HER2 overexpression and amplification as a potential therapeutic target in colorectal cancer: analysis of 3256 patients enrolled in the QUASAR, FOCUS and PICCOLO colorectal cancer trials (original) (raw)
Abstract
HER2 overexpression/amplification is linked to trastuzumab response in breast/gastric cancers. One suggested anti‐EGFR resistance mechanism in colorectal cancer (CRC) is aberrant MEK–AKT pathway activation through HER2 up‐regulation. We assessed HER2‐amplification/overexpression in stage II–III and IV CRC patients, assessing relationships to KRAS/BRAF and outcome. Pathological material was obtained from 1914 patients in the QUASAR stage II–III trial and 1342 patients in stage IV trials (FOCUS and PICCOLO). Tissue microarrays were created for HER2 immunohistochemistry. HER2‐amplification was assessed using FISH and copy number variation. KRAS/BRAF mutation status was assessed by pyrosequencing. Progression‐free survival (PFS) and overall survival (OS) data were obtained for FOCUS/PICCOLO and recurrence and mortality for QUASAR; 29/1342 (2.2%) stage IV and 25/1914 (1.3%) stage II–III tumours showed HER2 protein overexpression. Of the HER2‐overexpressing cases, 27/28 (96.4%) stage IV tumours and 20/24 (83.3%) stage II–III tumours demonstrated HER2 amplification by FISH; 41/47 (87.2%) also showed copy number gains. HER2‐overexpression was associated with KRAS/BRAF wild‐type (WT) status at all stages: in 5.2% WT versus 1.0% mutated tumours (p < 0.0001) in stage IV and 2.1% versus 0.2% in stage II–III tumours (p = 0.01), respectively. HER2 was not associated with OS or PFS. At stage II–III, there was no significant correlation between HER2 overexpression and 5FU/FA response. A higher proportion of HER2‐overexpressing cases experienced recurrence, but the difference was not significant. HER2‐amplification/overexpression is identifiable by immunohistochemistry, occurring infrequently in stage II–III CRC, rising in stage IV and further in KRAS/BRAF WT tumours. The value of HER2‐targeted therapy in patients with HER2‐amplified CRC must be tested in a clinical trial. © 2015 The Authors. Journal of Pathology published by John Wiley & Sons Ltd on behalf of Pathological Society of Great Britain and Ireland.
Keywords: HER2; colorectal cancer; amplification; overexpression, copy number variation
Introduction
Colorectal cancer (CRC) is one of the leading causes of cancer‐related deaths, with 1.36 million new cases and > 0.5 million deaths/year worldwide 1. In stage IV disease, cytotoxic agents such as fluorouracil, irinotecan and oxaliplatin have led to improved survival. Further improvements in outcomes have been seen with the introduction of targeted therapies. These include the use of monoclonal antibodies against the epidermal growth factor receptor (EGFR) in patients with KRAS wild‐type (WT) tumours, although only a minority of patients respond to this approach.
The HER family of tyrosine kinase receptors consists of EGFR, HER2 (ErbB2), HER3 and HER4. They are responsible for cell survival and proliferation via signalling through the RAS–RAF–ERK and PI3K–PTEN–AKT pathways 2. HER2 has been extensively studied in breast cancer, where gene amplification and overexpression of the protein is seen in approximately 20% cases and is associated with increased risk of recurrence and a poorer prognosis 3, 4, 5. Treatment of these patients with the anti‐HER2 monoclonal antibody trastuzumab (Herceptin) has led to improved survival in metastatic breast cancer 6.
HER2 overexpression has also been identified in other cancers, such as gastric cancer 7. FDA approval was given for the use of trastuzumab in _HER2_‐positive metastatic gastric cancers following the results of the ToGA trial, where median overall survival was 13.8 months in patients given trastuzumab with chemotherapy, compared to 11.1 months in those patients given chemotherapy alone 8. Several studies have assessed HER2 overexpression in CRC, with some reporting membranous expression, varying in the range 2.1–11% in 9, 10, 11, 12, 13, 14, 15, and others reporting cytoplasmic overexpression in the range 47.4–68.5% 12, 16, 17.
Topoisomerase II_α_ (TOP2A) alters the topology of DNA during transcription by causing transient double‐strand breaks. The TOP2A gene is located telomeric of HER2 on chromosome 17q. It is co‐amplified with HER2 in 35% of HER2‐positive breast cancers and is associated with response to anthracyclines and other agents, although evidence is mixed.
It is now accepted that in stage IV colorectal disease, RAS mutations confer resistance to anti‐EGFR therapy and BRAF mutations are an indicator of poor prognosis; however, even among patients with RAS WT tumours, only a minority respond. One suggested mechanism of drug resistance is aberrant signalling through the up‐regulation of other transmembrane receptors, such as HER2. Here we compared the frequency of HER2 protein overexpression between 1914 stage II–III CRC patients from the QUASAR clinical trial with 1342 patients from two CRC stage IV clinical trials, 888 from FOCUS and 454 from PICCOLO. HER2 protein expression was assessed in relation to HER2 amplification, as determined by FISH, KRAS and BRAF mutations, recurrence rates (in QUASAR patients) and finally to survival outcomes. Cases showing overexpression were also assessed for whole‐genome copy number variation, HER2 amplification and associated TOP2A amplification.
Materials and methods
Patients were recruited to the QUASAR (QUick And Simple And Reliable) trial during May 1994–December 2003, and were randomly assigned to receive 5FU/folinic acid chemotherapy (n = 1622) or to observation (n = 1617). Trial details are reported elsewhere 18. Tissue microarrays (TMAs) were constructed containing all cases with sufficient tumour for three cores/array. Ethical approval was obtained from both the West Midlands Multi‐Centre Research Ethics Committee (JR/MT/MREC/02/7/56a) and the Northern and Yorkshire Research Ethics Committee (08/H0903/62) for pathological substudies in QUASAR (ISRCTN82375386).
The MRC CR08 FOCUS trial [Fluorouracil, Oxaliplatin and CPT11 (irinotecan): Use and Sequencing] ISRCTN79877428 recruited 2135 first‐line advanced colorectal cancer (aCRC) patients during 2000–2003, comparing strategies of sequential or combination cytotoxic therapy, without targeted agents 19. Prospective consent was obtained to retrieve stored pathological material for research (MREC/99/3/1). From the trial repository of 1700 samples, 888 samples contained sufficient tumour material to provide three tumour cores for TMA construction.
PICCOLO (Panitumumab, Irinotecan and Cyclosporin in COLOrectal cancer therapy) assessed second‐line aCRC therapies, including the EGFR‐targeted agent panitumumab. It recruited 1198 patients during 2006–2010 20, 21. Patient consent was obtained to retrieve surplus stored pathological material for research (REC/06/Q0906/38). TMAs were made containing 582 patient samples, whose resection samples contained sufficient tumour material to provide three tumour cores. Following TMA sectioning and staining, 128 had missing cores or insufficient tumour within cores, leaving 454 patients suitable for assessment here.
HER2 immunohistochemistry
From each TMA, 5 µm sections were cut onto Superfrost Plus slides and dried overnight at 37°C. The slides were dewaxed, then incubated in 4% hydrogen peroxide solution in methanol for 20 min, prior to antigen retrieval in a pressure cooker for 2 min in unmasking solution (Vector Laboratories, Burlingame, CA, USA). The slides were incubated in a casein solution (Vector Laboratories) for 10 min and then incubated for 1 h at room temperature with the HER2 antibody (polyclonal rabbit anti‐human c‐erbB‐2 oncoprotein; cat no. A0485 from Dako, Ely, UK) at a dilution of 1:250. The slides were incubated for 30 min with secondary HRP‐conjugated anti‐rabbit (Envision+ System HRP‐labelled polymer, Dako), then developed using 3,3′‐diaminobenzidine (DAB). The slides were counterstained with Mayer's haematoxylin and lithium carbonate, dehydrated and mounted and then scanned using an Aperio scanner (Leica Microsystems, Milton Keynes, UK) for visualization and scoring. Tumour cores were deemed to show strong overexpression of HER2 if the majority of tumour cells displayed a ‘full basket weave’ pattern of membrane staining in > 90% of cell membranes; this level of expression is classified as 3+ using current consensus‐scoring guidelines 22. To confirm the TMA result and to assess the level of heterogeneity of HER2 expression, whole sections from the tumours with overexpression were also stained, according to the above protocol.
HER2 fluorescence in situ hybridization (FISH)
FISH was carried out using the HER2 Kit (Kreatech, Uckfield, UK) on 4 µm formalin‐fixed, paraffin‐embedded (FFPE) tissue sections from the tumours demonstrating HER2 overexpression by IHC, according to the manufacturer's instructions. Briefly, a rhodamine‐labelled HER2 probe was used to identify the number of target HER2 molecules, together with a FITC‐labelled control CEP17 probe, to identify normal gene copy number. The ratio of HER2 signal (red):CEP17 (green) was calculated in order to determine the presence or absence of HER2 amplification. The criterion for gene amplification was an average HER2:CEP17 ratio of ≥ 2 across at least 50 distinct, non‐overlapping tumour nuclei.
KRAS and BRAF pyrosequencing
For DNA extraction, the QIAamp DNA Micro kit (Qiagen, Manchester, UK) was used, employing the standard manufacturer's protocol. Primers for amplification and pyrosequencing were designed using Pyrosequencing Assay Design Software (Qiagen). KRAS codons 12–13 were amplified in one PCR reaction; KRAS codon 61 and BRAF codon 600 were amplified separately. Full details of PCR reaction conditions have been published elsewhere 23. PCR products were pyrosequenced on a PyroMark ID system (Qiagen), following the manufacturer's protocols.
Copy number variation analysis
Forty‐seven cases with overexpression/amplification of HER2 (25 stage II–III and 22 stage IV) were run on the OncoScan® FFPE Assay Kit (Affymetrix) to identify copy number alterations, according to the manufacturer's instructions. Array fluorescence intensity (CEL) files were generated automatically from DAT files using Affymetrix® GeneChip® Command Console® (AGCC) software v. 4.0. (Affymetrix). The CEL files were processed by OncoScan® Console software to generate OSCHP, which were loaded into Nexus Express for OncoScan® (BioDiscovery, Hawthorne, CA, USA) for analysis of clinically relevant copy number (CN) and loss of heterozygosity (LOH) events. In parallel, somatic mutations were visualised in a Somatic Mutation Viewer (Affymetrix) and cross‐referenced with the CN data for correlation with CN events.
Statistical analysis
Statistical analysis was carried out using STATA v. 12. The proportion of tumours with strong HER2 overexpression was compared between KRAS/BRAF WT and mutated tumours, using Fisher's exact test. Cox's proportional hazards models were used to test for the effect of HER2 strongly positive versus negative tumours on recurrence rates, progression‐free survival (PFS) and overall survival (OS), separately for each trial. Combined estimates were obtained by fixed effects meta‐analysis, testing for heterogeneity, in the two stage IV trials.
Results
HER2 immunohistochemistry
We observed strong membranous staining for HER2 (Figure 1A) in 25 of 1914 (1.3%) stage II–III tumours and 29 of 1342 (2.2%) stage IV tumours tested [18 of 888 (2.0%) in FOCUS; 11 of 454 (2.4%) in PICCOLO]. Weak membranous staining (Figure 1B) was observed in a further 53 of 1342 (3.9%) cases [44 of 888 (4.9%) in FOCUS, nine of 454 (2.0%) in PICCOLO tumours]. Corresponding whole‐tumour sections for the 29 stage IV tumours with strong membranous staining seen in the TMA cores were stained, and showed homogeneous overexpression across each tumour (Figure 1C).
Figure 1.
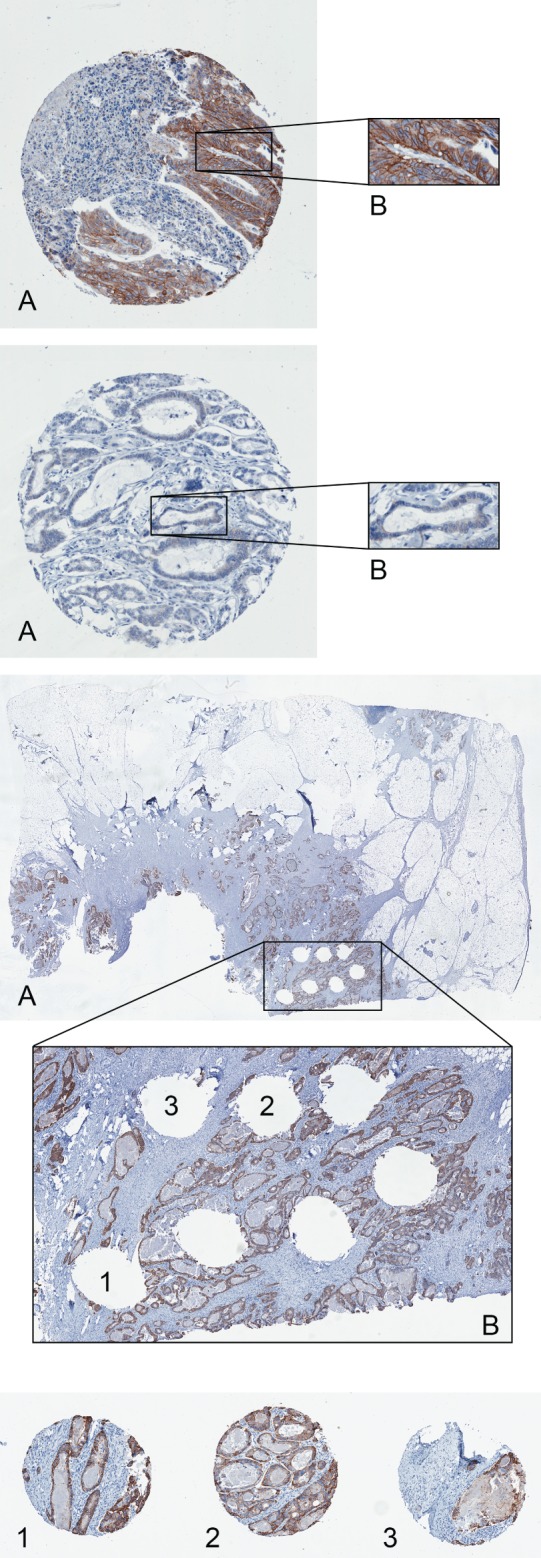
(A) Core from a colorectal tumour, demonstrating strong membranous overexpression of HER2; the cores were visually inspected at a native scanning resolution of ×20. (B) Core from a colorectal tumour, demonstrating weak membranous overexpression of HER2; the cores were visually inspected at a native scanning resolution of ×20. (C) Whole‐section staining was carried out to determine the homogeneity of HER2 overexpression across the tumour section; the location of each of the three cores that were stained on the TMA (1, 2 and 3) is also shown; the cores were visually inspected at a native scanning resolution of ×20
HER2 FISH analysis
Whole sections of the 54 cases with strong membranous staining by IHC were then subjected to HER2 FISH analysis; 24 of the 25 stage II–III tumours were successfully hybridized, with 20 of 24 (83.3%) showing HER2 amplification: 28 of 29 stage IV tumours were successfully hybridized, with 27 of 28 (96.4%) showing HER2 amplification (Figure 2); 10 of 53 stage IV tumours displaying weak membranous IHC staining were randomly selected for HER2 FISH analysis and none of these tumours showed HER2 amplification.
Figure 2.
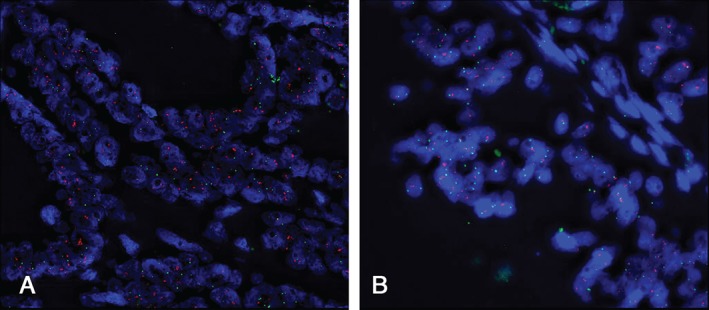
HER2 FISH analysis of two colorectal tumours, showing (A) amplification and (B) no amplification; red signals, HER2 probe; green signals, CEP17 probe; amplification is described as a HER2:CEP17 ratio of ≥ 2
HER2 copy number variation analysis by OncoScan
Forty‐seven cases where strong membranous staining of HER2 was observed were analysed for copy number variation of HER2 and TOP2A using OncoScan: 41 of 47 (87.2%) showed amplification of HER2, two of 47 (4.3%) showed partial amplification of HER2 and the remaining four showed no amplification of HER2; 17 of 47 (36.2%) showed co‐amplification of HER2 and TOP2A. HER2 amplification, as determined by FISH, correlated strongly with amplification, as determined by OncoScan analysis (Table 1).
Table 1.
Correlation between amplification of HER2, as assessed by FISH, and amplification, as determined by OncoScan (n = 47)
| OncoScan | |||
|---|---|---|---|
| Amplification | Partial amplification | No amplification | |
| FISH | n (%) | n (%) | n (%) |
| Amplification | 38 (80.9) | n/a | 2 (4.3) |
| No amplification | 2 (4.3) | 2 (4.3) | 1 (2.1) |
| Assay failure | 1 (2.1) | n/a | 1 (2.1) |
HER2 and KRAS/BRAF mutation status
HER2 overexpression was strongly associated with KRAS/BRAF WT status in all stages of disease. In the QUASAR trial, HER2 overexpression was observed in 17 of 811 (2.1%) KRAS/BRAF WT tumours compared with one of 421 (0.2%) tumours harbouring a KRAS or BRAF mutation (p = 0.01). In the stage IV tumours, strong HER2 staining was present in 24 of 466 (5.2%) WT cases, compared with 5 of 525 (1.0%) of those with a mutation (Fisher's exact test, p < 0.0001). This difference was observed in each of the stage IV trial datasets [FOCUS, 14 of 259 (5.4%) versus 4 of 284 (1.4%); PICCOLO, 10 of 207 (4.8%) versus 1 of 241 (0.4%)].
HER2 and recurrence rate, PFS and OS analysis
Clinical data was available in 1767 QUASAR cases. There were no significant correlations between membranous HER2 overexpression and either recurrence or overall survival (OS) (Figure 3). There appears to be a greater difference in recurrence between those cases with HER2 overexpression and those showing no overexpression, although this is not significant in the group receiving chemotherapy. The numbers of recurrences in the treated group are similar to the number of recurrences in the entire trial, and this is reflected in the relative p values.
Figure 3.
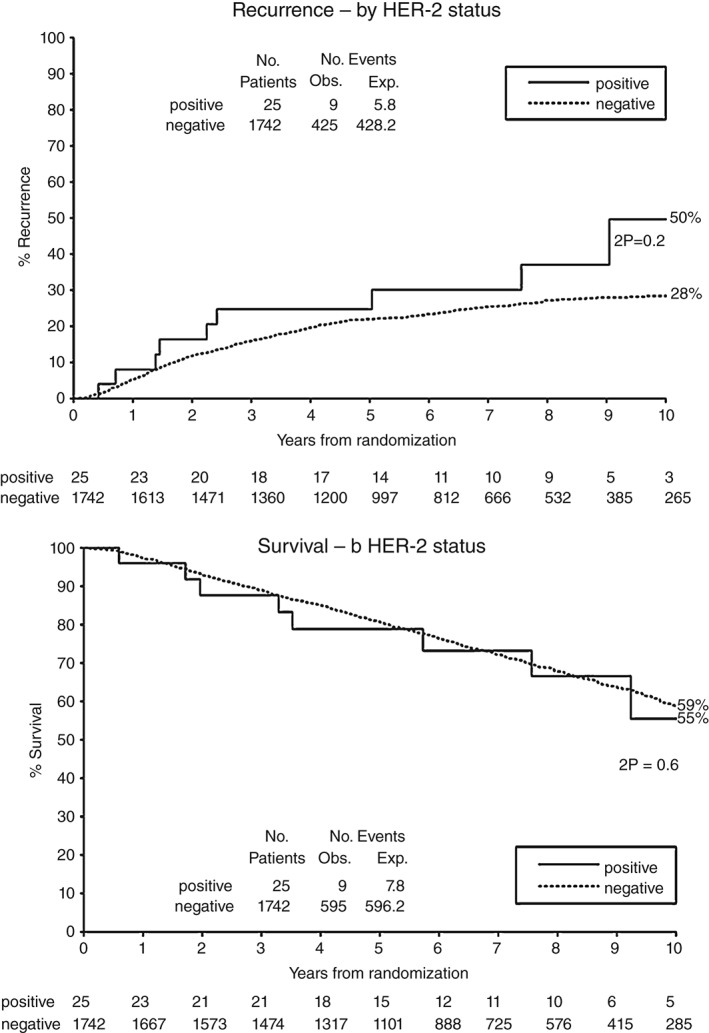
Kaplan–Meier estimates of recurrence and survival of patients in the QUASAR trial, with tumours showing overexpression or no expression of HER2 protein
Associations of HER2 expression with OS and progression‐free survival (PFS) were first assessed in the two stage IV trial datasets separately. In neither trial was there a significant association. In FOCUS, comparing patients with HER2 strongly positive tumours (n = 18) versus those with no visible membrane staining (n = 826) gave a hazard ratio (HR) for OS of 0.81 [95% confidence interval (CI) 0.50, 1.32; the HR for PFS was 0.68, CI 0.42, 1.12]. Corresponding estimates from the PICCOLO trial (n = 11 versus n = 434) were 0.98, 95% CI 0.54, 1.78 for OS and 0.80, 95% CI 0.44, 1.46 for PFS.
Combining the two datasets, there remains no evidence for an effect on outcome [HR for OS, 0.87, 95% CI 0.60, 1.27; p = 0.48; HR for PFS, 0.73, 95% CI 0.50, 1.07; p = 0.11], with no evidence for heterogeneity between the studies in either analysis (Figure 4). Restricting these survival analyses to patients with KRAS WT tumours, very similar results were obtained [HR for OS from meta‐analysis of the two trials, 0.99, 95% CI 0.65, 1.51; p = 0.95; HR for PFS, 0.83, 95% CI 0.54, 1.28; p = 0.40], with no evidence for heterogeneity between studies.
Figure 4.
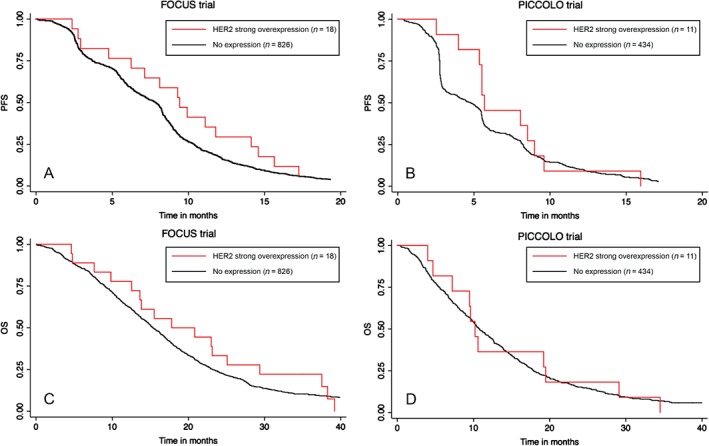
Kaplan–Meier estimates of progression‐free survival (PFS) and overall survival (OS) among patients in FOCUS (A and C, respectively) and PICCOLO (B and D, respectively), with tumours showing overexpression or no expression of HER2 protein
Discussion
It has recently been demonstrated in aCRC that, in addition to KRAS mutation status, mutations in other proteins downstream of EGFR, including NRAS and possibly also BRAF and PIK3CA, contribute to resistance to anti‐EGFR therapies. Data from trials such as PICCOLO 21 and PRIME 24 have provided strong supporting evidence for this. However, even tumours which are wild‐type for all these genes have a relatively high rate of drug‐resistance.
HER2 has the ability to form heterodimers with EGFR and HER3, and to activate the MAPK and AKT pathways. Overexpression of HER2 is therefore a potential mechanism for circumventing EGFR blockade, making it a potential biomarker of resistance to anti‐EGFR therapy and a potential selection criterion for novel HER2‐ or HER3‐targeted treatments. Bertotti et al 25 provided early evidence using HER2‐amplified tumour xenografts, which demonstrated resistance to cetuximab. Accurate characterization of HER2 expression in large patient populations is an important first step to its development as a biomarker in aCRC.
In this study we assessed HER2 expression by immunohistochemistry, identifying a subset of CRC with strong overexpression of membranous HER2. Overall, we observed this in just 25 of 1914 (1.3%) stage II–III tumours and 29 of 1342 (2.2%) of stage IV tumours, which, although a small proportion, still represents a sizeable number of patients in this common disease group. This low level of expression is almost identical to that reported by Lee et al 14 when reporting on HER2 overexpression in 94 consecutive stage IV CRCs. We found that 20 of 24 (83.3%) stage II–III patients and 27 of 28 (96.4%) stage IV patients with strong IHC staining also showed HER2 amplification by FISH.
Several previous studies have assessed HER2 using both IHC and FISH analyses, and also found that gene amplification is a consistent finding in tumours with strong (3+) membranous staining by IHC 9, 11, 15. These studies also identified cases with moderate/weak (2+) staining and equivocal amplification by FISH. However, of the 1342 stage IV patients, we found no cases falling into this intermediate staining category. We identified 53 tumours with visible but weak (1+) staining; 10 of these tumours, selected at random, underwent HER2 FISH analysis, with none demonstrating gene amplification. These results suggest that gene amplification is associated with strong membranous overexpression of HER2, and that IHC is a reliable technique for assessing HER2 in CRC. It remains unclear whether the HER2 status of these is representative of that of any corresponding metastases. Lee et al 14 addressed this very question and determined that in almost 15% of tumours, there was a lack of concordance between the HER2 status in the primary tumour and associated metastases. This may need to be taken into account if future clinical trials are realised.
In order to confirm the amplification of the HER2 gene seen in the FISH analysis, 47 cases with strong membranous overexpression of HER2 were analysed using the Affymetrix OncoScan FFPE Assay Kit: 43 of 47 (91.4%) showed full or partial amplification of HER2, suggesting that the OncoScan method is an alternative way of assessing copy number variation; only four of 47 cases (8.5%) showed no amplification of HER2, including one case where analysis by FISH failed and one case where FISH also showed no amplification. We were also able to access OncoScan data on 159 PICCOLO samples, which were negative for HER2 protein expression. None of these tumours demonstrated HER2 amplification. We feel this further evidences the strong correlation between protein expression and amplification, and is confirmation that the negative tumours are not in fact ‘false negatives’. These results suggest that HER2 expression is increased in a small subset of intermediate and aCRC, and that these cases may benefit from anti‐HER2 therapies such as trastuzumab and pertuzumab.
Seventeen of 47 cases (36.2%) showed co‐amplification of HER2 and TOP2A. These results support existing reports of HER2/TOP2A co‐amplification in the range 21–42% 26, 27, 28.
In all disease stages, but particularly in stage IV, we observed a significant association of HER2 overexpression with KRAS/BRAF WT tumours (p < 0.0001). This is consistent with the hypothesis that HER2 amplification may represent an alternative driver to MEK–AKT pathway activation in tumours without an activating mutation of a downstream oncogene. One might predict that these HER2‐amplified tumours would be resistant to cetuximab or panitumumab therapy, and that they may respond to targeted treatment against HER2, or a combination of therapies targeting HER2, EGFR and possibly also HER3. Such combinations have been reported to inhibit growth in cetuximab‐resistant cell lines 25, 29. A recent clinical study reported HER2 amplification in seven of 170 (4%) KRAS WT patients receiving cetuximab or panitumumab, and these patients had inferior PFS and OS compared to those without HER2 amplification; however, the small sample size limits interpretation of this result 30.
Recently presented data on the HERACLES trial, a phase II trial in _HER2_‐amplified, KRAS exon 2 WT mCRC patients of trastuzumab and lapatinib, achieved its primary endpoint of an objective response (OR) in > 30% of patients. This trial provided the first real evidence that patients with HER2 amplification may receive benefit from dual anti‐HER2 treatments, and certainly points to the incorporation of such therapies earlier in the treatment of mCRC patients 31. To ensure that the correct patients were enrolled into the HERACLES trial, work was carried out by Valtorta et al 32 to develop criteria to accurately define HER2 positivity in colorectal tumours. According to these criteria, HER2 positivity was defined as the presence of intense, homogeneous membranous staining in at least 50% of tumour cells, and these tumours also showed HER2 amplification by FISH. The study also reported a rate of HER2 amplification in KRAS WT tumours of just over 5%, consistent with our findings of 5.2%. This provides additional evidence that our negative tumours are indeed true negatives.
In the QUASAR trial, we did not find a significant correlation between HER2 expression and recurrence or survival. Furthermore, we did not find an association between HER2 expression and PFS or OS in the stage IV trials. Due to the rarity of HER2 overexpression, the power to detect a modest effect on outcome is low, but we can exclude a strong effect. This agrees with two previous studies, which failed to detect a prognostic biomarker effect for HER2 10, 13.
One area we have not investigated in this study is the presence of activating mutations in the HER2 gene. A recent functional analysis by Kloth et al 33 revealed a differential response to HER2‐targeted therapies in _HER2_‐mutant MSI CRC cell lines. A similar response was also shown by Kavuri et al 34, where treatment of _HER2_‐mut CRC cell‐lines with a single HER2‐targeted agent produced delayed tumour growth and treatment with dual agents (anti‐HER2 therapy plus a tyrosine kinase inhibitor) produced tumour regression. Bertotti et al 35 recently carried out exome sequence and copy number analyses of both patient‐derived xenografts and patient tumours, to investigate the effects of mutations in ERBB2, EGFR, FGFR1, PDGFRA and MAP2K1 in relation to resistance to anti‐EGFR therapies. Point mutations were detected in both the kinase domain and the ectodomain of HER2, and these correlated with cetuximab resistance. This provides early evidence of a role for somatic mutations in the activation of HER2 signalling, resulting in anti‐EGFR therapy resistance. With the advent of next‐generation sequencing, this is one area which ought to be investigated further, with the aim of defining a _HER2_‐mutant cohort who may gain benefit from HER2‐targeted therapies.
In conclusion, we have identified, within these large clinical trial cohorts, a small subset of patients with tumours overexpressing membranous HER2 and having amplification of the HER2 gene confirmed by FISH and microarray studies. HER2 overexpression is not a prognostic biomarker; however, it is strongly associated with RAS/RAF WT status and TOP2A amplification. This raises the hypothesis that HER2 amplification is an alternative driver of MEK–AKT pathway activation in CRC. We propose that HER2 should now be assessed as a putative biomarker of resistance to anti‐EGFR therapy in RAS/RAF WT patients, as a potential predictive biomarker for HER2‐targeted therapy and, if further studies confirm that TOP2A amplification is associated with anthracycline sensitivity, it might be worth screening for HER2 overexpression to triage for its presence in this disease.
Author contributions
SDR, KS, JB, MS and PQ conceived and designed the study; SDR, KS, PC, DC, GH, MT, JMF, AO and KGS carried out the data collection and assembly; data analysis and interpretation were performed by SDR, KS, PC, JB, HW, JMF, AO, KGS, SRB, MJ, DK, KH, RG and PQ. All authors were involved in the writing of the manuscript and also approving the final submitted version.
Acknowledgements
SDR, KS, GH, MT and PQ were funded by Yorkshire Cancer Research (YCR) and PC by Cancer Research UK (CRUK; Grant No. C37059/A11941). Infrastructure support was provided by grants from the Experimental Cancer Medicine Centre (ECMC) and the CRUK Cancer Centre. The FOCUS trial (Grant No. ISRCTN 79877428) was funded by the UK Medical Research Council. The PICCOLO trial (Grant No. ISRCTN 93248876) was funded by CRUK with support from Amgen. The QUASAR trial (Grant No. ISRCTN 82375386) was funded by the UK Medical Research Council. Affymetrix OncoScan chips were funded via the Technology Strategy Board Stratified Medicines Programme (Grant No. 101032). Thanks also to Mr Alex Wright for his assistance with the generation of the figures.
Conflicts of interest JMF, AO and KGS are all paid employees of Affymetrix UK Ltd and hold shares in Affymetrix (NASDAQ: AFFX). None of the other authors have any competing interests to declare.
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2.Wells A. EGF receptor. Int J Biochem Cell Biol 1999; 31: 637–643. [DOI] [PubMed] [Google Scholar]
- 3.Press MF, Bernstein L, Thomas PA, et al. HER‐2/neu gene amplification characterized by fluorescence in situ hybridization: poor prognosis in node‐negative breast carcinomas. J Clin Oncol 1997; 15: 2894–2904. [DOI] [PubMed] [Google Scholar]
- 4.Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER‐2/neu oncogene. Science 1987; 235: 177–182. [DOI] [PubMed] [Google Scholar]
- 5.Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER‐2/neu proto‐oncogene in human breast and ovarian cancer. Science 1989; 244: 707–712. [DOI] [PubMed] [Google Scholar]
- 6.Leonard DS, Hill AD, Kelly L, et al. Anti‐human epidermal growth factor receptor 2 monoclonal antibody therapy for breast cancer. Br J Surg 2002; 89: 262–271. [DOI] [PubMed] [Google Scholar]
- 7.Gravalos C, Jimeno A. HER2 in gastric cancer: a new prognostic factor and a novel therapeutic target. Ann Oncol 2008; 19: 1523–1529. [DOI] [PubMed] [Google Scholar]
- 8.Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of _HER2_‐positive advanced gastric or gastro‐oesophageal junction cancer (ToGA): a phase 3, open‐label, randomised controlled trial. Lancet 2010; 376: 687–697. [DOI] [PubMed] [Google Scholar]
- 9.Kavanagh DO, Chambers G, O'Grady L, et al. Is overexpression of HER‐2 a predictor of prognosis in colorectal cancer? BMC Cancer 2009; 9: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marx AH, Burandt EC, Choschzick M, et al. Heterogenous high‐level HER‐2 amplification in a small subset of colorectal cancers. Hum Pathol 2010; 41: 1577–1585. [DOI] [PubMed] [Google Scholar]
- 11.Ooi A, Takehana T, Li X, et al. Protein overexpression and gene amplification of HER‐2 and EGFR in colorectal cancers: an immunohistochemical and fluorescent in situ hybridization study. Mod Pathol 2004; 17: 895–904. [DOI] [PubMed] [Google Scholar]
- 12.Osako T, Miyahara M, Uchino S, et al. Immunohistochemical study of c‐erbB‐2 protein in colorectal cancer and the correlation with patient survival. Oncology 1998; 55: 548–555. [DOI] [PubMed] [Google Scholar]
- 13.Sclafani F, Roy A, Cunningham D, et al. HER2 in high‐risk rectal cancer patients treated in EXPERT‐C, a randomized phase II trial of neoadjuvant capecitabine and oxaliplatin (CAPOX) and chemoradiotherapy (CRT) with or without cetuximab. Ann Oncol 2013; 24: 3123–3128. [DOI] [PubMed] [Google Scholar]
- 14.Lee WS, Park YH, Lee JN, et al. Comparison of HER2 expression between primary colorectal cancer and their corresponding metastases. Cancer Med 2014; 3: 674–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nathanson DR, Culliford ATt, Shia J, et al. HER 2/neu expression and gene amplification in colon cancer. Int J Cancer 2003; 105: 796–802. [DOI] [PubMed] [Google Scholar]
- 16.Caruso ML, Valentini AM. Immunohistochemical p53 overexpression correlated to c‐erbB‐2 and cathepsin D proteins in colorectal cancer. Anticancer Res 1996; 16: 3813–3818. [PubMed] [Google Scholar]
- 17.Park SS, Kim SW. Activated Akt signaling pathway in invasive ductal carcinoma of the breast: correlation with HER2 overexpression. Oncol Rep 2007; 18: 139–143. [PubMed] [Google Scholar]
- 18.Quasar Collaborative G, Gray R, Barnwell J, et al. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet 2007; 370: 2020–2029. [DOI] [PubMed] [Google Scholar]
- 19.Seymour MT, Maughan TS, Ledermann JA, et al. Different strategies of sequential and combination chemotherapy for patients with poor prognosis advanced colorectal cancer (MRC FOCUS): a randomised controlled trial. Lancet 2007; 370: 143–152. [DOI] [PubMed] [Google Scholar]
- 20.Middleton G, Brown S, Lowe C, et al. A randomised phase III trial of the pharmacokinetic biomodulation of irinotecan using oral ciclosporin in advanced colorectal cancer: results of the Panitumumab, Irinotecan and Ciclosporin in COLOrectal cancer therapy trial (PICCOLO). Eur J Cancer 2013; 49: 3507–3516. [DOI] [PubMed] [Google Scholar]
- 21.Seymour MT, Brown SR, Middleton G, et al. Panitumumab and irinotecan versus irinotecan alone for patients with KRAS wild‐type, fluorouracil‐resistant advanced colorectal cancer (PICCOLO): a prospectively stratified randomised trial. Lancet Oncol 2013; 14: 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofmann M, Stoss O, Shi D, et al. Assessment of a HER2 scoring system for gastric cancer: results from a validation study. Histopathology 2008; 52: 797–805. [DOI] [PubMed] [Google Scholar]
- 23.Richman SD, Seymour MT, Chambers P, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol 2009; 27: 5931–5937. [DOI] [PubMed] [Google Scholar]
- 24.Douillard JY, Oliner KS, Siena S, et al. Panitumumab–FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013; 369: 1023–1034. [DOI] [PubMed] [Google Scholar]
- 25.Bertotti A, Migliardi G, Galimi F, et al. A molecularly annotated platform of patient‐derived xenografts ('xenopatients') identifies HER2 as an effective therapeutic target in cetuximab‐resistant colorectal cancer. Cancer Discov 2011; 1: 508–523. [DOI] [PubMed] [Google Scholar]
- 26.Fasching PA, Weihbrecht S, Haeberle L, et al. HER2 and TOP2A amplification in a hospital‐based cohort of breast cancer patients: associations with patient and tumor characteristics. Breast Cancer Res Treat 2014; 145: 193–203. [DOI] [PubMed] [Google Scholar]
- 27.Fountzilas G, Christodoulou C, Bobos M, et al. Topoisomerase II_α_ gene amplification is a favorable prognostic factor in patients with _HER2_‐positive metastatic breast cancer treated with trastuzumab. J Transl Med 2012; 10: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fountzilas G, Dafni U, Bobos M, et al. Evaluation of the prognostic role of centromere 17 gain and HER2/topoisomerase II_α_ gene status and protein expression in patients with breast cancer treated with anthracycline‐containing adjuvant chemotherapy: pooled analysis of two Hellenic Cooperative Oncology Group (HeCOG) phase III trials. BMC Cancer 2013; 13: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yonesaka K, Zejnullahu K, Okamoto I, et al. Activation of ERBB2 signaling causes resistance to the EGFR‐directed therapeutic antibody cetuximab. Sci Transl Med 2011; 3: 99ra86. [DOI] [PMC free article] [PubMed]
- 30.Martin V, Landi L, Molinari F, et al. HER2 gene copy number status may influence clinical efficacy to anti‐EGFR monoclonal antibodies in metastatic colorectal cancer patients. Br J Cancer 2013; 108: 668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salvatore Siena AS‐B, Lonardi S, Trusolino L, _et al._Trastuzumab and lapatinib in _HER2_‐amplified metastatic colorectal cancer patients (mCRC): the HERACLES trial. J Clin Oncol 2015; 33 (suppl); abstr 3508. [Google Scholar]
- 32.Valtorta E, Martino C, Sartore‐Bianchi A, et al. Assessment of a HER2 scoring system for colorectal cancer: results from a validation study. Mod Pathol 2015; 28: 1481–1491. [DOI] [PubMed] [Google Scholar]
- 33.Kloth M, Ruesseler V, Engel C, et al. Activating ERBB2/HER2 mutations indicate susceptibility to pan‐HER inhibitors in Lynch and Lynch‐like colorectal cancer. Gut 2015; DOI: 10.1136/gutjnl‐2014‐309026. [DOI] [PubMed] [Google Scholar]
- 34.Kavuri SM, Jain N, Galimi F, et al. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov 2015; 5: 832–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertotti A, Papp E, Jones S, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature 2015; 526: 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]