location of Anzick-1 to the south of the North American ice sheets and its clear affiliation with the ‘southern branch’ of Native Americans (taken broadly to include Amerindians from southern North America and Central and South America) (5), as determined with outgroup f3 statistics using SNP chip genotype data from present-day worldwide populations (47) (Fig. 4 and figs. S13 and S14). Divergence in North America would also be consistent with the known pre-Clovis age sites in the Americas, such as Monte Verde (14.6 KYA) (50). The most parsimonious model would be that both Amerindians and Athabascans are descendants of the same ancestral Native American population that entered the Americas then subsequently diversified. However, we cannot discount alternative and more complex scenarios, which could be tested with additional ancient samples.
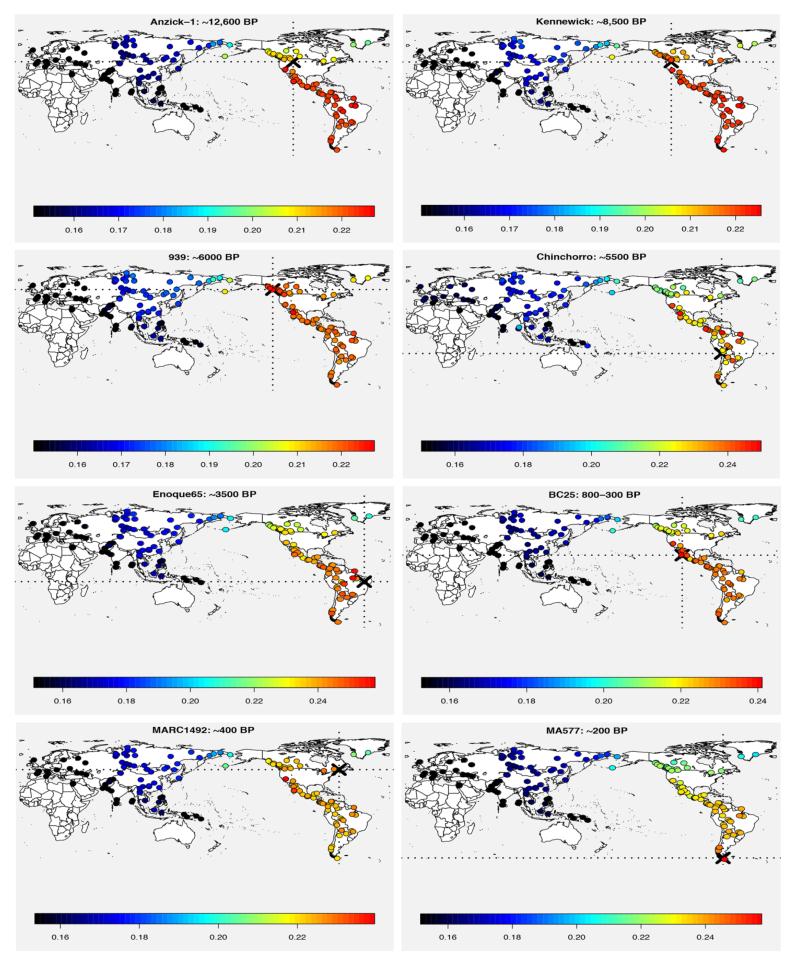
Fig. 4. Diversification within the Americas.
SNP chip genotype data-based outgroup f3 statistics (47) of the form f3(X, Ancient; Yoruba) were used to estimate the shared ancestry between ancient samples from the Americas and a large panel of worldwide present-day populations (X), including Athabascan and Amerindian groups from North America (table S3), some of which were masked for non-Native ancestry prior to the analysis (28). The outgroup f3 statistics are depicted as heat maps with the sampling location of the ancient sample marked by the dotted lines, and corresponding ranked plots with error bars are shown in fig. S14. BP refers to time before present. We find the Anzick-1 sample to share most ancestry with the ‘southern’ branch of Native Americans when using multiple northern Native Americans sequenced in this study, consistent with (5). The seven Holocene aged samples share most ancestry with Native Americans, with a general tendency to be genetically closer to present-day Native American populations from the same geographical region.
By the Clovis period (ca. 12.6 KYA), the ancestral Native American population had already diversified into 'northern' and 'southern' branches, with the former including ancestors of present-day Athabascans and northern Amerindian groups such as Chipewyan, Cree and Ojibwa and the latter including Amerindians from southern North America and Central and South America (Fig. 4 and fig. S14). We tested whether later gene flow from East Asian sources, such as the Inuit, might explain the genetic differences between these two branches. Using _D_-statistics on SNP chip genotype data (47) masked for non-Native ancestry, we observed a signal of gene flow between the Inuit and northwest Pacific Coast Amerindians such as Coastal Tsimshian and Nisga’a, residing in the same region as the northern Athabascans (28) (fig. S8B). However, this signal of admixture with the Inuit, also detected in Athabascans (figs. S6 and S8A), was not evident among northern Amerindian populations located further east such as Cree, Ojibwa and Chipewyan (28) (fig. S8C). This suggests that the observed difference between the ‘northern’ and ‘southern’ branches is not a consequence of post-split East Asian gene flow into the ‘northern branch’, and also provides a possible explanation as to why the ’southern branch’ Amerindians such as Karitiana are genetically closer to the northern Amerindians located further east than to northwest coast Amerindians and Athabascans (fig. S9).
In contrast to Anzick-1, several of the Holocene individuals from the Americas, including those sequenced in this study as well as the 8,500 year old Kennewick Man (51), are closely related to present-day Native American populations from the same geographical regions (Fig. 4 and figs. S13 and S14). This implies genetic continuity of ancient and modern populations in some parts of the Americas over at least the last 8.5 KYA, which is in agreement with recent results from Kennewick Man (51).
Evidence of more distant Old World gene flow into some Native Americans
When testing for gene flow between Athabascans and Inuit with masked SNP chip genotype data-based _D_-statistics (47) (fig. S8), we observed a weak tendency for the Inuit to be much closer to the Athabascans than to certain Amerindians like the North American Algonquin and Cree, and the Yaqui and Arhuaco of Central and South America (respectively), as compared to other Amerindians such as the Palikur and Surui of Brazil (fig. S8).
To further investigate this trend, we tested for additional gene flow from Eurasian populations into the Americas with _D_-statistics using the masked SNP chip genotype dataset (47). We found that some American populations, including the Aleutian Islanders, Surui, and Athabascans are closer to Australo-Melanesians compared to other Native Americans, such as North American Ojibwa, Cree and Algonquin, and the South American Purepecha, Arhuaco and Wayuu (fig. S10). The Surui are, in fact, one of closest Native American populations to East Asians and Australo-Melanesians, the latter including Papuans, non-Papuan Melanesians, Solomon Islanders, and South East Asian hunter-gatherers such as Aeta (fig. S10). We acknowledge that this observation is based on the analysis of a small fraction of the whole genome and SNP chip genotype datasets, especially for the Aleutian Islander data that is heavily masked due to recent admixture with Europeans (28), and that the trends in the data are weak.
Nonetheless, if it proves correct, these results suggest there may be a distant Old World signal related to Australo-Melanesians and East Asians in some Native Americans. The widely scattered and differential affinity of Native Americans to the Australo-Melanesians, ranging from a strong signal in the Surui to much weaker signal in northern Amerindians such as Ojibwa, points to this gene flow occurring after the initial peopling by Native American ancestors.
However, how this signal may have ultimately reached South America remains unclear. One possible means is along a northern route via the Aleutian Islanders, previously found to be closely related to the Inuit (39), who have a relatively greater affinity to East Asians, Oceanians and Denisovan than Native Americans in both whole genome and SNP chip genotype data-based _D_-tests (table S10 and figs. S10 and S11). On the basis of archaeological evidence and mtDNA data from ancient and modern samples, the Aleutian Islands are hypothesized to have been peopled as early as ca. 9 KYA by ‘Paleo-Aleuts’ who were succeeded by the ‘Neo-Aleuts’, with present-day Aleutian Islanders potentially resulting from admixture between these two populations (52, 53). Perhaps their complex genetic history included input from a population related to Australo-Melanesians through an East Asian continental route, and this genomic signal might have been subsequently transferred to parts of the Americas, including South America, through past gene flow events (Fig. 1). Evidence for this gene flow is supported by diCal2.0 and MSMC analyses showing a weak but recent gene flow into South Americans from populations related to present-day Northeast Asians (Koryak) (Fig. 2C and table S11C), who might be considered a proxy for the related Aleutian Islanders.
Testing the Paleoamerican model
The detection of an Australo-Melanesian genetic signal in the Americas, however subtle, returns the discussion to the Paleoamerican model, which hypothesizes, on the basis of cranial morphology, that two temporally and source-distinct populations colonized the Americas. The earlier population reportedly originated in Asia in the Late Pleistocene and gave rise to both Paleoamericans and present-day Australo-Melanesians, whose shared cranial morphological attributes are presumed to indicate their common ancestry (23). The Paleoamericans were, in turn, thought to have been largely replaced by ancestors of present-day Amerindians, whose crania resemble modern East Asians and who are argued to be descendants of later arriving Mongoloid populations (14, 23, 26, 54). The presence of Paleoamericans is inferred primarily from ancient archaeological specimens in North and South America, and a few relict populations of more recent age, which include the extinct Pericúes and Fuego-Patagonians (24, 25, 55).
The Paleoamerican hypothesis predicts that these groups should be genetically closer to Australo-Melanesians than other Amerindians. Previous studies of mtDNA and Y chromosome data obtained from Fuego-Patagonian and Paleo-american skeletons have identified haplogroups similar to those of modern Native Americans (55–57). Although these results indicate some shared maternal and paternal ancestry with contemporary Native Americans, uniparental markers can be misleading when drawing conclusions about the demographic history of populations. To conclusively identify the broader population of ancestors who may have contributed to the Paleoamerican gene pool, autosomal genomic data are required.
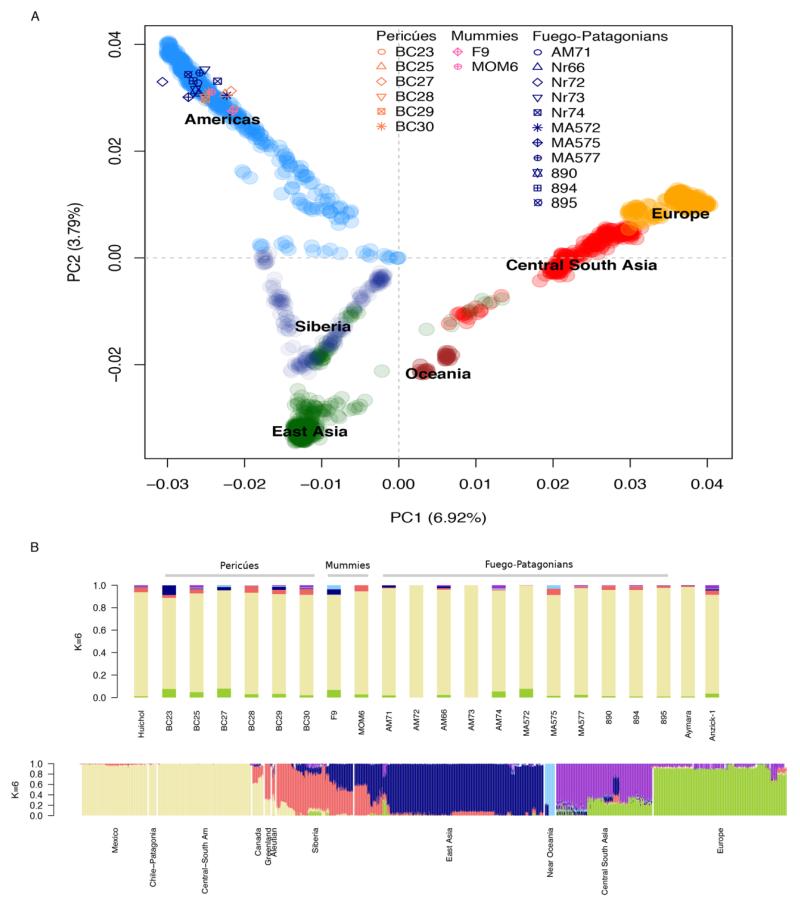
We, therefore, sequenced 17 ancient individuals affiliated to the now-extinct Pericúes from Mexico and Fuego-Patagonians from Chile and Argentina (28), who, on the basis of their distinctive skull morphologies, are claimed to be relicts of Paleoamericans (23, 27, 58, 59). Additionally, we sequenced two pre-Columbian mummies from northern Mexico (Sierra Tarahumara) to serve as morphological controls, since they are expected to fall within the range of Native American morphological cranial variation (28). We found that the ancient samples cluster with other Native American groups and are outside the range of Oceanian genetic variation (28) (Fig. 5 and figs. S32, S33, and S34). Similarly, outgroup f3 statistics (47) reveal low shared genetic ancestry between the ancient samples and Oceanians (28) (Figs. S36, S37), and genome-based and masked SNP chip genotype data-based _D_-statistics (46, 47) show no evidence for gene flow from Oceanians into the Pericúes or Fuego-Patagonians (28) (fig. S39).
Fig. 5. The Paleoamerican model.
(A) Principal Component Analysis plot of 19 ancient samples combined with a worldwide reference panel, including 1,823 individuals from (6). Our samples plot exclusively with American samples. For plots with other reference panels consisting of Native American populations, see fig. S32. (B) Population structure in the ancient Pericú, Mexican mummy and Fuego-Patagonian individuals from this study. Ancestry proportions are shown when assuming six ancestral populations (K = 6). The top bar shows the ancestry proportions of the 19 ancient individuals, Anzick-1 (5), and two present-day Native American genomes from this study (Huichol and Aymara). The plot at the bottom illustrates the ancestry proportions for 1,823 individuals from (6). Our samples show primarily Native American (ivory, >92%) and Siberian (red, ca. 5%) ancestry. For the plot with K =13, see fig. S33.
As the Paleoamerican model is based on cranial morphology (23, 27, 58, 59), we also measured craniometric data for the ancient samples and assessed their phenotypic affinities to supposed Paleoamericans, Amerindians and world-wide populations (28). The results revealed that the analyzed Fuego-Patagonians showed closest craniometric affinity to Arctic populations and the Paleoamericans, while the analyzed female Pericúes showed closest craniometric affinities to populations from North America, the Arctic region and Northern Japan (table S15). More importantly, our analyses demonstrated that the presumed ancestral ancient Paleoamerican reference sample from Lagoa Santa, Brazil (24) had closest affinities to Arctic and East Asian populations (table S15). Consequently, for the Fuego-Patagonians, the female Pericúes and the Lagoa Santa Paleoamerican sample, we were not able to replicate previous results (24) that report close similarity of Paleoamerican and Australo-Melanesian cranial morphologies. We note that male Pericúes samples displayed more craniometric affinities with populations from Africa and Australia relative to the female individuals of their population (fig. S41). The results of analyses based on craniometric data are, thus, highly sensitive to sample structure and the statistical approach and data filtering used (51). Our morphometric analyses suggest that these ancient samples are not true relicts of a distinct migration, as claimed, and hence do not support the Paleo-american model. Similarly, our genomic data also provide no support for an early migration of populations directly related to Australo-Melanesians into the Americas.
Discussion
That Native Americans diverged from their East Asian ancestors during the LGM and no earlier than 23 KYA provides an upper bound, and perhaps the climatic and environmental context, for the initial isolation of their ancestral population, and a maximum estimate for the entrance and subsequent spread into the Americas. This result is consistent with the model that people entered the Americas prior to the development of the Clovis complex and had reached as far as southern South America by 14.6 KYA. As archaeological evidence provides only a minimum age for human presence in the Americas, we can anticipate the possible discovery of sites that approach the time of the divergence of East Asians and Native Americans. However, our estimate for the initial divergence and entry of Native American ancestors does not support archaeological claims for an initial peopling significantly earlier than the LGM (8–10).
While our data cannot provide the precise geographical context for the initial peopling process, it has allowed us to more accurately estimate its temporal dynamics. This, in turn, has enabled us to re-assess the Beringian Incubation Model, which, based on mtDNA data and the timing and geographical distribution of archaeological sites, hypothesized a ca. 15,000 year-long period of isolation of ancestral Native Americans in Beringia during the LGM (19–21). Our results, along with recent findings of mtDNA haplogroup C1 in Iceland and ancient northwest Russia (60), do not fit with the proposed 15,000-year span of the Beringian Incubation Model (19–21). It is possible that a shorter period of isolation occurred (ca. 8 KYA), but whether it occurred in Siberia or Beringia will have to be determined by future ancient DNA and archaeological findings. Given the genetic continuity between Native Americans and some East Asian populations (figs. S4 and S5), other demographic factors, such as surfing during population expansions into unoccupied regions (61), may ultimately need to be taken into account to better understand the presence of a large number of high frequency private variants in the indigenous populations of the Americas.
The data presented here are consistent with a single initial migration of all Native Americans and with later gene flow from sources related to East Asians and, more distantly, Australo-Melanesians. From that single migration, there was a diversification of ancestral Native Americans leading to the formation of ‘northern’ and ‘southern’ branches, which appears to have taken place ca. 13 KYA within the Americas. This split is consistent with the patterns of uniparental genomic regions of mtDNA haplogroup X and some Y chromosome C haplotypes being present in northern, but not southern, populations in the Americas (18, 62). This diversification event coincides roughly with the opening of habitable routes along the coastal and the interior corridors into unglaciated North America some 16 KYA and 14 KYA, respectively (63, 64), suggesting a possible role of one or both these routes in the isolation and subsequent dispersal of Native Americans across the continent.
Methods
DNA was extracted from 31 present-day individuals from the Americas, Siberia and Oceania and 23 ancient samples from the Americas, and converted to Illumina libraries and shotgun-sequenced (28). Three of the ancient samples were radiocarbon dated, of which two were corrected for marine reservoir offset (28). SNP chip genotype data was generated from 79 present-day Siberians and Native Americans affiliated to 28 populations (28). Raw data from SNP chip and shotgun sequencing were processed using standard computational procedures (28). Error rate analysis, DNA damage analysis, contamination estimation, sex determination, mtDNA and Y chromosome haplogroup assignment, ADMIXTURE analysis, ancestry painting and admixture masking, Principal Component Analysis using SNP chip genotype data, TreeMix analysis on genomic sequence data, _D_-statistic and outgroup _f3_-statistic tests on SNP chip genotype and genomic sequence data, divergence time estimation using diCal2.0, an IBS tract method and MSMC, Climate-Informed Spatial Genetic Model analysis, and, craniometric analysis were performed as described (28).
Supplementary Material
Raghavan 2015 Americas SM
ACKNOWLEDGMENTS
We thank J. Valdés for providing craniometric measurements of the Pericúes at the National Museum of Anthropology in México; A. Monteverde from CINAH-Baja California Sur and V. Laborde at the Musée de l’Homme in Paris for providing documentation on Pericú and Fuego-Patagonian samples, respectively; T. Gilbert, M. McCoy, C. Sarkissian, M. Sikora, L. Orlando for helpful discussions and input; D. Yao and C. Barbieri for helping with the collection of the Aymara population sample; B. Henn and J. Kidd for providing early access to the Mayan sequencing data; Canadian Museum of History; Metlakatla and Lax Kw'alaams First Nations; Listuguj Mi’gmaq Band Council; A. Pye of TERRA Facility, Core Research Equipment & Instrument Training (CREAIT) Network at Memorial University; the Danish National High-throughput DNA Sequencing Centre (Copenhagen) for help with sequencing; and, Fondation Jean Dausset-Centre de'Etude du Polymorphism Humain (CEPH) for providing DNA for the Human Genome Diversity Project (HGDP) samples that were genome-sequenced in this study. This study was supported by several funding bodies; Lundbeck Foundation and the Danish National Research (Centre for GeoGenetics members), Wellcome Trust grant 098051 (S.S., A.B., Y.X., C.T.-S., M.S.S., R.D.), Marie Curie Intra-European Fellowship-FP7-People-PIEF-GA-2009-255503 and the Transforming Human Societies Research Focus Areas Fellowship from La Trobe University (C.V.), George Rosenkranz Prize for Health Care Research in Developing Countries and National Science Foundation award DMS-1201234 (M.C.A.A), Swiss National Science Foundation (PBSKP3_143529) (A.-S.M.), Ministerio de Ciencia e Innovación (MICINN) Project CGL2009-12703-C03-03 and MICINN (BES-2010-030127) (R.R.-V.), Consejo Nacional de Ciencia y Tecnología (Mexico) (J.V.M.M.), Biotechnology and Biological Sciences Research Council BB/H005854/1 (V.W., F.B., A.M.), European Research Council and Marie Curie Actions Grant 300554 (M.E.A.), Wenner-Gren Foundations and the Australian Research Council Future Fellowship FT0992258 (C.I.S.), European Research Council ERC-2011-AdG 295733 grant (Langelin) (D.P. and D.L.), Bernice Peltier Huber Charitable Trust (C.H., L.G.D.), Russian Foundation for Basic Research grant 13-06-00670 (E.B.), Russian Foundation for Basic Research grant 14-0400725 (E.K.), European Union European Regional Development Fund through the Centre of Excellence in Genomics to Estonian Biocentre and Estonian Institutional Research grant IUT24-1 (E.M., K.T., M.M., M.K., R.V.), Estonian Science Foundation grant 8973 (M.M.), Stanford Graduate Fellowship (J.R.H.), Washington State University (B.M.K.), French National Research Agency grant ANR-14-CE31-0013-01 (F-X.R), European Research Council grant 261213 (T.K.), National Science Foundation BCS- 1025139 (R.S.M.), Social Science Research Council of Canada (K.-A.P., V.G.), National Institutes of Health grants R01-GM094402 (M.S., Y.S.S.); R01 - AI17892 (P.J.N., P.P.); 2R01HG003229-09 (R.N., C.D.B.), Packard Fellowship for Science and Engineering (Y.S.S.), Russian Science Fund grant 14-04-00827 and Presidium of Russian Academy of Sciences Molecular and Cell Biology Programme (O.B.), and Russian Foundation for Basic Research grant 14-06-00384 (Y.B.). Informed consent was obtained for the sequencing of the modern individuals, with ethical approval from The National Committee on Health Research Ethics, Denmark (H-3-2012-FSP21). SNP chip genotype data and whole genome data for select present-day individuals are available only for demographic research under data access agreement with E.W. (see Tables S1 and S4 for a list of these samples). Raw reads from the ancient and the remainder of the present-day genomes are available for download through European Nucleotide Archive (ENA) accession no. PRJEB9733, and the corresponding alignment files are available at http://www.cbs.dtu.dk/suppl/NativeAmerican/. The remainder of the SNP chip genotype data can be accessed through Gene Expression Omnibus (GEO) series accession no. GSE70987 and at www.ebc.ee/free_data. C.D.B. is on the advisory board of Personalis, Inc.; Identify Genomics; Etalon DX; and Ancestry.com.
Footnotes
The authors declare no competing financial interests.
REFERENCES AND NOTES
- 1.Dillehay TD. The late Pleistocene cultures of South America. Evol. Anthropol. 1999;7:206–216. doi:10.1002/(SICI)1520-6505(1999)7:6<206::AID-EVAN5>3.0.CO;2-G. [Google Scholar]
- 2.Jenkins DL, Davis LG, Stafford TW, Jr., Campos PF, Hockett B, Jones GT, Cummings LS, Yost C, Connolly TJ, Yohe RM, 2nd, Gibbons SC, Raghavan M, Rasmussen M, Paijmans JL, Hofreiter M, Kemp BM, Barta JL, Monroe C, Gilbert MT, Willerslev E. Clovis age Western Stemmed projectile points and human coprolites at the Paisley Caves. Science. 2012;337:223–228. doi: 10.1126/science.1218443. [DOI] [PubMed] [Google Scholar]
- 3.Meltzer DJ. First Peoples in a New World: Colonizing Ice Age America. University of California Press; Berkeley: 2009. [Google Scholar]
- 4.Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, Moltke I, Rasmussen S, Stafford TW, Jr., Orlando L, Metspalu E, Karmin M, Tambets K, Rootsi S, Mägi R, Campos PF, Balanovska E, Balanovsky O, Khusnutdinova E, Litvinov S, Osipova LP, Fedorova SA, Voevoda MI, DeGiorgio M, Sicheritz-Ponten T, Brunak S, Demeshchenko S, Kivisild T, Villems R, Nielsen R, Jakobsson M, Willerslev E. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature. 2014;505:87–91. doi: 10.1038/nature12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasmussen M, Anzick SL, Waters MR, Skoglund P, DeGiorgio M, Stafford TW, Jr., Rasmussen S, Moltke I, Albrechtsen A, Doyle SM, Poznik GD, Gudmundsdottir V, Yadav R, Malaspinas A-S, White SS, 5th, Allentoft ME, Cornejo OE, Tambets K, Eriksson A, Heintzman PD, Karmin M, Korneliussen TS, Meltzer DJ, Pierre TL, Stenderup J, Saag L, Warmuth VM, Lopes MC, Malhi RS, Brunak S, Sicheritz-Ponten T, Barnes I, Collins M, Orlando L, Balloux F, Manica A, Gupta R, Metspalu M, Bustamante CD, Jakobsson M, Nielsen R, Willerslev E. The genome of a Late Pleistocene human from a Clovis burial site in western Montana. Nature. 2014;506:225–229. doi: 10.1038/nature13025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reich D, Patterson N, Campbell D, Tandon A, Mazieres S, Ray N, Parra MV, Rojas W, Duque C, Mesa N, García LF, Triana O, Blair S, Maestre A, Dib JC, Bravi CM, Bailliet G, Corach D, Hünemeier T, Bortolini MC, Salzano FM, Petzl-Erler ML, Acuña-Alonzo V, Aguilar-Salinas C, Canizales-Quinteros S, Tusié-Luna T, Riba L, Rodríguez-Cruz M, Lopez-Alarcón M, Coral-Vazquez R, Canto-Cetina T, Silva-Zolezzi I, Fernandez-Lopez JC, Contreras AV, Jimenez-Sanchez G, Gómez-Vázquez MJ, Molina J, Carracedo A, Salas A, Gallo C, Poletti G, Witonsky DB, Alkorta-Aranburu G, Sukernik RI, Osipova L, Fedorova SA, Vasquez R, Villena M, Moreau C, Barrantes R, Pauls D, Excoffier L, Bedoya G, Rothhammer F, Dugoujon JM, Larrouy G, Klitz W, Labuda D, Kidd J, Kidd K, Di Rienzo A, Freimer NB, Price AL, Ruiz-Linares A. Reconstructing Native American population history. Nature. 2012;488:370–374. doi: 10.1038/nature11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waters MR, Forman SL, Jennings TA, Nordt LC, Driese SG, Feinberg JM, Keene JL, Halligan J, Lindquist A, Pierson J, Hallmark CT, Collins MB, Wiederhold JE. The Buttermilk Creek complex and the origins of Clovis at the Debra L. Friedkin site, Texas. Science. 2011;331:1599–1603. doi: 10.1126/science.1201855. [DOI] [PubMed] [Google Scholar]
- 8.Santos GM, Bird MI, Parenti F, Fifield LK, Guidon N, Hausladen PA. A revised chronology of the lowest occupation layer of Pedra Furada Rock Shelter, Piauí, Brazil: The Pleistocene peopling of the Americas. Quat. Sci. Rev. 2003;22:2303–2310. doi: 10.1016/S0277-3791(03)00205-1. [DOI] [Google Scholar]
- 9.Holen SR, Holen K, K . In: Paleoamerican Odyssey. Graf KE, Ketron CV, Waters MR, editors. Texas A&M University Press; College Station: 2014. pp. 429–444. [Google Scholar]
- 10.Boëda E, Clemente-Conte I, Fontugne M, Lahaye C, Pino M, Felice GD, Guidon N, Hoeltz S, Lourdeau A, Pagli M, Pessis A-M, Viana S, Da Costa A, Douville E. A new late Pleistocene archaeological sequence in South America: The Vale da Pedra Furada (Piauí, Brazil) Antiquity. 2014;88:927–941. doi: 10.1017/S0003598X00050845. [DOI] [Google Scholar]
- 11.Owsley DW, Jantz RL. Kennewick Man: The Scientific Investigation of an Ancient American Skeleton. Texas A&M University Press; College Station: 2014. [Google Scholar]
- 12.Achilli A, Perego UA, Bravi CM, Coble MD, Kong QP, Woodward SR, Salas A, Torroni A, Bandelt HJ. The phylogeny of the four pan-American MtDNA haplogroups: Implications for evolutionary and disease studies. PLOS ONE. 2008;3:e1764. doi: 10.1371/journal.pone.0001764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Battaglia V, Grugni V, Perego UA, Angerhofer N, Gomez-Palmieri JE, Woodward SR, Achilli A, Myres N, Torroni A, Semino O. The first peopling of South America: New evidence from Y-chromosome haplogroup Q. PLOS ONE. 2013;8:e71390. doi: 10.1371/journal.pone.0071390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brace CL, Nelson AR, Seguchi N, Oe H, Sering L, Qifeng P, Yongyi L, Tumen D. Old World sources of the first New World human inhabitants: A comparative craniofacial view. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10017–10022. doi: 10.1073/pnas.171305898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perego UA, Achilli A, Angerhofer N, Accetturo M, Pala M, Olivieri A, Kashani B. Hooshiar, Ritchie KH, Scozzari R, Kong Q-P, Myres NM, Salas A, Semino O, Bandelt H-J, Woodward SR, Torroni A. Distinctive Paleo-Indian migration routes from Beringia marked by two rare mtDNA haplogroups. Curr. Biol. 2009;19:1–8. doi: 10.1016/j.cub.2008.11.058. [DOI] [PubMed] [Google Scholar]
- 16.Perego UA, Angerhofer N, Pala M, Olivieri A, Lancioni H, Kashani B. Hooshiar, Carossa V, Ekins JE, Gómez-Carballa A, Huber G, Zimmermann B, Corach D, Babudri N, Panara F, Myres NM, Parson W, Semino O, Salas A, Woodward SR, Achilli A, Torroni A. The initial peopling of the Americas: A growing number of founding mitochondrial genomes from Beringia. Genome Res. 2010;20:1174–1179. doi: 10.1101/gr.109231.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fagundes NJR, Kanitz R, Bonatto SL. A reevaluation of the Native American mtDNA genome diversity and its bearing on the models of early colonization of Beringia. PLOS ONE. 2008;3:e3157. doi: 10.1371/journal.pone.0003157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zegura SL, Karafet TM, Zhivotovsky LA, Hammer MF. High-resolution SNPs and microsatellite haplotypes point to a single, recent entry of Native American Y chromosomes into the Americas. Mol. Biol. Evol. 2004;21:164–175. doi: 10.1093/molbev/msh009. [DOI] [PubMed] [Google Scholar]
- 19.Tamm E, Kivisild T, Reidla M, Metspalu M, Smith DG, Mulligan CJ, Bravi CM, Rickards O, Martinez-Labarga C, Khusnutdinova EK, Fedorova SA, Golubenko MV, Stepanov VA, Gubina MA, Zhadanov SI, Ossipova LP, Damba L, Voevoda MI, Dipierri JE, Villems R, Malhi RS. Beringian standstill and spread of Native American founders. PLOS ONE. 2007;2:e829. doi: 10.1371/journal.pone.0000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitchen A, Miyamoto MM, Mulligan CJ. A three-stage colonization model for the peopling of the Americas. PLOS ONE. 2008;3:e1596. doi: 10.1371/journal.pone.0001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulligan CJ, Kitchen A, Miyamoto MM. Updated three-stage model for the peopling of the Americas. PLOS ONE. 2008;3:e3199. doi: 10.1371/journal.pone.0003199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schroeder KB, Schurr TG, Long JC, Rosenberg NA, Crawford MH, Tarskaia LA, Osipova LP, Zhadanov SI, Smith DG. A private allele ubiquitous in the Americas. Biol. Lett. 2007;3:218–223. doi: 10.1098/rsbl.2006.0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González-José R, González-Martín A, Hernández M, Pucciarelli HM, Sardi M, Rosales A, Van Der Molen S. Craniometric evidence for Palaeoamerican survival in Baja California. Nature. 2003;425:62–65. doi: 10.1038/nature01816. [DOI] [PubMed] [Google Scholar]
- 24.Neves WA, Hubbe M. Cranial morphology of early Americans from Lagoa Santa, Brazil: Implications for the settlement of the New World. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18309–18314. doi: 10.1073/pnas.0507185102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neves W, et al. In: Paleoamerican Odyssey. Graf KE, Ketron CV, Waters MR, editors. Texas A&M University Press; College Station: 2014. pp. 397–412. [Google Scholar]
- 26.González-José R, Bortolini MC, Santos FR, Bonatto SL. The peopling of America: Craniofacial shape variation on a continental scale and its interpretation from an interdisciplinary view. Am. J. Phys. Anthropol. 2008;137:175–187. doi: 10.1002/ajpa.20854. [DOI] [PubMed] [Google Scholar]
- 27.Lahr MM. Patterns of modern human diversification: Implications for Amerindian origins. Am. J. Phys. Anthropol. 1995;38(S21):163–198. doi: 10.1002/ajpa.1330380609. [DOI] [Google Scholar]
- 28.Materials and methods are available as supplementary materials on Science Online.
- 29.Rasmussen M, Li Y, Lindgreen S, Pedersen JS, Albrechtsen A, Moltke I, Metspalu M, Metspalu E, Kivisild T, Gupta R, Bertalan M, Nielsen K, Gilbert MT, Wang Y, Raghavan M, Campos PF, Kamp HM, Wilson AS, Gledhill A, Tridico S, Bunce M, Lorenzen ED, Binladen J, Guo X, Zhao J, Zhang X, Zhang H, Li Z, Chen M, Orlando L, Kristiansen K, Bak M, Tommerup N, Bendixen C, Pierre TL, Grønnow B, Meldgaard M, Andreasen C, Fedorova SA, Osipova LP, Higham TF, Ramsey CB, Hansen TV, Nielsen FC, Crawford MH, Brunak S, Sicheritz-Pontén T, Villems R, Nielsen R, Krogh A, Wang J, Willerslev E. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757–762. doi: 10.1038/nature08835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yunusbayev B, Metspalu M, Metspalu E, Valeev A, Litvinov S, Valiev R, Akhmetova V, Balanovska E, Balanovsky O, Turdikulova S, Dalimova D, Nymadawa P, Bahmanimehr A, Sahakyan H, Tambets K, Fedorova S, Barashkov N, Khidiyatova I, Mihailov E, Khusainova R, Damba L, Derenko M, Malyarchuk B, Osipova L, Voevoda M, Yepiskoposyan L, Kivisild T, Khusnutdinova E, Villems R. The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia. PLOS Genet. 2015;11:e1005068. doi: 10.1371/journal.pgen.1005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardona A, Pagani L, Antao T, Lawson DJ, Eichstaedt CA, Yngvadottir B, Shwe MT, Wee J, Romero IG, Raj S, Metspalu M, Villems R, Willerslev E, Tyler-Smith C, Malyarchuk BA, Derenko MV, Kivisild T. Genome-wide analysis of cold adaptation in indigenous Siberian populations. PLOS ONE. 2014;9:e98076. doi: 10.1371/journal.pone.0098076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 33.Moreno-Estrada A, Gravel S, Zakharia F, McCauley JL, Byrnes JK, Gignoux CR, Ortiz-Tello PA, Martínez RJ, Hedges DJ, Morris RW, Eng C, Sandoval K, Acevedo-Acevedo S, Norman PJ, Layrisse Z, Parham P, Martínez-Cruzado JC, Burchard EG, Cuccaro ML, Martin ER, Bustamante CD. Reconstructing the population genetic history of the Caribbean. PLOS Genet. 2013;9:e1003925. doi: 10.1371/journal.pgen.1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreno-Estrada A, Gignoux CR, Fernández-López JC, Zakharia F, Sikora M, Contreras AV, Acuña-Alonzo V, Sandoval K, Eng C, Romero-Hidalgo S, Ortiz-Tello P, Robles V, Kenny EE, Nuño-Arana I, Barquera-Lozano R, Macín-Pérez G, Granados-Arriola J, Huntsman S, Galanter JM, Via M, Ford JG, Chapela R, Rodriguez-Cintron W, Rodríguez-Santana JR, Romieu I, Sienra-Monge JJ, del Rio Navarro B, London SJ, Ruiz-Linares A, Garcia-Herrera R, Estrada K, Hidalgo-Miranda A, Jimenez-Sanchez G, Carnevale A, Soberón X, Canizales-Quinteros S, Rangel-Villalobos H, Silva-Zolezzi I, Burchard EG, Bustamante CD. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science. 2014;344:1280–1285. doi: 10.1126/science.1251688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verdu P, Pemberton TJ, Laurent R, Kemp BM, Gonzalez-Oliver A, Gorodezky C, Hughes CE, Shattuck MR, Petzelt B, Mitchell J, Harry H, William T, Worl R, Cybulski JS, Rosenberg NA, Malhi RS. Patterns of admixture and population structure in native populations of Northwest North America. PLOS Genet. 2014;10:e1004530. doi: 10.1371/journal.pgen.1004530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLOS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, Sudmant PH, Alkan C, Fu Q, Do R, Rohland N, Tandon A, Siebauer M, Green RE, Bryc K, Briggs AW, Stenzel U, Dabney J, Shendure J, Kitzman J, Hammer MF, Shunkov MV, Derevianko AP, Patterson N, Andrés AM, Eichler EE, Slatkin M, Reich D, Kelso J, Pääbo S. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raghavan M, DeGiorgio M, Albrechtsen A, Moltke I, Skoglund P, Korneliussen TS, Grønnow B, Appelt M, Gulløv HC, Friesen TM, Fitzhugh W, Malmström H, Rasmussen S, Olsen J, Melchior L, Fuller BT, Fahrni SM, Stafford T, Jr., Grimes V, Renouf MA, Cybulski J, Lynnerup N, Lahr MM, Britton K, Knecht R, Arneborg J, Metspalu M, Cornejo OE, Malaspinas AS, Wang Y, Rasmussen M, Raghavan V, Hansen TV, Khusnutdinova E, Pierre T, Dneprovsky K, Andreasen C, Lange H, Hayes MG, Coltrain J, Spitsyn VA, Götherström A, Orlando L, Kivisild T, Villems R, Crawford MH, Nielsen FC, Dissing J, Heinemeier J, Meldgaard M, Bustamante C, O’Rourke DH, Jakobsson M, Gilbert MT, Nielsen R, Willerslev E. The genetic prehistory of the New World Arctic. Science. 2014;345:1255832–1255832. doi: 10.1126/science.1255832. [DOI] [PubMed] [Google Scholar]
- 40.Scally A, Durbin R. Revising the human mutation rate: Implications for understanding human evolution. Nat. Rev. Genet. 2012;13:745–753. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- 41.Sheehan S, Harris K, Song YS. Estimating variable effective population sizes from multiple genomes: A sequentially markov conditional sampling distribution approach. Genetics. 2013;194:647–662. doi: 10.1534/genetics.112.149096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinrücken M, Paul JS, Song YS. A sequentially Markov conditional sampling distribution for structured populations with migration and recombination. Theor. Popul. Biol. 2013;87:51–61. doi: 10.1016/j.tpb.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris K, Nielsen R. Inferring demographic history from a spectrum of shared haplotype lengths. PLOS Genet. 2013;9:e1003521. doi: 10.1371/journal.pgen.1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schiffels S, Durbin R. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 2014;46:919–925. doi: 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eriksson A, Betti L, Friend AD, Lycett SJ, Singarayer JS, von Cramon-Taubadel N, Valdes PJ, Balloux F, Manica A. Late Pleistocene climate change and the global expansion of anatomically modern humans. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16089–16094. doi: 10.1073/pnas.1209494109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, Hansen NF, Durand EY, Malaspinas AS, Jensen JD, Marques-Bonet T, Alkan C, Prüfer K, Meyer M, Burbano HA, Good JM, Schultz R, Aximu-Petri A, Butthof A, Höber B, Höffner B, Siegemund M, Weihmann A, Nusbaum C, Lander ES, Russ C, Novod N, Affourtit J, Egholm M, Verna C, Rudan P, Brajkovic D, Kucan Z, Gusic I, Doronichev VB, Golovanova LV, Lalueza-Fox C, de la Rasilla M, Fortea J, Rosas A, Schmitz RW, Johnson PL, Eichler EE, Falush D, Birney E, Mullikin JC, Slatkin M, Nielsen R, Kelso J, Lachmann M, Reich D, Pääbo S. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffecker JF, Elias SA. Human Ecology of Beringia. Columbia University Press; New York: 2007. [Google Scholar]
- 49.Oppenheimer S, Bradley B, Stanford D. Solutrean hypothesis: Genetics, the mammoth in the room. World Archaeol. 2014;46:752–774. doi: 10.1080/00438243.2014.966273. [DOI] [Google Scholar]
- 50.Dillehay TD. Monte Verde, A Late Pleistocene Settlement in Chile: The archaeological context and interpretation. Smithsonian Institution Press; Washington D.C.: 1997. [DOI] [PubMed] [Google Scholar]
- 51.Rasmussen M, Sikora M, Albrechtsen A, Korneliussen TS, Moreno-Mayar JV, Poznik GD, Zollikofer CP, de León M. S. Ponce, Allentoft ME, Moltke I, Jónsson H, Valdiosera C, Malhi RS, Orlando L, Bustamante CD, Stafford TW, Jr., Meltzer DJ, Nielsen R, Willerslev E. The ancestry and affiliations of Kennewick Man. Nature. 2015 doi: 10.1038/nature14625. doi: 10.1038/nature14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davis RS, Knecht RA. Continuity and change in the eastern Aleutian archaeological sequence. Hum. Biol. 2010;82:507–524. doi: 10.3378/027.082.0503. [DOI] [PubMed] [Google Scholar]
- 53.Crawford MH, Rubicz RC, Zlojutro M. Origins of Aleuts and the genetic structure of populations of the archipelago: Molecular and archaeological perspectives. Hum. Biol. 2010;82:695–717. doi: 10.3378/027.082.0511. [DOI] [PubMed] [Google Scholar]
- 54.Hubbe M, Neves WA, Harvati K. Testing evolutionary and dispersion scenarios for the settlement of the new world. PLOS ONE. 2010;5:e11105. doi: 10.1371/journal.pone.0011105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chatters JC, Kennett DJ, Asmerom Y, Kemp BM, Polyak V, Blank AN, Beddows PA, Reinhardt E, Arroyo-Cabrales J, Bolnick DA, Malhi RS, Culleton BJ, Erreguerena PL, Rissolo D, Morell-Hart S, Stafford TW., Jr. Late Pleistocene human skeleton and mtDNA link Paleoamericans and modern Native Americans. Science. 2014;344:750–754. doi: 10.1126/science.1252619. [DOI] [PubMed] [Google Scholar]
- 56.García-Bour J, Pérez-Pérez A, Alvarez S, Fernández E, López-Parra AM, Arroyo-Pardo E, Turbón D. Early population differentiation in extinct aborigines from Tierra del Fuego-Patagonia: Ancient mtDNA sequences and Y-chromosome STR characterization. Am. J. Phys. Anthropol. 2004;123:361–370. doi: 10.1002/ajpa.10337. [DOI] [PubMed] [Google Scholar]
- 57.Perez SI, Bernal V, Gonzalez PN, Sardi M, Politis GG. Discrepancy between cranial and DNA data of early Americans: Implications for American peopling. PLOS ONE. 2009;4:e5746. doi: 10.1371/journal.pone.0005746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hernández M, Fox CL, García-Moro C. Fueguian cranial morphology: The adaptation to a cold, harsh environment. Am. J. Phys. Anthropol. 1997;103:103–117. doi: 10.1002/(SICI)1096-8644(199705)103:1<103::AID-AJPA7>3.0.CO;2-X. doi:10.1002/(SICI)1096-8644(199705)103:1<103::AID-AJPA7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 59.González-José R, Dahinten SL, Luis MA, Hernández M, Pucciarelli HM. Craniometric variation and the settlement of the Americas: Testing hypotheses by means of R-matrix and matrix correlation analyses. Am. J. Phys. Anthropol. 2001;116:154–165. doi: 10.1002/ajpa.1108. [DOI] [PubMed] [Google Scholar]
- 60.Der Sarkissian C, Brotherton P, Balanovsky O, Templeton JE, Llamas B, Soubrier J, Moiseyev V, Khartanovich V, Cooper A, Haak W, Genographic Consortium Mitochondrial genome sequencing in Mesolithic North East Europe Unearths a new sub-clade within the broadly distributed human haplogroup C1. PLOS ONE. 2014;9:e87612. doi: 10.1371/journal.pone.0087612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Excoffier L, Ray N. Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol. Evol. 2008;23:347–351. doi: 10.1016/j.tree.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 62.Achilli A, Perego UA, Lancioni H, Olivieri A, Gandini F, Kashani B. Hooshiar, Battaglia V, Grugni V, Angerhofer N, Rogers MP, Herrera RJ, Woodward SR, Labuda D, Smith DG, Cybulski JS, Semino O, Malhi RS, Torroni A. Reconciling migration models to the Americas with the variation of North American native mitogenomes. Proc. Natl. Acad. Sci. U.S.A. 2013;110:14308–14313. doi: 10.1073/pnas.1306290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dixon EJ. Late Pleistocene colonization of North America from Northeast Asia: New insights from large-scale paleogeographic reconstructions. Quat. Int. 2013;285:57–67. doi: 10.1016/j.quaint.2011.02.027. [DOI] [Google Scholar]
- 64.Mandryk CAS, Josenhans H, Fedje DW, Mathewes RW. Late Quaternary paleoenvironments of Northwestern North America: Implications for inland versus coastal migration routes. Quat. Sci. Rev. 2001;20:301–314. doi: 10.1016/S0277-3791(00)00115-3. [DOI] [Google Scholar]
- 65.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, Chen Z, Chu J, Carcassi C, Contu L, Du R, Excoffier L, Ferrara GB, Friedlaender JS, Groot H, Gurwitz D, Jenkins T, Herrera RJ, Huang X, Kidd J, Kidd KK, Langaney A, Lin AA, Mehdi SQ, Parham P, Piazza A, Pistillo MP, Qian Y, Shu Q, Xu J, Zhu S, Weber JL, Greely HT, Feldman MW, Thomas G, Dausset J, Cavalli-Sforza LL. A human genome diversity cell line panel. Science. 2002;296:261b–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 67.Cui Y, Lindo J, Hughes CE, Johnson JW, Hernandez AG, Kemp BM, Ma J, Cunningham R, Petzelt B, Mitchell J, Archer D, Cybulski JS, Malhi RS. Ancient DNA analysis of mid-holocene individuals from the Northwest Coast of North America reveals different evolutionary paths for mitogenomes. PLOS ONE. 2013;8:e66948. doi: 10.1371/journal.pone.0066948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Archer D. The Lucy Island Archaeological Project. Victoria: 2011. Unpublished report on file with the British Columbia Archaeology Branch. [Google Scholar]
- 69.McLaren D. Sea level change and archaeological site locations on the Dundas Island Archipelago of North Coastal British Columbia. University of Victoria; 2008. PhD dissertation. [Google Scholar]
- 70.Cybulski JS. “Human Remains from Lucy Island, British Columbia, Site GbTp 1, 1984/85,” ms. 2360. Canadian Museum of Civilization Library Archives; Gatineau, Canada: 1986. [Google Scholar]
- 71.Chisholm BS, Nelson DE, Schwarcz HP. Marine and terrestrial protein in prehistoric diets on the British Columbia coast. Curr. Anthropol. 1983;24:396–398. doi: 10.1086/203018. [DOI] [Google Scholar]
- 72.Cybulski JS. In: Human Variation in the Americas: The Integration of Archaeology and Biological Anthropology. Auerbach BM, editor. Center for Archaeological Investigations; Carbondale: 2010. pp. 77–112. [Google Scholar]
- 73.Cybulski JS. In: Violence and Warfare Among Hunter-Gatherers. Allen MW, Jones TL, editors. Left Coast Press; Walnut Creek: 2014. pp. 333–350. [Google Scholar]
- 74.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010;2010:t5448. doi: 10.1101/pdb.prot5448. doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 75.Maricic T, Whitten M, Pääbo S. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLOS ONE. 2010;5:e14004. doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arriaza B. Beyond Death: The Chinchorro Mummies of Ancient Chile. Smithsonian Insitiution Press; 1995. [Google Scholar]
- 77.Arriaza BT, et al. Chemical and mineral characterization of gray sediments used to model Chinchorro bodies. Chungara. 2012;44:177–194. [Google Scholar]
- 78.Arriaza B, Standen V, Reinhard K, Araújo A, Heukelbach J, Dittmar K. On head lice and social interaction in archaic Andean coastal populations. Int. J. Paleopathol. 2013;3:257–268. doi: 10.1016/j.ijpp.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 79.Orlando L, Ginolhac A, Zhang G, Froese D, Albrechtsen A, Stiller M, Schubert M, Cappellini E, Petersen B, Moltke I, Johnson PL, Fumagalli M, Vilstrup JT, Raghavan M, Korneliussen T, Malaspinas AS, Vogt J, Szklarczyk D, Kelstrup CD, Vinther J, Dolocan A, Stenderup J, Velazquez AM, Cahill J, Rasmussen M, Wang X, Min J, Zazula GD, Seguin-Orlando A, Mortensen C, Magnussen K, Thompson JF, Weinstock J, Gregersen K, Røed KH, Eisenmann V, Rubin CJ, Miller DC, Antczak DF, Bertelsen MF, Brunak S, Al-Rasheid KA, Ryder O, Andersson L, Mundy J, Krogh A, Gilbert MT, Kjær K, Sicheritz-Ponten T, Jensen LJ, Olsen JV, Hofreiter M, Nielsen R, Shapiro B, Wang J, Willerslev E. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499:74–78. doi: 10.1038/nature12323. [DOI] [PubMed] [Google Scholar]
- 80.Clarke PD. A la recherché de La Petite-Rochelle: Memory and Identity in Restigouche. Acadiensis (Frederict.) 1999;XXVIII:3–40. [Google Scholar]
- 81.Clarke PD. Land of East Wind: Mise en Forme d’une Memoire Mi’gmaq. Can. Rev. Sociol. 2000;37:167–195. doi: 10.1111/j.1755-618X.2000.tb01263.x. [DOI] [Google Scholar]
- 82.Leonard K. Archaeology of the Restigouche River, New Brunswick: A Summary. Mi’gmawei Mawiomi Secretariat; 2002. Wesgijinua’luet Research Title Project. [Google Scholar]
- 83.Leonard K. Archaeology of the New Brunswick Sites of Gespegewagji. Mi’gmawei Mawiomi Secretariat; 2002. Wesgijinua’luet Research Title Project. [Google Scholar]
- 84.Martijn C. An Archaeological Survey of the Northeast Coast of New Brunswick 1968 (Restigouche and Gloucester Counties) Historical Resources Administration; Fredericton, New Brunswick: 1968. [Google Scholar]
- 85.Turnbull CJ. Old Mission Point 1973: Report for an Archaeological Survey of Canada Salvage Contract. Archaeological Survey of Canada; Ottawa: 1974. [Google Scholar]
- 86.Turnbull CJ. The Richibucto Burial Site (CeDf-18), New Brunswick. New Brunswick: 1981. Manuscript on file with Archaeological Services Unit. [Google Scholar]
- 87.Garlie TN. An Ethnohistorical and Archaeological Review regarding Aboriginal Mortuary Remains reported from Nova Scotia and New Brunswick and the Potential for Future Research. Memorial University; 1992. Unpublished Honours Essay. [Google Scholar]
- 88.Pike KA. Bearing Identity: A Biocultural Analysis of Human Remains from Old Mission Point (ClDq-1), New Brunswick. Memorial University; 2014. Unpublished Master of Arts Thesis. [Google Scholar]
- 89.Turnbull CJ, Turnbull SW. Preliminary Report of the 1973 Excavations at Old Mission Point (ClDq-1) New Brunswick. Archaeological Survey of Canada; Ottawa: 1973. [Google Scholar]
- 90.Petersen JB, Sanger D. In: Prehistory of the Maritime Provinces: Past and Present Research. Deal M, Blair S, editors. Council of Maritime Premiers; Fredericton: 1991. pp. 113–170. [Google Scholar]
- 91.Svensson EM, Anderung C, Baubliene J, Persson P, Malmström H, Smith C, Vretemark M, Daugnora L, Götherström A. Tracing genetic change over time using nuclear SNPs in ancient and modern cattle. Anim. Genet. 2007;38:378–383. doi: 10.1111/j.1365-2052.2007.01620.x. [DOI] [PubMed] [Google Scholar]
- 92.Pijoan GV, Romero A, Mansilla J. Los Pericues de Baja California Sur en Perspectiva Tafonómica. Colección Científica INAH. 2010;560:67. [Google Scholar]
- 93.García-Bour J, Pérez-Pérez A, Alvarez S, Fernández E, López-Parra AM, Arroyo-Pardo E, Turbón D. Early population differentiation in extinct aborigines from Tierra del Fuego-Patagonia: Ancient mtDNA sequences and Y-chromosome STR characterization. Am. J. Phys. Anthropol. 2004;123:361–370. doi: 10.1002/ajpa.10337. [DOI] [PubMed] [Google Scholar]
- 94.Lalueza C, Pérez-Pérez A, Prats E, Cornudella L, Turbón D. Lack of founding Amerindian mitochondrial DNA lineages in extinct aborigines from Tierra del Fuego-Patagonia. Hum. Mol. Genet. 1997;6:41–46. doi: 10.1093/hmg/6.1.41. [DOI] [PubMed] [Google Scholar]
- 95.Moraga ML, Rocco P, Miquel JF, Nervi F, Llop E, Chakraborty R, Rothhammer F, Carvallo P. Mitochondrial DNA polymorphisms in Chilean aboriginal populations: Implications for the peopling of the southern cone of the continent. Am. J. Phys. Anthropol. 2000;113:19–29. doi: 10.1002/1096-8644(200009)113:1<19::AID-AJPA3>3.0.CO;2-X. doi:10.1002/1096-8644(200009)113:1<19::AID-AJPA3>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 96.Yang DY, Eng B, Waye JS, Dudar JC, Saunders SR. Improved DNA extraction from ancient bones using silica-based spin columns. Am. J. Phys. Anthropol. 1998;105:539–543. doi: 10.1002/(SICI)1096-8644(199804)105:4<539::AID-AJPA10>3.0.CO;2-1. doi:10.1002/(SICI)1096-8644(199804)105:4<539::AID-AJPA10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 97.Rohland N, Hofreiter M. Ancient DNA extraction from bones and teeth. Nat. Protoc. 2007;2:1756–1762. doi: 10.1038/nprot.2007.247. [DOI] [PubMed] [Google Scholar]
- 98.Gilbert MTP, Wilson AS, Bunce M, Hansen AJ, Willerslev E, Shapiro B, Higham TF, Richards MP, O’Connell TC, Tobin DJ, Janaway RC, Cooper A. Ancient mitochondrial DNA from hair. Curr. Biol. 2004;14:R463–R464. doi: 10.1016/j.cub.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 99.Behar DM, Yunusbayev B, Metspalu M, Metspalu E, Rosset S, Parik J, Rootsi S, Chaubey G, Kutuev I, Yudkovsky G, Khusnutdinova EK, Balanovsky O, Semino O, Pereira L, Comas D, Gurwitz D, Bonne-Tamir B, Parfitt T, Hammer MF, Skorecki K, Villems R. The genome-wide structure of the Jewish people. Nature. 2010;466:238–242. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- 100.Behar DM, Metspalu M, Baran Y, Kopelman NM, Yunusbayev B, Gladstein A, Tzur S, Sahakyan H, Bahmanimehr A, Yepiskoposyan L, Tambets K, Khusnutdinova EK, Kushniarevich A, Balanovsky O, Balanovsky E, Kovacevic L, Marjanovic D, Mihailov E, Kouvatsi A, Triantaphyllidis C, King RJ, Semino O, Torroni A, Hammer MF, Metspalu E, Skorecki K, Rosset S, Halperin E, Villems R, Rosenberg NA. No evidence from genome-wide data of a Khazar origin for the Ashkenazi Jews. Hum. Biol. 2013;85:859–900. doi: 10.3378/027.085.0604. [DOI] [PubMed] [Google Scholar]
- 101.Fedorova SA, Reidla M, Metspalu E, Metspalu M, Rootsi S, Tambets K, Trofimova N, Zhadanov SI, Kashani B. Hooshiar, Olivieri A, Voevoda MI, Osipova LP, Platonov FA, Tomsky MI, Khusnutdinova EK, Torroni A, Villems R. Autosomal and uniparental portraits of the native populations of Sakha (Yakutia): Implications for the peopling of Northeast Eurasia. BMC Evol. Biol. 2013;13:127. doi: 10.1186/1471-2148-13-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kenny EE, Timpson NJ, Sikora M, Yee MC, Moreno-Estrada A, Eng C, Huntsman S, Burchard EG, Stoneking M, Bustamante CD, Myles S. Melanesian blond hair is caused by an amino acid change in TYRP1. Science. 2012;336:554. doi: 10.1126/science.1217849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Migliano AB, Romero IG, Metspalu M, Leavesley M, Pagani L, Antao T, Huang DW, Sherman BT, Siddle K, Scholes C, Hudjashov G, Kaitokai E, Babalu A, Belatti M, Cagan A, Hopkinshaw B, Shaw C, Nelis M, Metspalu E, Mägi R, Lempicki RA, Villems R, Lahr MM, Kivisild T. Evolution of the pygmy phenotype: Evidence of positive selection fro genome-wide scans in African, Asian, and Melanesian pygmies. Hum. Biol. 2013;85:251–284. doi: 10.3378/027.085.0313. [DOI] [PubMed] [Google Scholar]
- 104.Pierron D, Razafindrazaka H, Pagani L, Ricaut FX, Antao T, Capredon M, Sambo C, Radimilahy C, Rakotoarisoa JA, Blench RM, Letellier T, Kivisild T. Genome-wide evidence of Austronesian-Bantu admixture and cultural reversion in a hunter-gatherer group of Madagascar. Proc. Natl. Acad. Sci. U.S.A. 2014;111:936–941. doi: 10.1073/pnas.1321860111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rasmussen M, Guo X, Wang Y, Lohmueller KE, Rasmussen S, Albrechtsen A, Skotte L, Lindgreen S, Metspalu M, Jombart T, Kivisild T, Zhai W, Eriksson A, Manica A, Orlando L, De La Vega FM, Tridico S, Metspalu E, Nielsen K, Ávila-Arcos MC, Moreno-Mayar JV, Muller C, Dortch J, Gilbert MT, Lund O, Wesolowska A, Karmin M, Weinert LA, Wang B, Li J, Tai S, Xiao F, Hanihara T, van Driem G, Jha AR, Ricaut FX, de Knijff P, Migliano AB, Romero I. Gallego, Kristiansen K, Lambert DM, Brunak S, Forster P, Brinkmann B, Nehlich O, Bunce M, Richards M, Gupta R, Bustamante CD, Krogh A, Foley RA, Lahr MM, Balloux F, Sicheritz-Pontén T, Villems R, Nielsen R, Wang J, Willerslev E. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334:94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Verdu P, Pemberton TJ, Laurent R, Kemp BM, Gonzalez-Oliver A, Gorodezky C, Hughes CE, Shattuck MR, Petzelt B, Mitchell J, Harry H, William T, Worl R, Cybulski JS, Rosenberg NA, Malhi RS. Patterns of admixture and population structure in native populations of Northwest North America. PLOS Genet. 2014;10:e1004530. doi: 10.1371/journal.pgen.1004530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yunusbayev B, Metspalu M, Järve M, Kutuev I, Rootsi S, Metspalu E, Behar DM, Varendi K, Sahakyan H, Khusainova R, Yepiskoposyan L, Khusnutdinova EK, Underhill PA, Kivisild T, Villems R. The Caucasus as an asymmetric semipermeable barrier to ancient human migrations. Mol. Biol. Evol. 2012;29:359–365. doi: 10.1093/molbev/msr221. [DOI] [PubMed] [Google Scholar]
- 109.Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, Dermitzakis E, Bonnen PE, Altshuler DM, Gibbs RA, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Yu F, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Gibbs RA, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Dermitzakis E, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Bonnen PE, Gibbs RA, Gonzaga-Jauregui C, Keinan A, Price AL, Yu F, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Schaffner SF, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Pollack S, Price AL, Schaffner SF, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE, International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. Article published online before print in May 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: A web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moorjani P, Thangaraj K, Patterson N, Lipson M, Loh PR, Govindaraj P, Berger B, Reich D, Singh L. Genetic evidence for recent population mixture in India. Am. J. Hum. Genet. 2013;93:422–438. doi: 10.1016/j.ajhg.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Thornton T, Tang H, Hoffmann TJ, Ochs-Balcom HM, Caan BJ, Risch N. Estimating kinship in admixed populations. Am. J. Hum. Genet. 2012;91:122–138. doi: 10.1016/j.ajhg.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Skoglund P, Malmström H, Raghavan M, Storå J, Hall P, Willerslev E, Gilbert MT, Götherström A, Jakobsson M. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science. 2012;336:466–469. doi: 10.1126/science.1216304. [DOI] [PubMed] [Google Scholar]
- 116.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lindgreen S. AdapterRemoval: Easy cleaning of next-generation sequencing reads. BMC Res. Notes. 2012;5:337. doi: 10.1186/1756-0500-5-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 121.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am. J. Hum. Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gronau I, Hubisz MJ, Gulko B, Danko CG, Siepel A. Bayesian inference of ancient human demography from individual genome sequences. Nat. Genet. 2011;43:1031–1034. doi: 10.1038/ng.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 124.Schubert M, Ginolhac A, Lindgreen S, Thompson JF, Al-Rasheid KA, Willerslev E, Krogh A, Orlando L. Improving ancient DNA read mapping against modern reference genomes. BMC Genomics. 2012;13:178. doi: 10.1186/1471-2164-13-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Daley T, Smith AD. Predicting the molecular complexity of sequencing libraries. Nat. Methods. 2013;10:325–327. doi: 10.1038/nmeth.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Briggs AW, Stenzel U, Johnson PL, Green RE, Kelso J, Prüfer K, Meyer M, Krause J, Ronan MT, Lachmann M, Pääbo S. Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14616–14621. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sawyer S, Krause J, Guschanski K, Savolainen V, Pääbo S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLOS ONE. 2012;7:e34131. doi: 10.1371/journal.pone.0034131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Allentoft ME, et al. The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils. Proc. R. Soc. B Biol. Sci. 2012 doi: 10.1098/rspb.2012.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pedersen JS, Valen E, Velazquez AM, Parker BJ, Rasmussen M, Lindgreen S, Lilje B, Tobin DJ, Kelly TK, Vang S, Andersson R, Jones PA, Hoover CA, Tikhonov A, Prokhortchouk E, Rubin EM, Sandelin A, Gilbert MT, Krogh A, Willerslev E, Orlando L. Genome-wide nucleosome map and cytosine methylation levels of an ancient human genome. Genome Res. 2014;24:454–466. doi: 10.1101/gr.163592.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA, 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fu Q, Mittnik A, Johnson PL, Bos K, Lari M, Bollongino R, Sun C, Giemsch L, Schmitz R, Burger J, Ronchitelli AM, Martini F, Cremonesi RG, Svoboda J, Bauer P, Caramelli D, Castellano S, Reich D, Pääbo S, Krause J. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr. Biol. 2013;23:553–559. doi: 10.1016/j.cub.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gelman A, Rubin DB. Inference from Iterative Simulation Using Multiple Sequences. Stat. Sci. 1992;7:457–472. doi: 10.1214/ss/1177011136. [DOI] [Google Scholar]
- 134.Plummer M, Best N, Cowles K, Vines K. CODA: Convergence diagnosis and output analysis for MCMC. R. News. 2006;6:7–11. [Google Scholar]
- 135.Skoglund P, Storå J, Götherström A, Jakobsson M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J. Archaeol. Sci. 2013;40:4477–4482. doi: 10.1016/j.jas.2013.07.004. [DOI] [Google Scholar]
- 136.Kloss-Brandstätter A, Pacher D, Schönherr S, Weissensteiner H, Binna R, Specht G, Kronenberg F. HaploGrep: A fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum. Mutat. 2011;32:25–32. doi: 10.1002/humu.21382. [DOI] [PubMed] [Google Scholar]
- 137.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 138.Behar DM, van Oven M, Rosset S, Metspalu M, Loogväli EL, Silva NM, Kivisild T, Torroni A, Villems R. A “Copernican” reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 2012;90:675–684. doi: 10.1016/j.ajhg.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Derenko MV, Malyarchuk BA, Dambueva IK, Shaikhaev GO, Dorzhu CM, Nimaev DD, Zakharov IA. Mitochondrial DNA variation in two South Siberian Aboriginal populations: Implications for the genetic history of North Asia. Hum. Biol. 2000;72:945–973. [PubMed] [Google Scholar]
- 140.Derenko M, Malyarchuk B, Grzybowski T, Denisova G, Dambueva I, Perkova M, Dorzhu C, Luzina F, Lee HK, Vanecek T, Villems R, Zakharov I. Phylogeographic analysis of mitochondrial DNA in northern Asian populations. Am. J. Hum. Genet. 2007;81:1025–1041. doi: 10.1086/522933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pimenoff VN, Comas D, Palo JU, Vershubsky G, Kozlov A, Sajantila A. Northwest Siberian Khanty and Mansi in the junction of West and East Eurasian gene pools as revealed by uniparental markers. Eur. J. Hum. Genet. 2008;16:1254–1264. doi: 10.1038/ejhg.2008.101. [DOI] [PubMed] [Google Scholar]
- 142.Derbeneva OA, Starikovskaya EB, Wallace DC, Sukernik RI. Traces of early Eurasians in the Mansi of northwest Siberia revealed by mitochondrial DNA analysis. Am. J. Hum. Genet. 2002;70:1009–1014. doi: 10.1086/339524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Derbeneva OA, Starikovskaia EB, Volod’ko NV, Wallace DC, Sukernik RI. [Mitochondrial DNA variation in Kets and Nganasans and the early peoples of Northern Eurasia] Genetika. 2002;38:1554–1560. [PubMed] [Google Scholar]
- 144.Karmin M, Saag L, Vicente M, Sayres M. A. Wilson, Järve M, Talas UG, Rootsi S, Ilumäe AM, Mägi R, Mitt M, Pagani L, Puurand T, Faltyskova Z, Clemente F, Cardona A, Metspalu E, Sahakyan H, Yunusbayev B, Hudjashov G, DeGiorgio M, Loogväli EL, Eichstaedt C, Eelmets M, Chaubey G, Tambets K, Litvinov S, Mormina M, Xue Y, Ayub Q, Zoraqi G, Korneliussen TS, Akhatova F, Lachance J, Tishkoff S, Momynaliev K, Ricaut FX, Kusuma P, Razafindrazaka H, Pierron D, Cox MP, Sultana GN, Willerslev R, Muller C, Westaway M, Lambert D, Skaro V, Kovačevic L, Turdikulova S, Dalimova D, Khusainova R, Trofimova N, Akhmetova V, Khidiyatova I, Lichman DV, Isakova J, Pocheshkhova E, Sabitov Z, Barashkov NA, Nymadawa P, Mihailov E, Seng JW, Evseeva I, Migliano AB, Abdullah S, Andriadze G, Primorac D, Atramentova L, Utevska O, Yepiskoposyan L, Marjanovic D, Kushniarevich A, Behar DM, Gilissen C, Vissers L, Veltman JA, Balanovska E, Derenko M, Malyarchuk B, Metspalu A, Fedorova S, Eriksson A, Manica A, Mendez FL, Karafet TM, Veeramah KR, Bradman N, Hammer MF, Osipova LP, Balanovsky O, Khusnutdinova EK, Johnsen K, Remm M, Thomas MG, Tyler-Smith C, Underhill PA, Willerslev E, Nielsen R, Metspalu M, Villems R, Kivisild T. A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome Res. 2015;25:459–466. doi: 10.1101/gr.186684.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.van Oven M, Van Geystelen A, Kayser M, Decorte R, Larmuseau MH. Seeing the wood for the trees: A minimal reference phylogeny for the human Y chromosome. Hum. Mutat. 2014;35:187–191. doi: 10.1002/humu.22468. [DOI] [PubMed] [Google Scholar]
- 146.ISOGG http://www.isogg.org.
- 147.Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Karafet TM, Osipova LP, Gubina MA, Posukh OL, Zegura SL, Hammer MF. High levels of Y-chromosome differentiation among native Siberian populations and the genetic signature of a boreal hunter-gatherer way of life. Hum. Biol. 2002;74:761–789. doi: 10.1353/hub.2003.0006. [DOI] [PubMed] [Google Scholar]
- 149.Dulik MC, Zhadanov SI, Osipova LP, Askapuli A, Gau L, Gokcumen O, Rubinstein S, Schurr TG. Mitochondrial DNA and Y chromosome variation provides evidence for a recent common ancestry between Native Americans and Indigenous Altaians. Am. J. Hum. Genet. 2012;90:229–246. doi: 10.1016/j.ajhg.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rootsi S, Kivisild T, Benuzzi G, Help H, Bermisheva M, Kutuev I, Barać L, Peričić M, Balanovsky O, Pshenichnov A, Dion D, Grobei M, Zhivotovsky LA, Battaglia V, Achilli A, Al-Zahery N, Parik J, King R, Cinnioğlu C, Khusnutdinova E, Rudan P, Balanovska E, Scheffrahn W, Simonescu M, Brehm A, Goncalves R, Rosa A, Moisan J-P, Chaventre A, Ferak V, Füredi S, Oefner PJ, Shen P, Beckman L, Mikerezi I, Terzić R, Primorac D, Cambon-Thomsen A, Krumina A, Torroni A, Underhill PA, Santachiara-Benerecetti AS, Villems R, Magri C, Semino O. Phylogeography of Y-chromosome haplogroup I reveals distinct domains of prehistoric gene flow in europe. Am. J. Hum. Genet. 2004;75:128–137. doi: 10.1086/422196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jakobsson M, Rosenberg NA. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 153.Maples BK, Gravel S, Kenny EE, Bustamante CD. RFMix: A discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 2013;93:278–288. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Shen Y, Sun W, Wang H, Wang Y, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallée C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PI, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe’er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Altshuler D, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Tsunoda T, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Zeng C, Zhao H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CD, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Gibbs RA, Belmont JW, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Wheeler DA, Yakub I, Gabriel SB, Onofrio RC, Richter DJ, Ziaugra L, Birren BW, Daly MJ, Altshuler D, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L’Archevêque P, Bellemare G, Saeki K, Wang H, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, Stewart J, International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PL, Maricic T, Good JM, Marques-Bonet T, Alkan C, Fu Q, Mallick S, Li H, Meyer M, Eichler EE, Stoneking M, Richards M, Talamo S, Shunkov MV, Derevianko AP, Hublin JJ, Kelso J, Slatkin M, Pääbo S. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Durand EY, Patterson N, Reich D, Slatkin M. Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 2011;28:2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Busing FMTA, Meijer E, van der Leeden R. Delete-m Jackknife for Unequal m. Stat. Comput. 1999;9:3–8. doi: 10.1023/A:1008800423698. [DOI] [Google Scholar]
- 158.Paul JS, Song YS. A principled approach to deriving approximate conditional sampling distributions in population genetics models with recombination. Genetics. 2010;186:321–338. doi: 10.1534/genetics.110.117986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Paul JS, Steinrücken M, Song YS. An accurate sequentially Markov conditional sampling distribution for the coalescent with recombination. Genetics. 2011;187:1115–1128. doi: 10.1534/genetics.110.125534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wiuf C, Hein J. Recombination as a point process along sequences. Theor. Popul. Biol. 1999;55:248–259. doi: 10.1006/tpbi.1998.1403. [DOI] [PubMed] [Google Scholar]
- 161.McVean GA, Cardin NJ. Approximating the coalescent with recombination. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:1387–1393. doi: 10.1098/rstb.2005.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Marjoram P, Wall JD. Fast “coalescent” simulation. BMC Genet. 2006;7:16. doi: 10.1186/1471-2156-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu S, Lorenzen ED, Fumagalli M, Li B, Harris K, Xiong Z, Zhou L, Korneliussen TS, Somel M, Babbitt C, Wray G, Li J, He W, Wang Z, Fu W, Xiang X, Morgan CC, Doherty A, O’Connell MJ, McInerney JO, Born EW, Dalén L, Dietz R, Orlando L, Sonne C, Zhang G, Nielsen R, Willerslev E, Wang J. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell. 2014;157:785–794. doi: 10.1016/j.cell.2014.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Harris K, Nielsen R. Error-prone polymerase activity causes multinucleotide mutations in humans. Genome Res. 2014;24:1445–1454. doi: 10.1101/gr.170696.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Staab PR, Zhu S, Metzler D, Lunter G. scrm: Efficiently simulating long sequences using the approximated coalescent with recombination. Bioinformatics. 2015;31:1680–1682. doi: 10.1093/bioinformatics/btu861. doi: 10.1093/bioinformatics/btu861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Harritt RK. Paleo-Eskimo beginnings in North America: A new discovery at Kuzitrin Lake, Alaska. Etud. Inuit. 1998;22:61–81. [Google Scholar]
- 167.Kong A, Thorleifsson G, Gudbjartsson DF, Masson G, Sigurdsson A, Jonasdottir A, Walters GB, Jonasdottir A, Gylfason A, Kristinsson KT, Gudjonsson SA, Frigge ML, Helgason A, Thorsteinsdottir U, Stefansson K. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 168.Dyke AS, Moore A, Robertson L. Deglaciation of North America. Geological Survey of Canada Open File. 2003:1574. [Google Scholar]
- 169.Wegmann D, Leuenberger C, Neuenschwander S, Excoffier L. ABCtoolbox: A versatile toolkit for approximate Bayesian computations. BMC Bioinformatics. 2010;11:116. doi: 10.1186/1471-2105-11-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Liu H, Prugnolle F, Manica A, Balloux F. A geographically explicit genetic model of worldwide human-settlement history. Am. J. Hum. Genet. 2006;79:230–237. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rosenberg NA, Mahajan S, Ramachandran S, Zhao C, Pritchard JK, Feldman MW. Clines, clusters, and the effect of study design on the inference of human population structure. PLOS Genet. 2005;1:e70. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 173.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLOS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Oksanen J, et al. vegan: Community Ecology Package. 2013 http://cran.r-project.org/web/packages/vegan/index.html.
- 175.Home RStudio. available at http://www.rstudio.com/
- 176.Sikora M, Carpenter ML, Moreno-Estrada A, Henn BM, Underhill PA, Sánchez-Quinto F, Zara I, Pitzalis M, Sidore C, Busonero F, Maschio A, Angius A, Jones C, Mendoza-Revilla J, Nekhrizov G, Dimitrova D, Theodossiev N, Harkins TT, Keller A, Maixner F, Zink A, Abecasis G, Sanna S, Cucca F, Bustamante CD. Population genomic analysis of ancient and modern genomes yields new insights into the genetic ancestry of the Tyrolean Iceman and the genetic structure of Europe. PLOS Genet. 2014;10:e1004353. doi: 10.1371/journal.pgen.1004353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Neves W. The origin of the first Americans: An analysis based on the cranial morphology of early South American human remains. Am. J. Phys. Anthropol. 1990;81:274. [Google Scholar]
- 178.Neves W, Blum M. “Luzia” is not alone: Further evidence of a non-mongoloid settlement of the new world. Curr. Res. Pleistocene. 2001;18:73–77. [Google Scholar]
- 179.González-José R, Neves W, Lahr MM, González S, Pucciarelli H, Martínez M. Hernández, Correal G. Late Pleistocene/Holocene craniofacial morphology in Mesoamerican Paleoindians: Implications for the peopling of the New World. Am. J. Phys. Anthropol. 2005;128:772–780. doi: 10.1002/ajpa.20165. [DOI] [PubMed] [Google Scholar]
- 180.Neves WA, Hubbe M, Correal G. Human skeletal remains from Sabana de Bogotá, Colombia: A case of Paleoamerican morphology late survival in South America? Am. J. Phys. Anthropol. 2007;133:1080–1098. doi: 10.1002/ajpa.20637. [DOI] [PubMed] [Google Scholar]
- 181.Perez SI, Bernal V, Gonzalez PN, Sardi M, Politis GG. Discrepancy between cranial and DNA data of early Americans: Implications for American peopling. PLOS ONE. 2009;4:e5746. doi: 10.1371/journal.pone.0005746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pucciarelli HM, Perez SI, Politis GG. Early Holocene human remains from the Argentinean Pampas: Additional evidence for distinctive cranial morphology of early South Americans. Am. J. Phys. Anthropol. 2010;143:298–305. doi: 10.1002/ajpa.21347. [DOI] [PubMed] [Google Scholar]
- 183.Neves W, Pucciarelli H. The Zhoukoudian Upper Cave skull 101 as seen from the Americans. J. Hum. Evol. 1998;34:219–222. doi: 10.1006/jhev.1997.0183. [DOI] [PubMed] [Google Scholar]
- 184.Powell JF, Neves WA. Craniofacial morphology of the first Americans: Pattern and process in the peopling of the New World. Am. J. Phys. Anthropol. 1999;110(Suppl 29):153–188. doi: 10.1002/(sici)1096-8644(1999)110:29+<153::aid-ajpa6>3.3.co;2-c. doi:10.1002/(SICI)1096-8644(1999)110:29+<153::AID-AJPA6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 185.Steele DG, Powell JF. Paleobiology of the first Americans. Evol. Anthropol. Issues News Rev. 1993;2:138–146. doi: 10.1002/evan.1360020409. [DOI] [Google Scholar]
- 186.Powell JF. The first Americans: race, evolution and the origin of native Americans. Cambridge University Press; Cambridge: 2005. [Google Scholar]
- 187.Gonçalves VF, Stenderup J, Rodrigues-Carvalho C, Silva HP, Gonçalves-Dornelas H, Líryo A, Kivisild T, Malaspinas AS, Campos PF, Rasmussen M, Willerslev E, Pena SD. Identification of Polynesian mtDNA haplogroups in remains of Botocudo Amerindians from Brazil. Proc. Natl. Acad. Sci. U.S.A. 2013;110:6465–6469. doi: 10.1073/pnas.1217905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Neves W, Pucciarelli H. Morphological affinities of the first Americans: An exploratory analysis based on early South American human remains. J. Hum. Evol. 1991;21:261–273. doi: 10.1016/0047-2484(91)90107-7. [DOI] [Google Scholar]
- 189.Dillehay TD. Probing deeper into first American studies. Proc. Natl. Acad. Sci. U.S.A. 2009;106:971–978. doi: 10.1073/pnas.0808424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.van Vark GN, Kuizenga D, Williams FL. Kennewick and Luzia: Lessons from the European Upper Paleolithic. Am. J. Phys. Anthropol. 2003;121:181–184. doi: 10.1002/ajpa.10176. discussion 185–188. [DOI] [PubMed] [Google Scholar]
- 191.Jantz RL, Owsley DW. Reply to Van Vark et al.: Is European Upper Paleolithic cranial morphology a useful analogy for early Americans? Am. J. Phys. Anthropol. 2003;121:185–188. doi: 10.1002/ajpa.10188. [DOI] [PubMed] [Google Scholar]
- 192.Bookstein FL. Morphometric Tools for Landmark Data. Cambridge University Press; Cambridge: 1991. [Google Scholar]
- 193.Roseman CC, Weaver TD. Multivariate apportionment of global human craniometric diversity. Am. J. Phys. Anthropol. 2004;125:257–263. doi: 10.1002/ajpa.10424. [DOI] [PubMed] [Google Scholar]
- 194.Betti L, Balloux F, Amos W, Hanihara T, Manica A. Distance from Africa, not climate, explains within-population phenotypic diversity in humans. Proc. R. Soc. B-Biol. Sci. 2009;276:809–814. doi: 10.1098/rspb.2008.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hubbe M, Hanihara T, Harvati K. Climate signatures in the morphological differentiation of worldwide modern human populations. Anat. Rec. 2009;292:1720–1733. doi: 10.1002/ar.20976. [DOI] [PubMed] [Google Scholar]
- 196.Betti L, Balloux F, Hanihara T, Manica A. The relative role of drift and selection in shaping the human skull. Am. J. Phys. Anthropol. 2010;141:76–82. doi: 10.1002/ajpa.21115. [DOI] [PubMed] [Google Scholar]
- 197.Howells WW. Skull Shapes and the Map: Craniometric Analyses in the Dispersion of Modern Homo. Harvard University Press; Cambridge: 1989. Peabody Museum of Archaeology and Ethnology. [Google Scholar]
- 198.Howells WW. Cranial Variation in Man: A Study by Multivariate Analysis of Patterns of Difference Among Recent Human Populations. Harvard University Press; 1973. [Google Scholar]
- 199.Morimoto N, de León M. S. Ponce, Zollikofer CP. Phenotypic variation in infants, not adults, reflects genotypic variation among chimpanzees and bonobos. PLOS ONE. 2014;9:e102074. doi: 10.1371/journal.pone.0102074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Howells WW. Howells’ craniometric data on the Internet. Am. J. Phys. Anthropol. 1996;101:441–442. doi: 10.1002/ajpa.1331010302. [DOI] [PubMed] [Google Scholar]
- 201.Neves WA, Hubbe M, Okumura MM, González-José R, Figuti L, Eggers S, De Blasis PA. A new early Holocene human skeleton from Brazil: Implications for the settlement of the New World. J. Hum. Evol. 2005;48:403–414. doi: 10.1016/j.jhevol.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 202.Neves WA, Hubbe M, Piló LB. Early Holocene human skeletal remains from Sumidouro Cave, Lagoa Santa, Brazil: History of discoveries, geological and chronological context, and comparative cranial morphology. J. Hum. Evol. 2007;52:16–30. doi: 10.1016/j.jhevol.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 203.Roseman CC. Detecting interregionally diversifying natural selection on modern human cranial form by using matched molecular and morphometric data. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12824–12829. doi: 10.1073/pnas.0402637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, Li H, Mallick S, Dannemann M, Fu Q, Kircher M, Kuhlwilm M, Lachmann M, Meyer M, Ongyerth M, Siebauer M, Theunert C, Tandon A, Moorjani P, Pickrell J, Mullikin JC, Vohr SH, Green RE, Hellmann I, Johnson PL, Blanche H, Cann H, Kitzman JO, Shendure J, Eichler EE, Lein ES, Bakken TE, Golovanova LV, Doronichev VB, Shunkov MV, Derevianko AP, Viola B, Slatkin M, Reich D, Kelso J, Pääbo S. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Greenberg JH, Turner CG, Zegura SL. The settlement of the Americas: A comparison of the linguistic, dental, and genetic evidence. Curr. Anthropol. 1986;27:477–497. doi: 10.1086/203472. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raghavan 2015 Americas SM