Production and Characterization of Synthetic Carboxysome Shells with Incorporated Luminal Proteins (original) (raw)
Synthetic carboxysome shells, containing all of the key component proteins provide a platform for fundamental studies of shell permeability and metabolic modeling of organelle function.
Abstract
Spatial segregation of metabolism, such as cellular-localized CO2 fixation in C4 plants or in the cyanobacterial carboxysome, enhances the activity of inefficient enzymes by selectively concentrating them with their substrates. The carboxysome and other bacterial microcompartments (BMCs) have drawn particular attention for bioengineering of nanoreactors because they are self-assembling proteinaceous organelles. All BMCs share an architecturally similar, selectively permeable shell that encapsulates enzymes. Fundamental to engineering carboxysomes and other BMCs for applications in plant synthetic biology and metabolic engineering is understanding the structural determinants of cargo packaging and shell permeability. Here we describe the expression of a synthetic operon in Escherichia coli that produces carboxysome shells. Protein domains native to the carboxysome core were used to encapsulate foreign cargo into the synthetic shells. These synthetic shells can be purified to homogeneity with or without luminal proteins. Our results not only further the understanding of protein-protein interactions governing carboxysome assembly, but also establish a platform to study shell permeability and the structural basis of the function of intact BMC shells both in vivo and in vitro. This system will be especially useful for developing synthetic carboxysomes for plant engineering.
A key enzyme in photosynthesis is the CO2 fixation enzyme ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco). Rubisco not only fixes CO2, resulting in carbon assimilation, but it can also fix O2, leading to photorespiration. Suppressing the unwanted oxygenase activity of Rubisco by sequestering Rubisco with a source of CO2 is Nature’s solution to this substrate discrimination problem. While C4 plants compartmentalize CO2 fixation in specific cells (Hibberd et al., 2008; Parry et al., 2011), cyanobacteria have evolved a specialized organelle composed entirely of protein to encapsulate Rubisco—the carboxysome.
The carboxysome is just one type of bacterial microcompartment (BMC), widespread, functionally diverse bacterial organelles (Axen et al., 2014). All BMCs consist of an enzymatic core surrounded by a selectively permeable protein shell (Kerfeld et al., 2005; Tanaka et al., 2008; Chowdhury et al., 2014; Kerfeld and Erbilgin, 2015). While the encapsulated enzymes differ among functionally distinct BMCs, they share an architecturally similar shell composed of three types of proteins: BMC-H, BMC-T, and BMC-P forming hexamers, pseudohexamers, and pentamers, respectively (Kerfeld and Erbilgin, 2015). These constitute the building blocks of a self-assembling, apparently icosahedral shell with a diameter ranging from 40 to 400 nm (Shively et al., 1973a,b, 1998; Price and Badger, 1991; Bobik et al., 1999; Iancu et al., 2007, 2010; Petit et al., 2013; Erbilgin et al., 2014). Recent studies have also shown that in the biogenesis of BMCs an encapsulation peptide (EP) (Fan and Bobik, 2011; Kinney et al., 2012; Aussignargues et al., 2015; Jakobson et al., 2015), a short (approximately 18 residues) amphipathic α-helix mediates interactions between a subset of core protein and the shell (Fan and Bobik, 2011; Choudhary et al., 2012; Kinney et al., 2012; Lawrence et al., 2014; Lin et al., 2014; Aussignargues et al., 2015). Indeed, because they are self-assembling organelles composed entirely of protein, BMCs hold great promise for diverse applications in bioengineering and development of bionanomaterials (Frank et al., 2013; Chowdhury et al., 2014; Chessher et al., 2015; Kerfeld and Erbilgin, 2015); the key features of BMCs include selective permeability, spatial colocalization of enzymes, the establishment of private cofactor pools, and the potentially beneficial effects of confinement on protein stability. For example, introducing carboxysomes into plants could provide a saltational enhancement of crop photosynthesis (Price et al., 2013; Zarzycki et al., 2013; Lin et al., 2014; McGrath and Long, 2014).
The β-carboxysome, which sequesters form 1B Rubisco, has been an important model system for the study of the structural basis of carboxysome function, assembly, and engineering (Kerfeld et al., 2005; Tanaka et al., 2008; Cameron et al., 2013; Aussignargues et al., 2015; Cai et al., 2015). Beta-carboxysomes assemble from the inside out (Cameron et al., 2013; Gonzalez-Esquer et al., 2015). Two proteins that are absolutely conserved and unique to β-carboxysomes, CcmM and CcmN, play essential roles in this process: CcmM crosslinks Rubisco through its C-terminal Rubisco small subunit-like domains (SSLDs; pfam00101); CcmM and CcmN interact through their N-terminal domains; and C-terminal EP of CcmN interacts with the carboxysome shell.
Here we describe a system for producing synthetic β-carboxysome shells and encapsulating nonnative cargo. We constructed a synthetic operon composed of ccmK1, ccmK2, ccmL, and ccmO, genes encoding, respectively, two BMC-H proteins, a BMC-P protein, and a BMC-T protein of the carboxysome shell of the halotolerant cyanobacterium, Halothece sp. PCC 7418 (Halo hereafter). Recombinant shells composed of all four proteins were produced and purified. We also demonstrated that the terminal α-helices of CcmK1 and CcmK2 are not, as had been proposed (Samborska and Kimber, 2012), required for the shell formation, and that the synthetic shell is a single-layered protein membrane. Cargo could be targeted to the interior of the synthetic shells using either the EP of CcmN or the N-terminal domain of CcmM; the latter observation provides new insight into the organization of the β-carboxysome. Our results not only further the understanding of protein-protein interactions governing carboxysome assembly but also provide a platform to study carboxysome shell permeability. These results will be useful in guiding the design and optimization of carboxysomes and other BMCs for introduction into plants.
RESULTS
Expression of a Synthetic Operon Results in Recombinant Carboxysome Shells That Can Be Purified
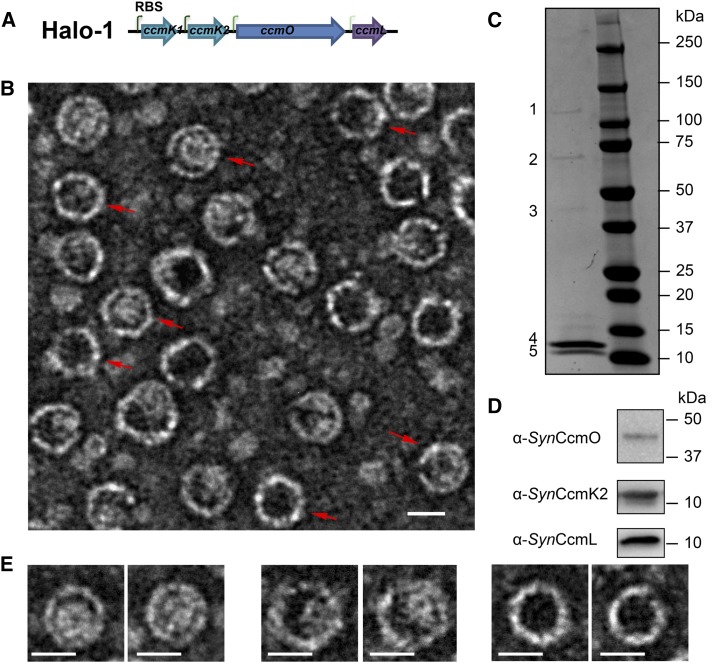
Halo is a halotolerant cyanobacterium, with optimal growth under laboratory conditions with up to 20% salinity at 38°C (Garcia-Pichel et al., 1998). Enzymes and protein complexes encoded by halophiles often exhibit enhanced stability relative to their nonhalophilic counterparts (Jaenicke and Böhm, 1998) and are thus valuable for applications in biotechnology (Margesin and Schinner, 2001). Therefore, we reasoned that carboxysome components of Halo may be good candidates for heterologous production of synthetic shells. The carboxysome genes of Halo are found in the main ccm locus (ccmK1, ccmK2, ccmL, ccmM, and ccmN) and three satellite loci. As in many other β-cyanobacteria, the essential ccmO gene is not encoded in the main ccm locus but elsewhere in the genome. A synthetic operon, Halo-1, was designed in an attempt to produce synthetic carboxysome shells heterologously (Fig. 1A). In order to mimic shell protein ratios consistent with the current model of the β-carboxysome shell (Tanaka et al., 2008; Cameron et al., 2013) and currently available transcriptomic data for ccm genes expression (Schwarz et al., 2011; Vijayan et al., 2011; Billis et al., 2014), a strong ribosomal binding site (RBS) was added preceding the coding regions of ccmK1 and ccmK2, with a medium and a low strength RBS preceding ccmO and ccmL, respectively.
Figure 1.
Synthetic operon design and purification of synthetic carboxysome shells. A, The synthetic operon for expression of the four Halo carboxysome shell genes. Each gene has its own RBS, shown as bent green arrows. The dark to faint color represents the strong to the weak strength of the RBS. B, Synthetic carboxysome shells purified from E. coli and negatively stained for TEM. Shells with obvious hexagonal profiles are marked with red arrows. C, Separation of purified shell components on SDS-PAGE. Bands 1–5 were analyzed using MS fingerprint analysis. D, Immunoblots on purified shell components separated by SDS-PAGE, developed with antibodies raised against Synechococcus elongatus PCC7942 (anti_syn_) CcmO, CcmK2, and CcmL. E, Examples of the different appearances of purified Halo carboxysome shells. Bars indicate 20 nm.
The synthetic operon Halo-1 was cloned into an Escherichia coli vector under a T7 promoter, and expression was induced by addition of IPTG. Synthetic Halo carboxysome shells could be purified after detergent lysis of E. coli cells via differential centrifugations and anion-exchange chromatography (see “Materials and Methods” for details). The purified shells were negatively stained and imaged using transmission electron microscopy (TEM). The shells measured 24.70 ± 1.43 nm in diameter (n = 1507), and many displayed polyhedral profiles consistent with icosahedral symmetry (Fig. 1B, red arrows). The purified shells were resolved into five bands with sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE; Fig. 1C). The presence of CcmO, CcmK1, CcmK2, or CcmL proteins was confirmed by colorimetric immunoblots (Fig. 1D), the latter two comigrating as a single band (the calculated molecular masses of CcmK2 and CcmL are 10.8 kD and 10.3 kD, respectively) (Fig. 1C, band 5). The identity of each protein band was also verified unambiguously by mass spectrometry (MS) fingerprinting analysis: band 3 and band 4 correspond to CcmO and CcmK1, respectively. Band 5 was a mixture of CcmK2 and CcmL. Two other bands (bands 1 and 2) were consistently resolved on SDS-PAGE from synthetic shell preparations. Band 1 was identified as the LacZ protein of E. coli. A closer look at the TEM images reveals that not all purified Halo shells appear empty (Fig. 1E), possibly due to the presence of impurities (e.g., LacZ) captured during assembly. Band 2, interestingly, was identified as a mixture of CcmO, CcmK1, and CcmL, which may represent a subcomplex formed for the construction of the vertices of the shell.
Fluorescent Proteins Can Be Targeted to Synthetic Carboxysome Shells Using the EP of CcmN
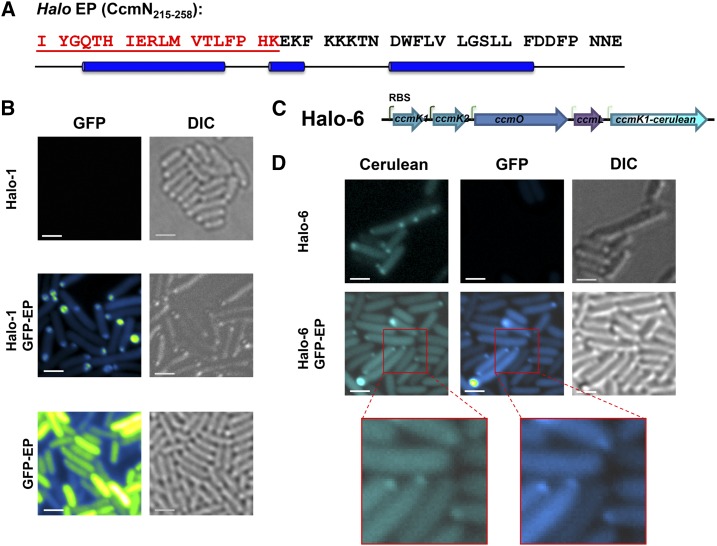
One of the requirements for adapting carboxysomes and other BMC architectures for applications in bioengineering is the ability to target protein(s) of choice for compartmentalization. CcmN orthologs contain an EP on the C terminus, following a poorly conserved linker region (Kinney et al., 2012; Aussignargues et al., 2015). The 18-amino-acid-long EP from Synechococcus elongatus PCC 7942 (Syn hereafter) (Supplemental Fig. S1, blue box) is essential for the interaction between CcmN and CcmK2 both in vivo and in vitro (Kinney et al., 2012). Compared to EPs from many other cyanobacteria, the predicted EP of Halo CcmN (residues 215–258) is unusual (Supplemental Fig. S1, red box): it is 26 residues longer than the experimentally characterized CcmN EP of Syn (Supplemental Fig. S1, blue box). Residues 215 to 232 resemble the canonical EP, followed by a C-terminal extension that is found among some CcmN orthologs (Kinney et al., 2012). The predicted secondary structure for the EP of Halo CcmN is unusual in that it consists of three α-helices (Fig. 2A and Supplemental Fig. S2).
Figure 2.
Fluorescent proteins can be targeted to the synthetic carboxysome shells. A, The amino-acid sequence of the extended encapsulation peptide region from Halo CcmN. Residues in the region corresponding to the experimentally characterized EP of CcmN are shown in red. The blue cylinders denote regions predicted to form α-helices. B, GFP was fused to the EP sequence and visualized without or without coexpression of the Halo shells. Pseudo-color (green-fire-blue in the ImageJ lookup table (Schneider et al., 2012)) was applied for GFP signal. C, The synthetic operon Halo-6 contains a fusion gene, _ccmK1_-cerulean, downstream from ccmL. D, Visualization of cerulean-labeled Halo carboxysome shells (cyan) and encapsulated GFP proteins (green-fire-blue). Bars indicate 2 µm.
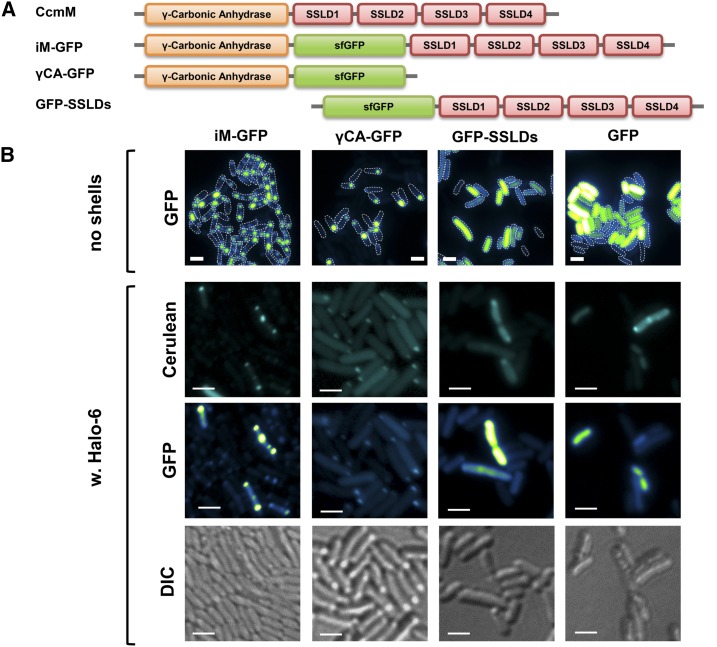
To test the feasibility for using the Halo CcmN or its EP for targeting protein into the synthetic Halo shells, GFP was used as cargo. Full-length CcmN was fused to the C terminus of GFP, but the resulting fusion protein was insoluble and formed inclusion bodies in vivo. The observed insolubility may be attributable to the predilection of recombinant CcmN to aggregate when expressed without its interaction partner, CcmM (data not shown). In contrast, when CcmN211-258 was fused to the C terminus of GFP, we observed diffuse fluorescent signal throughout the E. coli cytosol, indicating the fusion protein was soluble (Fig. 2B). When the GFP-EP was coexpressed with the Halo shells, the signal from GFP-EP is no longer diffuse, but localized to the cell poles (Fig. 2B).
To further confirm the colocalization of GFP-EP and Halo shells, an open reading frame encoding a cerulean fluorescent protein (CFP) was fused in-frame to the ccmK1 gene and inserted into the Halo-1 construct with a low-strength RBS following the ccmL gene (Fig. 2C). For imaging, the resulting synthetic operon (Halo-6) was expressed alone or coexpressed with the GFP-EP (Fig. 2D). CFP-labeled synthetic Halo shells appeared as fluorescent puncta in E. coli cells (Fig. 2D). When GFP-EP was coexpressed with the CFP-labeled shell, although the puncta were frequently small, the colocalization of both signals was unambiguous (Fig. 2D).
Synthetic Carboxysome Shells with Cargo Can Be Purified
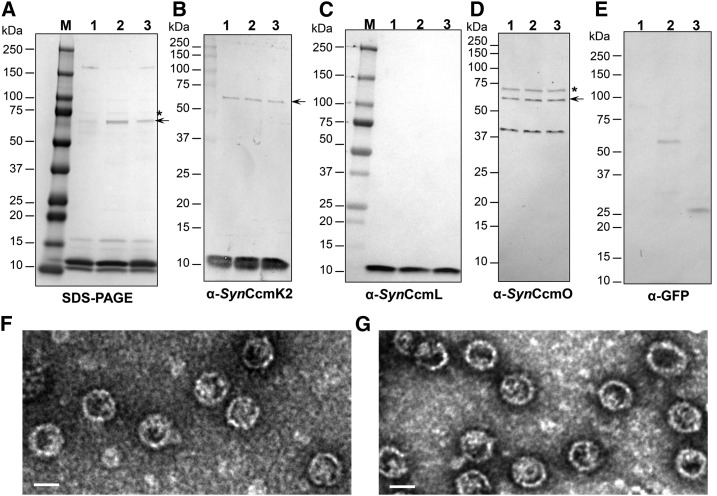
We purified Halo shells from the strain coexpressing Halo-1 and GFP-EP or GFP-CcmN. Purified Halo shells with cargo were negatively stained and imaged with TEM (Fig. 3, F and G). They are homogenous and similar in size and appearance to Halo shells without cargo (Φ23.58 ± 1.80 nm, n = 2315 for Halo shells with GFP-EP; Φ24.50 ± 2.27 nm, n = 1663 for Halo shells with GFP-CcmN). To confirm the presence of cargo, purified shells were analyzed by SDS-PAGE and immunoblotting using chemiluminescent detection. As expected, separation patterns of both samples on SDS-PAGE were very similar to that of empty Halo shells (Fig. 3A). Immunoblots confirmed the presence of all four shell proteins (Fig. 3, B–D). Although GFP fusion proteins were not clearly observed on the SDS-PAGE, immunoblots developed with anti-GFP antibodies indicate the presence of GFP fusions at the expected _M_r values (Fig. 3E). The band previously identified as a mixture of CcmK1, CcmO, and CcmL in MS analysis was also confirmed by anti_Syn_CcmK2 and anti_Syn_CcmO antibodies (Fig. 3, B and D, black arrows). Interestingly, a band with an observed molecular mass slightly less than 75 kD was also recognized by anti_Syn_CcmO antibodies and was present in all three samples regardless of absence or presence of cargo (Fig. 3D, asterisk). This band is hardly visible upon Coomassie blue-stained SDS-PAGE but obvious in immunoblots. It may be an oligomer of CcmO.
Figure 3.
Purification of synthetic carboxysome shells with cargo. SDS-PAGE (A) and immunoblots (B–E) of purified synthetic carboxysome shells with GFP-CcmN (lane 2) or GFP-EP (lane 3) are compared to Halo shells without cargo (lane 1). Antibodies used in (B–E) are α-_Syn_CcmK2, α-_Syn_CcmL, α-_Syn_CcmO and α-GFP, respectively. The band indicated with an arrow in (A) was also recognized by α-_Syn_CcmK2 and α-_Syn_CcmO antibodies. Αnti-_Syn_CcmO antibodies recognized a band slightly smaller than the 75-kD marker (indicated by asterisks in A and D). Visualization of negatively stained Halo shells with GFP-CcmN (F) and GFP-EP (G) under TEM. Bars indicate 20 nm.
The Synthetic Carboxysome Shell Is Composed of Single-Layer
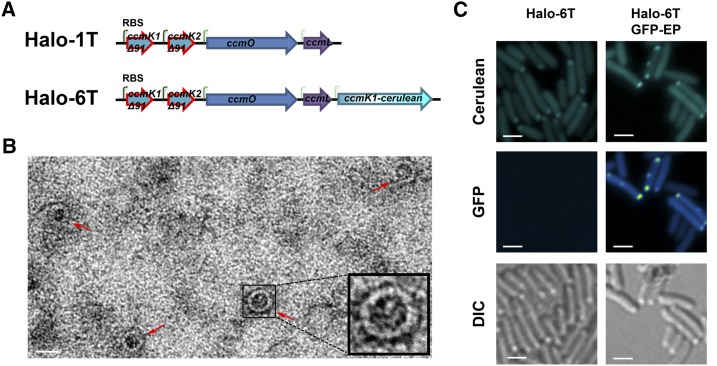
Based on measurements from micrographs of Halo shells, the thickness of the shell is 2.8 ± 0.5 nm (n = 192). This suggests that the synthetic carboxysome shells are composed of only a monolayer of shell proteins. However, because of the possible errors in TEM-based measurements, we examined the impact of truncation of the C-terminal α-helix (αd-helix; Supplemental Fig. S3B, shown in red), which has been described as crucial for the dimerization of shell proteins that is proposed to lead to the formation of a double-layered facet in the carboxysome of Thermosynechococcus elongatus (Samborska and Kimber, 2012). Truncated variants of Halo CcmK1 and CcmK2 were designed based on sequence alignments with T. elongatus CcmK1 and CcmK2 (Supplemental Fig. S3A). Deletion of residues beyond Pro-90, which precedes the αd-helix in both CcmK1 and CcmK2, should completely abolish dimerization (Supplemental Fig. S3C). The truncated ccmK1 and ccmK2 genes were synthesized and used for replacement of full-length ccmK1 and ccmK2 in the synthetic operons Halo-1 and Halo-6. The resultant operons were named Halo-1T and Halo-6T (T for truncated), respectively (Fig. 4A). Expression of Halo-1T produced shells, and TEM on an enrichment fraction showed that the thickness of the shells with truncated shell proteins were similar to wild type: 2.5 ± 0.5 nm (n = 57; Fig. 4B), implying that the synthetic carboxysome shells are single-layered. Coexpression of Halo-6T and GFP-EP resulted in colocalization of fluorescently labeled shells and cargo (Fig. 4C), suggesting the truncated shells retain the ability to encapsulate cargo and the αd-helix is not involved in the interaction between the EP and the Halo shell proteins CcmK1 and CcmK2.
Figure 4.
Synthetic carboxysome shell formation with CcmK1 and CcmK2 truncations. A, Synthetic operons Halo-1T and Halo-6T are variants of Halo-1 and Halo-6, respectively, in which the full-length ccmK1 or ccmK2 gene was replaced by truncated versions. B, Visualization of cerulean-labeled mutant Halo shells (cyan) without or with encapsulated GFP (green-fire-blue). Bars indicate 2 µm. C, An enrichment of mutant Halo shells (red arrows) was negatively stained and visualized by TEM. The bar indicates 20 nm. Inlet: an enlarged mutant shell.
The γ-CA Domain of CcmM Interacts with the Synthetic Carboxysome Shell
The absolutely conserved β-carboxysome protein, CcmM, is composed of an N-terminal γ-CA domain and three to five copies of SSLDs. Previous studies indicated that the SSLDs of CcmM are required for the nucleation of Rubisco and, therefore, crucial for the β-carboxysome formation (Long et al., 2007; Long et al., 2010; Cameron et al., 2013). However, the potential structural role of the N-terminal γ-CA domain in the interaction between the carboxysome core and the shell has not been investigated. The synthetic Halo shell system provides a background to probe for a direct interaction between CcmM and the shell without the influence of Rubisco. A superfolding GFP (sfGFP) variant was used as a reporter and fused to CcmM between the γ-CA domain and four SSLDs to generate a recombinant protein, iM-GFP (Fig. 5A). The sfGFP was also fused to the γ-CA domain or the four SSLDs of Halo CcmM to generate γCA-GFP or GFP-SSLDs, respectively (Fig. 5A). Interestingly, different localization patterns were observed, even when these fusion proteins were expressed in isolation (Fig. 5B, top row). When both the γ-CA domain and SSLDs were present, multiple fluorescent puncta per cell were observed; in contrast, no or at most one polar punctum per cell was apparent when only the γ-CA domain was present. The sfGFP-labeled SSLDs resulted in diffuse fluorescent signal, similar to what was observed in the negative control (sfGFP alone). Furthermore, a similar trend was observed when these fusion proteins were coexpressed with fluorescently labeled Halo shells (Halo-6) (Fig. 5B). Although the expression level of fluorescently labeled Halo shells seemed to vary, CFP-puncta were observed in all cases, indicative of the formation of Halo shells. Colocalization of GFP signal with the CFP signal was observed in the case of iM-GFP or γCA-GFP, with multiple and single puncta per cell, respectively. Collectively, in vivo labeling results suggested that there is a direct protein-protein interaction between CcmM and the shell proteins in the absence of CcmN and that this interaction is mediated via the N-terminal γ-CA domain of CcmM.
Figure 5.
CcmM interacts with synthetic carboxysome shells through its γ-CA domain. A, Halo CcmM contains a γ-CA domain followed by four SSLD domains. A sfGFP was inserted between the γ-CA domain and the first SSLD domain to generate the fusion protein iM-GFP. Deletion of four SSLD domains of iM-GFP results in γCA-GFP, and deletion of the γ-CA domain results in GFP-SSLDs. B, Visualization of sfGFP-labeled full-length or partial CcmM (green-fire-blue) without or with the presence of cerulean-labeled Halo shells (cyan). Bars indicate 2 µm.
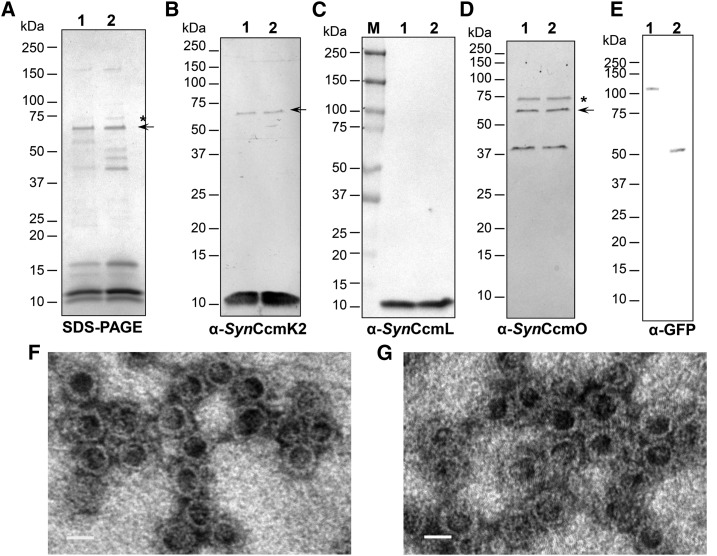
To further test if iM-GFP and γCA-GFP are sequestered within Halo shells, we purified shells from strains expressing both types of cargo. Isolated Halo shells with iM-GFP or γCA-GFP have the expected composition on SDS-PAGE: CcmK1, CcmK2, CcmL, and CcmO were present as well as the bands for the mixture of CcmK1/CcmL/CcmO and the putative oligomer of CcmO (Fig. 6, A–D). The fusion proteins iM-GFP and γCA-GFP were also detected by immunoblots at their expected _M_r values (Fig. 6E), suggesting iM-GFP and γCA-GFP are indeed sequestered and coisolated with Halo shells. Purified samples were also imaged with TEM, and similar shell structures were evident in both cases (Fig. 6, F and G).
Figure 6.
Purification of synthetic carboxysome shells with full-length and truncated CcmM. SDS-PAGE (A) and immunoblots (B–E) of purified Halo shells with iM-GFP (lane 1) or γCA-GFP (lane 2). Antibodies used in (B–E) are α-_Syn_CcmK2, α-_Syn_CcmL, α-_Syn_CcmO, and α-GFP, respectively. The band indicated with an arrow in (A) was also recognized by α-_Syn_CcmK2 and α-_Syn_CcmO antibodies. Αnti-_Syn_CcmO antibodies recognized a band slightly smaller than the 75-kD marker (indicated by asterisks in A and D). Visualization of negatively stained synthetic carboxysome shells with iM-GFP (F) and γCA-GFP (G) under TEM. Bars indicate 20 nm.
DISCUSSION
The synthetic Halo carboxysome shells reported here demonstrate that β-carboxysome shells can be formed with the four key shell proteins in the absence of cargo in E. coli. Empty β-carboxysome shells, to our knowledge, have not been observed in vivo. We demonstrate targeting of cargo to the synthetic carboxysome shells via an EP, albeit requiring detection by immunoblotting. This method of detection has also been required in other (noncarboxysome) synthetic shell systems (Parsons et al., 2010; Choudhary et al., 2012; Lassila et al., 2014). The inability to directly visualize cargo proteins on SDS-PAGE may hint at the importance of cross linking among luminal proteins for filling the cores of shells (Cameron et al., 2013; Gonzalez-Esquer et al., 2015) in the natural encapsulation process.
Our data also provide, to our knowledge, new insights into the details of β-carboxysome assembly. The highly purified Halo shells can be resolved into at least four discernible bands that are identified by both immunoblotting and MS. Characterization by TEM indicates that the facets are formed by a single layer of shell proteins. Although which side of the BMC-H hexamer faces the cytosol is still an open question, our data suggests the αd-helix of shell proteins CcmK1 and CcmK2, which is located on the concave side of CcmK1 or CcmK2 hexamer, is not involved in the EP-mediated process. Interestingly, immunoblots also consistently revealed two bands that are SDS-resistant complexes of shell proteins in all of the shell preparations. One appears to be an oligomer of CcmO and the other a complex of CcmO/CcmK1/CcmL. Notably, structural information on CcmO is still unavailable, although this absolutely essential carboxysome protein (Marco et al., 1994; Martinez et al., 1997; Rae et al., 2012) is assumed to be a trimer (pseudohexamer). The observation of a robust complex of CcmO/CcmK1/CcmL may represent a shell assembly intermediate. In any case, these data indicate that our model for the carboxysome shell, which is based on the structures of CcmL and CcmK proteins and the assumption of icosahedral symmetry (Kerfeld et al., 2005; Tanaka et al., 2008), is perhaps too simplistic.
Furthermore, we demonstrate two different types of interaction between domains of carboxysomal luminal proteins and the synthetic shells. These include an unusual variant of the canonical EP. In contrast to previously experimentally characterized EPs that consist of a single amphipathic α-helix (Fan et al., 2012; Kinney et al., 2012; Lawrence et al., 2014), the EP of Halo is predicted to comprise at least two α-helices (Fig. 2A). Similar observations have been reported for the EPs of some glycyl radical enzyme-associated BMCs (Zarzycki et al., 2015). We also demonstrate interaction between the γ-CA domain of CcmM and shell components of β-carboxysome. These observations not only provide strategies for encapsulation into the shell but provide, to our knowledge, new details on the internal organization of the carboxysome. CcmM is essential to β-carboxysome assembly as it is required for nucleating Rubisco in procarboxysome formation; its multiple SSLDs cross link Rubisco molecules (Long et al., 2007; Long et al., 2010; Cameron et al., 2013). On the other hand, the γ-CA domain of CcmM interacts with CcmN, which also interacts with the shell through CcmN’s C-terminal EP (Kinney et al., 2012). Here we showed that CcmM, in the absence of both Rubisco and CcmN, associates with the shell. This allows further refinement of the model for the interior organization of the carboxysome core and its interactions with the shell. Collectively, observations from the synthetic carboxysome shell system reported here provide new insight into the organization of the interior and of the shell of the β-carboxysome.
The synthetic carboxysome shells, containing all of the key component proteins, establish a platform for fundamental studies of shell permeability; this is essential for metabolic modeling of organelle function (McGrath and Long, 2014). For carboxysomes, the shell functions as the interface between the first step of CO2 fixation and the rest of cyanobacterial metabolism. More generally, the successful production and purification of synthetic Halo shells provides, to our knowledge, a new system for repurposing BMCs for applications in biotechnology applications and development of nanomaterials.
MATERIALS AND METHODS
Constructions of Expression Vectors
The synthetic operon Halo-1 was constructed as follows (Fig. 1A): the ccmK1, ccmK2, ccmO, and ccmL genes of Halo genome (GenBank no. ID NC_019779.1) were placed after the promoter sequence. A nonnative, 79-bp linker region was added between any two adjacent genes, each including a unique restriction site and a ribosomal binding site (RBS). A strong RBS was added preceding the coding region of ccmK1 or ccmK2, and there are a medium and a low strength RBS preceding ccmO and ccmL, respectively. Each coding sequence was codon-optimized for expression in Escherichia coli and the gene synthesized (GenScript Biotech, Piscataway Township, NJ). The Halo-1 operon was cloned into a Bgl-brick compatible vector pETBb3 at _Eco_RI and _Bam_HI sites. Another DNA fragment containing the truncated ccmK1 and ccmK2 genes with _Eco_RI and _Nco_I flanking regions on either side was also synthesized. This fragment was subcloned into pHalo-1 at _Eco_RI and _Nco_I sites to replace the nontruncated ccmK1 and ccmK2 via standard digestion and ligation procedures, and the resulting operon was named “Halo-2”. pHalo-6 and pHalo-7 were constructed based on pHalo-1 and pHalo-2, respectively, using a ligation-free method (In-Fusion Cloning kit: Clontech, Mountain View, CA) following the manufacturer’s protocol. DNA fragments encoding GFP-CcmN and iM-GFP fusion were also synthesized at GenScript with codon optimization for E. coli and subcloned into a low-copy number Bgl-brick compatible vector pMCLBb to generated pFC227 and pFC243. pFC223 was generated by a PCR-based approach using pFC227 as a template. pFC244, pFC245, and pFC246 were generated using a PCR-based approach with pFC243 as a template. All the constructs and primers used are listed in Supplemental Table S1 and Supplemental Table S2, respectively.
Strains and Growth Conditions
Recombinant protein and synthetic operon expression was carried out in E. coli BL21 (DE3) strains (Invitrogen, Carlsbad, CA). Precultures were grown overnight in LB broth (EMD Millipore, Billerica, MA) media at 30°C shaken at 200 rpm with appropriate antibiotics (100 μg/mL Ampicillin or 34 μg/mL Chloramphenicol). Then a 1:100 dilution was made in fresh LB, and cultures were grown at 37°C and shaken at 180 rpm. To induce protein expression, IPTG (Gold Biotechnology, St. Louis, MO) was added to a final concentration of 0.5 mm when cultures had grown to OD600 ≈ 0.7–0.8. Growth of cultures was continued at 37°C for 4 h before harvesting. After 15-min centrifugation at 5000_g_, cell pellets were weighed and stored in −20°C until purification.
Synthetic Carboxysome Shell Purification
The frozen cell pellet was thawed in a room-temperature water bath. A quantity of 2.5 mL/g of B-PER II reagent (Pierce Protein Biology, Thermo Scientific, Waltham, MA) was added to the cell pellet, and the pellet was resuspended by pipetting; 200 µL of RNase A at 10 mg/mL was then added per 6–8 g of cells. rLysome (Novagen, Merck, Darmstadt, Germany) at 30 kU/µL was added at 1.4 µL/g cell. The cell lysate was incubated on a rocker vigorously for 30 min at room temperature. Cell debris was removed with centrifugation set at 27,000_g_ for 18 min, 4°C. Then Benzonase nuclease (25 U/µL; Novagen, Merck) was added to the clear cell lysate at 7.1 µL/g cell, and the cell lysate was incubated on a rocker for another 45 min at room temperature. Extracts were ultracentrifuged on 7 mL of 30% Suc cushion made in TBS 20/50 pH 7.4 (20 mm Tris-HCl, pH 7.4; 50 mm NaCl) at 42,000 rpm in a Ti-70 rotor for 4 h at 4°C. The supernatant was carefully removed, and the soft glassy pellet was resuspended in 1 mL of ice-cold TBS 20/50 pH7.4. The resuspended sample containing shells was briefly centrifuged at 1200_g_ for 5 min at 4°C, and the supernatant was loaded on a 20–70% Suc gradient made in TBS 20/50 pH 7.4 buffer. The gradient was run in an SW-28 rotor at 23,000 rpm at 4°C for 16 h. After the ultracentrifugation, 10 fractions (4 mL each) were recovered from the gradient from top to bottom. The last fraction was used to resuspend the pellet at the bottom of the tube. A sample of each fraction was run using sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and fractions with shell proteins were pooled. The pooled sample was the shell enrichment sample, and could be used for transmission electron microscopy (TEM). The enriched sample was then loaded on a Mono Q 10/100 column (GE Healthcare, Port Washington, NY); fractions were collected from the TBS 20:0 to TBS 20:1000 pH 7.4 gradient. Based on SDS-PAGE of Mono Q fractions, samples containing shells were pooled. A final clean-up step included dilution of pooled sample in TBS 20:350 pH 7.4 to 40 mL total followed by an ultracentrifugation run at 42,000 rpm in a Ti-70 rotor for 4 h at 4°C. The final pellet was resuspended in 200 µL of TBS 20:50 pH 7.4.
SDS-PAGE and Immunoblots
Protein samples were separated on precast 4–20% or 10–20% SDS polyacrylamide gradient gels (BioRad, Hercules, CA) to analyze their composition. Polypeptide bands were visualized by staining with Gel Code Blue (Pierce Protein Biology, Thermo Fisher Scientific). For immunoblotting, the proteins were transferred onto a 0.45-μm-pore-size nitrocellulose membrane in a Mini Trans-Blot electrophoretic transfer cell (Bio-Rad). The blot was blocked with immunoblot blocking buffer (5% nonfat dry milk in PBS pH 7.4 with 0.1% Triton X-100) for 45 min. The appropriate primary antibody (raised in rabbits to recombinant protein) was incubated with the blot for 1 h at room temperature to probe the presence of the target antigen. After rinsing with PBS buffer, immunoblot blocking buffer, and PBS buffer for 15 min each, the blot was incubated with goat anti-rabbit IgG antibody conjugated with alkaline phosphatase or horseradish peroxidase at 1:10,000 dilution for 1 h at room temperature. The blot was developed with one-step NBT-BCIP solution for colorimetric detection of alkaline phosphatase activity or SuperSignal West Pico Chemiluminescent Substrate for horseradish peroxidase detection (Pierce Protein Biology, Thermo Fisher Scientific). Images of stained gels and immunoblots were captured and documented using a ChemiDoc imaging system (Bio-Rad). Densitometry analysis was performed using the ImageLab program (Bio-Rad).
Sequence Alignment and Bioinformatics
Multiple sequence alignment was performed using Clustal X 2.1 (Larkin et al., 2007). Pairwise alignment was performed using the on-line global alignment program, Needle, at http://www.ebi.ac.uk/Tools/emboss/ (Li et al., 2015). The secondary motif prediction was performed using Quick2D at http://toolkit.tuebingen.mpg.de/quick2_d with the PSIPRED algorithm (Jones, 1999). Protein homology models were built using the Swiss-model interface (http://swissmodel.expasy.org//SWISS-MODEL.html) (Arnold et al., 2006). Ab initio protein structure prediction for the encapsulation peptide region (CcmN211-258) was performed with the QUARK server (http://zhanglab.ccmb.med.umich.edu/QUARK/) (Xu and Zhang, 2012, 2013). All the structure figures were prepared with PyMOL (The PyMOL Molecular Graphics System, V. 1.5.0.3; Schrödinger, New York, NY).
Fluorescence Microscopy and TEM
Induced cells prior to harvesting were used for fluorescence microscopy imaging. Two microliters of cells were spotted on a thin agar pad and air-dried before imaging with an Axioplan 2 microscope or LSM710 (Carl Zeiss, Jena, Germany) using a 100× oil immersion objective. Images were visualized and analyzed with ImageJ 1.4.8 (National Institutes of Health, Bethesda, MD; Schneider et al., 2012). Purified synthetic shells were spotted on formvar/carbon-coated copper grids (no. FCF300-Cu; Electron Microscopy Sciences, Hatfield, PA) and negatively stained with 2% uranyl acetate for 60 s. Images were taken on a model no. 1200 EX TEM (JEOL USA, Peabody, MA). Particle analysis was done using ImageJ 1.4.8 (National Institutes of Health).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession number NC_019779.1.
Supplemental Data
The following supplemental materials are available.
- Supplemental Table S1. E. coli plasmids used in this study.
- Supplemental Table S2. Oligonucleotides used in cloning.
- Supplemental Figure S1. Multiple sequence alignment of CcmN from Halo with the 50 most closely related orthologs.
- Supplemental Figure S2. Fold prediction for the Halo EP.
- Supplemental Figure S3. Homology model of Halo CcmK1 and CcmK2.
Supplementary Material
Supplemental Data
Acknowledgments
We thank the UC Berkeley Electron Microscopy Lab for assistance with TEM. We also thank Dr. Denise Schichnes and Dr. Steven Ruzin at UC Berkeley Biological Imaging Facility for advice and assistance with fluorescent microscopy, and Dr. Markus Sutter for advice during the construction of the Halo-1 operon and development of the Halo shell purification. We thank Dr. Manuel Sommer for critical reading of the manuscript.
Glossary
BMC
bacterial microcompartments
CFP
cerulean fluorescent protein
EP
encapsulation peptide
RBS
ribosomal binding site
SDS-PAGE
sodium dodecyl-sulfate polyacrylamide gel electrophoresis
sfGFP
superfolding GFP
SSLD
small subunit-like domain
TEM
transmission electron microscopy
Footnotes
[OPEN]
Articles can be viewed without a subscription.
References
- Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Aussignargues C, Paasch BC, Gonzalez-Esquer R, Erbilgin O, Kerfeld CA (2015) Bacterial microcompartment assembly: the key role of encapsulation peptides. Commun Integr Biol 8: e1039755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axen SD, Erbilgin O, Kerfeld CA (2014) A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLOS Comput Biol 10: e1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billis K, Billini M, Tripp HJ, Kyrpides NC, Mavromatis K (2014) Comparative transcriptomics between Synechococcus PCC 7942 and Synechocystis PCC 6803 provide insights into mechanisms of stress acclimation. PLoS One 9: e109738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC (1999) The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B(12)-dependent 1, 2-propanediol degradation. J Bacteriol 181: 5967–5975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F, Sutter M, Bernstein SL, Kinney JN, Kerfeld CA (2015) Engineering bacterial microcompartment shells: chimeric shell proteins and chimeric carboxysome shells. ACS Synth Biol 4: 444–453 [DOI] [PubMed] [Google Scholar]
- Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA (2013) Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 155: 1131–1140 [DOI] [PubMed] [Google Scholar]
- Chessher A, Breitling R, Takano E (2015) Bacterial microcompartments: biomaterials for synthetic biology-based compartmentalization strategies. ACS Biomater Sci Eng. 1: 345–351 [DOI] [PubMed] [Google Scholar]
- Choudhary S, Quin MB, Sanders MA, Johnson ET, Schmidt-Dannert C (2012) Engineered protein nano-compartments for targeted enzyme localization. PLoS One 7: e33342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury C, Sinha S, Chun S, Yeates TO, Bobik TA (2014) Diverse bacterial microcompartment organelles. Microbiol Mol Biol Rev 78: 438–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbilgin O, McDonald KL, Kerfeld CA (2014) Characterization of a planctomycetal organelle: a novel bacterial microcompartment for the aerobic degradation of plant saccharides. Appl Environ Microbiol 80: 2193–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Bobik TA (2011) The N-terminal region of the medium subunit (PduD) packages adenosylcobalamin-dependent diol dehydratase (PduCDE) into the Pdu microcompartment. J Bacteriol 193: 5623–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Cheng S, Sinha S, Bobik TA (2012) Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments. Proc Natl Acad Sci USA 109: 14995–15000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Lawrence AD, Prentice MB, Warren MJ (2013) Bacterial microcompartments moving into a synthetic biological world. J Biotechnol 163: 273–279 [DOI] [PubMed] [Google Scholar]
- Garcia-Pichel F, Nübel U, Muyzer G (1998) The phylogeny of unicellular, extremely halotolerant cyanobacteria. Arch Microbiol 169: 469–482 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Esquer CR, Shubitowski TB, Kerfeld CA (2015) Streamlined construction of the cyanobacterial CO2-fixing organelle via protein domain fusions for use in plant synthetic biology. Plant Cell 27: 2637–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd JM, Sheehy JE, Langdale JA (2008) Using C4 photosynthesis to increase the yield of rice-rationale and feasibility. Curr Opin Plant Biol 11: 228–231 [DOI] [PubMed] [Google Scholar]
- Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, Martino A, Jensen GJ (2007) The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J Mol Biol 372: 764–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu CV, Morris DM, Dou Z, Heinhorst S, Cannon GC, Jensen GJ (2010) Organization, structure, and assembly of alpha-carboxysomes determined by electron cryotomography of intact cells. J Mol Biol 396: 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenicke R, Böhm G (1998) The stability of proteins in extreme environments. Curr Opin Struct Biol 8: 738–748 [DOI] [PubMed] [Google Scholar]
- Jakobson CM, Kim EY, Slininger MF, Chien A, Tullman-Ercek D (2015) Localization of proteins to the propanediol utilization microcompartment by non-native signal sequences is mediated by a common hydrophobic motif. J Biol Chem 1: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292: 195–202 [DOI] [PubMed] [Google Scholar]
- Kerfeld CA, Erbilgin O (2015) Bacterial microcompartments and the modular construction of microbial metabolism. Trends Microbiol 23: 22–34 [DOI] [PubMed] [Google Scholar]
- Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO (2005) Protein structures forming the shell of primitive bacterial organelles. Science 309: 936–938 [DOI] [PubMed] [Google Scholar]
- Kinney JN, Salmeen A, Cai F, Kerfeld CA (2012) Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J Biol Chem 287: 17729–17736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lassila JK, Bernstein SL, Kinney JN, Axen SD, Kerfeld CA (2014) Assembly of robust bacterial microcompartment shells using building blocks from an organelle of unknown function. J Mol Biol 426: 2217–2228 [DOI] [PubMed] [Google Scholar]
- Lawrence AD, Frank S, Newnham S, Lee MJ, Brown IR, Xue WF, Rowe ML, Mulvihill DP, Prentice MB, Howard MJ, Warren MJ (2014) Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth Biol 3: 454–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, Park YM, Buso N, Lopez R (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43(W1): W580–W584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Devonshire J, Hines KM, Parry MA, Hanson MR (2014) β-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J 79: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long BM, Badger MR, Whitney SM, Price GD (2007) Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA. J Biol Chem 282: 29323–29335 [DOI] [PubMed] [Google Scholar]
- Long BM, Tucker L, Badger MR, Price GD (2010) Functional cyanobacterial beta-carboxysomes have an absolute requirement for both long and short forms of the CcmM protein. Plant Physiol 153: 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco E, Martinez I, Ronen-Tarazi M, Orus MI, Kaplan A (1994) Inactivation of ccmO in Synechococcus sp. strain PCC 7942 results in a mutant requiring high levels of CO2. Appl Environ Microbiol 60: 1018–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margesin R, Schinner F (2001) Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles 5: 73–83 [DOI] [PubMed] [Google Scholar]
- Martinez I, Orus MI, Marco E (1997) Carboxysome structure and function in a mutant of Synechococcus that requires high levels of CO2 for growth. Plant Physiol Biochem 35: 137–146 [Google Scholar]
- McGrath JM, Long SP (2014) Can the cyanobacterial carbon-concentrating mechanism increase photosynthesis in crop species? A theoretical analysis. Plant Physiol 164: 2247–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry MAJ, Reynolds M, Salvucci ME, Raines C, Andralojc PJ, Zhu XG, Price GD, Condon AG, Furbank RT (2011) Raising yield potential of wheat. II. Increasing photosynthetic capacity and efficiency. J Exp Bot 62: 453–467 [DOI] [PubMed] [Google Scholar]
- Parsons JB, Frank S, Bhella D, Liang M, Prentice MB, Mulvihill DP, Warren MJ (2010) Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament-associated organelle movement. Mol Cell 38: 305–315 [DOI] [PubMed] [Google Scholar]
- Petit E, LaTouf WG, Coppi MV, Warnick TA, Currie D, Romashko I, Deshpande S, Haas K, Alvelo-Maurosa JG, Wardman C, Schnell DJ, Leschine SB, et al. (2013) Involvement of a bacterial microcompartment in the metabolism of fucose and rhamnose by Clostridium phytofermentans. PLoS One 8: e54337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Badger MR (1991) Evidence for the role of carboxysomes in the cyanobacterial CO2-concentrating mechanism. Can J Bot Revue Can Bot 69: 963–973 [Google Scholar]
- Price GD, Pengelly JJL, Forster B, Du J, Whitney SM, von Caemmerer S, Badger MR, Howitt SM, Evans JR (2013) The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species. J Exp Bot 64: 753–768 [DOI] [PubMed] [Google Scholar]
- Rae BD, Long BM, Badger MR, Price GD (2012) Structural determinants of the outer shell of β-carboxysomes in Synechococcus elongatus PCC 7942: roles for CcmK2, K3-K4, CcmO, and CcmL. PLoS One 7: e43871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samborska B, Kimber MS (2012) A dodecameric CcmK2 structure suggests β-carboxysomal shell facets have a double-layered organization. Structure 20: 1353–1362 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D, Nodop A, Hüge J, Purfürst S, Forchhammer K, Michel KP, Bauwe H, Kopka J, Hagemann M (2011) Metabolic and transcriptomic phenotyping of inorganic carbon acclimation in the Cyanobacterium Synechococcus elongatus PCC 7942. Plant Physiol 155: 1640–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively JM, Ball F, Brown DH, Saunders RE (1973a) Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 182: 584–586 [DOI] [PubMed] [Google Scholar]
- Shively JM, Ball FL, Kline BW (1973b) Electron microscopy of the carboxysomes (polyhedral bodies) of Thiobacillus neapolitanus. J Bacteriol 116: 1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively JM, Bradburne CE, Aldrich HC, Bobik TA, Mehlman JL, Jin S, Baker SH (1998) Sequence homologs of the carboxysomal polypeptide CsoS1 of the thiobacilli are present in cyanobacteria and enteric bacteria that form carboxysomes-polyhedral bodies. Can J Bot Revue Can Bot 76: 906–916 [Google Scholar]
- Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO (2008) Atomic-level models of the bacterial carboxysome shell. Science 319: 1083–1086 [DOI] [PubMed] [Google Scholar]
- Vijayan V, Jain IH, O’Shea EK (2011) A high resolution map of a cyanobacterial transcriptome. Genome Biol 12: R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Zhang Y (2012) Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 80: 1715–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Zhang Y (2013) Toward optimal fragment generations for ab initio protein structure assembly. Proteins 81: 229–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarzycki J, Axen SD, Kinney JN, Kerfeld CA (2013) Cyanobacterial-based approaches to improving photosynthesis in plants. J Exp Bot 64: 787–798 [DOI] [PubMed] [Google Scholar]
- Zarzycki J, Erbilgin O, Kerfeld CA (2015) Bioinformatic characterization of glycyl radical enzyme-associated bacterial microcompartments. Appl Environ Microbiol 81: 8315–8329 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data