Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells (original) (raw)
Here, Guo et al. investigate how autophagy maintains mitochondrial function in Ras-driven cancer cells and why autophagy is critical for their survival in starvation. By analyzing mitochondrial genomes from Atg7 wild-type and _Atg7_-deficient Kras-driven lung tumors, the authors demonstrated that autophagy supports nucleotide pools, which enables survival of Ras-driven cancer cells in starvation.
Keywords: ROS, Ras-driven cancer, amino acid, autophagy, energy charge, mitochondrial metabolism, nucleotide
Abstract
Autophagy degrades and is thought to recycle proteins, other macromolecules, and organelles. In genetically engineered mouse models (GEMMs) for _Kras_-driven lung cancer, autophagy prevents the accumulation of defective mitochondria and promotes malignancy. Autophagy-deficient tumor-derived cell lines are respiration-impaired and starvation-sensitive. However, to what extent their sensitivity to starvation arises from defective mitochondria or an impaired supply of metabolic substrates remains unclear. Here, we sequenced the mitochondrial genomes of wild-type or autophagy-deficient (_Atg7_−/−) _Kras_-driven lung tumors. Although Atg7 deletion resulted in increased mitochondrial mutations, there were too few nonsynonymous mutations to cause generalized mitochondrial dysfunction. In contrast, pulse-chase studies with isotope-labeled nutrients revealed impaired mitochondrial substrate supply during starvation of the autophagy-deficient cells. This was associated with increased reactive oxygen species (ROS), lower energy charge, and a dramatic drop in total nucleotide pools. While starvation survival of the autophagy-deficient cells was not rescued by the general antioxidant N-acetyl-cysteine, it was fully rescued by glutamine or glutamate (both amino acids that feed the TCA cycle and nucleotide synthesis) or nucleosides. Thus, maintenance of nucleotide pools is a critical challenge for starving _Kras_-driven tumor cells. By providing bioenergetic and biosynthetic substrates, autophagy supports nucleotide pools and thereby starvation survival.
Macroautophagy (referred to here as autophagy) is a cellular self-digestion pathway evolutionarily conserved from yeast to mammals that degrades and is thought to recycle proteins and organelles essential to survive starvation (Mortimore and Poso 1987). Yeast cells rely on autophagy to survive nitrogen starvation, hence identifying the autophagy-related (Atg) genes and elucidating the autophagy pathway (Tsukada and Ohsumi 1993). Neonatal mice require autophagy to maintain serum amino acid levels and survival during physiological neonatal starvation (Kuma et al. 2004; Komatsu et al. 2005). Adult mice require autophagy to sustain serum glucose levels during fasting, which prevents cachexia and fatal hypoglycemia (Karsli-Uzunbas et al. 2014). Thus, autophagy buffers metabolic stress, likely by degrading intracellular components to generate amino acids, nucleotides, fatty acids, and sugars that are recycled into metabolic and biosynthetic pathways (Rabinowitz and White 2010). Loss of autophagy in mice also causes neurodegeneration as well as muscle and liver damage over time, indicating a long-term role for autophagy in selective tissue maintenance as well as in acute response to nutrient limitation (Mizushima and Levine 2010; Karsli-Uzunbas et al. 2014). The specific autophagy substrates that are important, the products that they produce, and the pathways and functions that they are used for are not known.
Evidence suggests that autophagy has a context-dependent role in cancer. Autophagy suppresses tumor initiation in mouse models by eliminating SQSTM1/p62 and also likely suppressing chronic tissue damage and inflammation (White 2012; White et al. 2015). In established tumors, autophagy promotes survival of metabolic stress in the microenvironment such as hypoxic tumor regions (Degenhardt et al. 2006). Some tumors have high levels of basal autophagy that may be due to elevated nutrient requirements (Guo et al. 2011; Yang et al. 2011; White 2012; White et al. 2015) or direct activation of the autophagy machinery by oncogenic signaling (Perera et al. 2015; Wong et al. 2015), indicating that some tumors may be autophagy-dependent.
In genetically engineered mouse models (GEMMs) for _Kras_-driven non-small-cell lung cancer (NSCLC), deletion of Atg7 causes tumor cells to accumulate autophagy substrates (particularly mitochondria), suppresses tumor growth, and promotes tumor cell death. Importantly, Atg7 deficiency alters the fate of _Kras_-induced carcinomas to oncocytomas (Guo et al. 2013), which are rare, predominantly benign tumors characterized by the accumulation of defective mitochondria (Joshi et al. 2015). Thus, Atg7 is required for NSCLC growth, survival, and malignancy. Moreover, acute, systemic genetic ablation of Atg7 in mice with established NSCLC promotes tumor regression prior to damage to most normal tissues, indicating that tumors can be selectively autophagy-dependent (Karsli-Uzunbas et al. 2014). Deletion of the essential autophagy gene Atg5 in a GEMM for NSCLC also attenuates _Kras_-driven lung tumor growth, suggesting that anti-tumor activity is likely due to loss of autophagy (Rao et al. 2014).
In other GEMMs for cancer, autophagy promotes the growth of Braf _V600E_-induced lung tumors (Strohecker and White 2014) and _Pten_-deficient prostate tumors (Santanam et al. 2016) and suppresses senescence in melanomas driven by Braf V600E and loss of Pten (Xie et al. 2015). Genetic loss of autophagy impairs the progression of _Kras_-driven premalignant pancreatic intraepithelial neoplasia (PANIN) lesions to invasive cancer (Rosenfeldt et al. 2013; Yang et al. 2014). Atg7 deficiency in intestinal epithelial cells in Apc+/− mice leads to intestinal dysbiosis required for an anti-cancer immune response, which prevents intestinal tumor initiation and growth (Levy et al. 2015). Thus, these and other GEMMs have identified autophagy-dependent cancers that are generalizable to tumors driven by distinct oncogenic events and not related to the origin of tissue or tumor type.
The mechanism by which autophagy promotes tumorigenesis in some cases is in part due to inhibition of p53, but the underlying mechanisms are not known (Guo et al. 2013; Rosenfeldt et al. 2013; Strohecker and White 2014; Wei et al. 2014; Yang et al. 2014). In contrast, deletion of essential autophagy genes in human cancer cell lines by genome editing does not impair tumor growth in immune-compromised mice (Eng et al. 2016). This suggests that the immune system may suppress growth of autophagy-deficient tumors or that there is selection for compensatory mechanisms that overcome autophagy deficiency. Determining the mechanism by which autophagy promotes tumor cell growth and malignancy will begin to resolve these and other possibilities.
Oncogenic mutation or loss of tumor suppressors promotes glycolysis for rapid energy production to meet the high metabolic demand of cancer cell growth (Vander Heiden et al. 2009). However, most tumor cells still rely on mitochondria metabolism for their proliferation and tumorigenesis (Gaglio et al. 2011; Fan et al. 2013). Mitochondria are the hub for bioenergetic processes, in which substrates taken from the cytoplasm are used to drive fatty acid oxidation, the tricarboxylic acid (TCA) cycle, the electron transport chain (ETC), and respiration. Mitochondria also function as a platform for biosynthesis of amino acids, lipids, nucleotides, heme, and iron sulfur clusters as well as NADPH for their own antioxidant defense (Wallace 2012). Elimination of cancer cell mitochondrial DNA (mtDNA) impairs tumorigenicity, which is restored by the acquisition of mtDNA from host cells to tumor cells (Tan et al. 2015). Loss of the mitochondrial transcription factor A (TFAM) gene disrupts mitochondrial function and impedes _Kras_-driven tumorigenesis (Weinberg et al. 2010). Impaired mitochondrial respiration by suppressing the functionally coupled mitochondrial pyrimidine biosynthesis enzyme dihydroorotate dehydrogenase (DHODH) leads to the depletion of pyrimidines, which triggers the activation of p53 and induction of p53-dependent apoptosis (Khutornenko et al. 2010). Therefore, maintaining functional mitochondrial metabolism is necessary for cancer cell survival and proliferation (Zong et al. 2016).
A continuous inflow of four-carbon (4C) units to balance the outflow of such units to amino acids and other biosynthetic products is essential to sustain mitochondrial metabolism (Zong et al. 2016). The replenishment of TCA cycle metabolites can be achieved by the carboxylation of pyruvate or catabolism of glutamine or other amino acids (DeBerardinis et al. 2007; Yuneva et al. 2007; Fan et al. 2009; Comerford et al. 2014; Mashimo et al. 2014). 4C units enter the TCA cycle and generate citrate by condensing with two-carbon (2C) units, which are supplied by acetyl-CoA. Citrate can be metabolized further to generate all classical TCA cycle intermediates or exported to the cytosol for lipid synthesis and protein modifications. Moreover, mitochondria also play a central role in the metabolism of one-carbon (1C) units, which are required for purine, thymidine, and methionine synthesis (Zong et al. 2016).
Mitochondria-mediated biosynthetic pathways are important for cell proliferation. Aspartate supplementation or overexpression of an aspartate transporter allows cells without ETC activity to proliferate, indicating that enabling aspartate synthesis is an essential role of mitochondria in cell proliferation (Birsoy et al. 2015). Glutamine autonomously synthesized by glutamine synthetase-positive glioma cells or supplied by astrocytes is required for the growth of glutamine-restricted glioblastoma by fueling de novo purine synthesis (Tardito et al. 2015). The mitochondrial 1C pathway is consistently overexpressed in cancer and supports serine-mediated purine and thymidine synthesis. Particularly, in nutrient-poor conditions, mitochondrial 1C metabolism is required for tumorigenesis (Ducker et al. 2016). Mitochondria-mediated serine catabolism also supports tumor growth by maintaining mitochondrial redox balance to support tumor cell survival and growth (Ye et al. 2014).
Autophagy deficiency causes the accumulation of dysfunctional mitochondria in tumor cells and impairs tumorigenesis, suggesting that autophagy may promote tumorigenesis and malignancy by maintaining mitochondrial quality and possibly also providing substrates for mitochondrial metabolism (White 2015). However, whether autophagy recycles substrates, the identity of specific substrates, and the metabolic pathways and functions that they are used for are not known.
Here we address how autophagy maintains mitochondrial function in Ras-driven cancer cells and why autophagy is critical for their survival in starvation. We found that deletion of Atg7 in tumors increased the frequency of mitochondrial genome variation, but the heteroplasmic mitochondrial mutations did not account for metabolic impairment. Pulse-chase studies with isotope-labeled nutrients showed that Atg7 deficiency reduced metabolite recycling in starvation, specifically TCA cycle intermediates, glutamate, aspartate, and α-ketoglutarate (α-KG), indicating that substrate limitation in autophagy-deficient tumor cells impaired mitochondrial metabolism. Dysfunctional mitochondrial metabolism caused by autophagy deficiency was associated with increased reactive oxygen species (ROS), lower energy charge, and a dramatic drop in total nucleotide pools in starvation. Supplementation of glutamine or nucleosides was sufficient to maintain energy charge, sustain nucleotide pools, and rescue death of starved _Atg7_-deficient tumor cells. [U13C5]-glutamine (Gln) tracing showed that glutamine was metabolized to replenish TCA cycle intermediates for nucleotide synthesis and energy and redox homeostasis in starvation. Thus, the role of autophagy in Ras-driven tumor cells is to recycle substrates to prevent fatal nucleotide pool depletion and energy crisis in starvation.
Results
Autophagy contributes to mitochondrial genome maintenance but is not essential to maintain proper coding of mitochondrial proteins
To address whether defective clearance of mitochondrial genomes in _Atg7_-deficient tumors contributed to metabolic defects, the mtDNA from Atg7 wild-type and _Atg7-_deficient _Kras_-driven mouse lung tumors was sequenced (∼24,000× coverage; N = 8 mice per group) (Supplemental Table S1). Sequence variants were found more commonly in the _Atg7_-deficient tumors (P = 0.0267) (Supplemental Table S2). Using an allele frequency cutoff of 3%, among the eight wild-type tumors, there was only one sequence variant, while among the eight _Atg7_−/− tumors, five showed at least a single sequence variant, with a total of eight variants detected (Supplemental Table S2). The variant allele frequency in _Atg7_-deficient tumors ranged from 3% to 26% (Supplemental Table S3). The mutations that were found were all either G>A or T>C on the light strand (corresponding to C>T and A>G on the heavy strand), which is consistent with mitochondrial mutations detected across human cancers and postulated to result from mtDNA replication error (Supplemental Table S3; Ju et al. 2014). Critically, all variants in protein-coding regions were synonymous (Supplemental Table S3). Thus, while Atg7 suppresses mtDNA allelic variation, loss of mitochondrial genome quality control is unlikely to be the reason for defective mitochondrial function in _Atg_7-deficient tumors.
Autophagy mediates substrate recycling
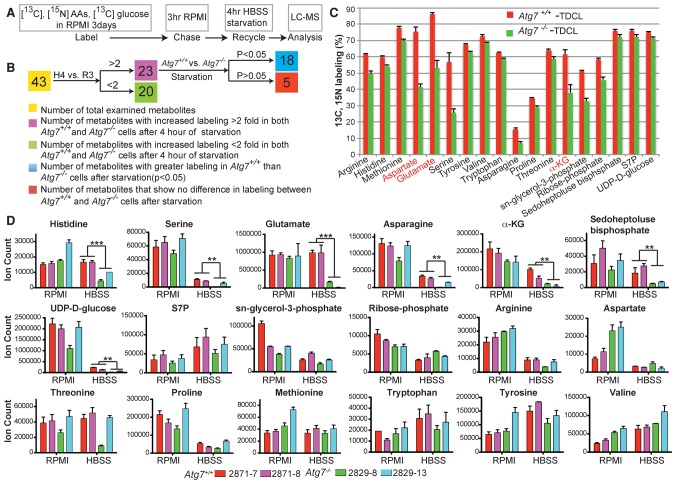
To elucidate the substrates provided by autophagy-mediated degradation and recycling in starvation, _Kras_-driven Atg7 wild-type and _Atg7-_deficient tumor-derived cell lines (TDCLs) were cultured in RPMI medium with uniformly 13C-labeled and 15N-labeled amino acids and 13C-labeled glucose for 3 d to label endogenous components, including proteins and metabolites. This was followed by a chase with unlabeled RPMI medium for 3 h to replace labeled with unlabeled metabolites. The cells were then subjected to starvation in Hanks’ balanced salt solution (HBSS) without glucose for 4 h prior to significant cell death induction, and the metabolites were examined by liquid chromatography-mass spectrometry (LC-MS) for recycling as indicated by the presence of 13C-labeled and 15N-labeled metabolites from the breakdown of intracellular macromolecules (Fig. 1A). Analysis focused on 43 abundant central metabolites whose labeling we could reliably measure across these conditions, including amino acids and nucleotides as well as glycolytic, pentose phosphate, and TCA intermediates.
Figure 1.
Autophagy mediates substrate recycling in starvation. (A) Schematic of 13C and15N tracer studies. (B) The schematic chart shows different metabolite recycling distributions between Atg7+/+ and _Atg7_−/− TDCLs in starvation. (R3) The time point after cells were cultured in unlabeled RPMI medium for 3 h; (H4) the time point after 4 h of HBSS to induce degradation of intracellular components for recycling. (C) 13C and 15N tracing shows substrates of autophagy-mediated recycling in _Kras_-driven TDCLs in starvation. _t_-test, false discovery rate [FDR]-adjusted _P_-value < 0.05. Red indicates TCA cycle intermediates and TCA-derived amino acids. The error bar indicates ±SEM. n = 3. (D) Pool sizes of metabolites identified from 1C in nutrient-rich (RPMI) and nutrient-depleted (HBSS) conditions. (S7P) Sedoheptulose 7-phosphate. The error bar indicates ±SEM. n = 3. (**) P < 0.01; (***) P < 0.001, _t_-test.
Three days of growth in 13C-labeled and 15N-labeled nutrients resulted in extensive and equivalent labeling of intracellular metabolites in both Atg7 wild-type and _Atg7-_deficient TDCLs (Supplemental Table S4.1). After the 3-h chase with unlabeled medium, most metabolites were unlabeled as expected due to their production from nutrients in RPMI (Supplemental Table S4.2). Starvation following the chase significantly increased labeling of 43 examined metabolites, with 23 increasing in labeling by at least twofold (Supplemental Table S4.3), consistent with labeled intracellular components being degraded and recycled into metabolic pathways. Of these 23 metabolites that were derived substantially from macromolecule degradation, 18 showed significant differences in labeling between Atg7 wild-type and _Atg7-_deficient cells, with greater labeling in the wild-type cells in all cases (Fig. 1B; Supplemental Table S4.3). These 18 metabolites included amino acids, pentose phosphate intermediates, UDP-glucose, glycerol-phosphate, and α-KG (Fig. 1C; Supplemental Table S4.3). For some of these metabolites, including many essential amino acids, there was substantial formation from macromolecular degradation even in the absence of autophagy, indicating that other degradative processes also contribute to recycling in starvation (Fig. 1C; Supplemental Table S4.3). For others, including the key TCA cycle intermediate α-KG and TCA-derived amino acids aspartate and glutamate, the contribution of autophagy was large, consistent with autophagy playing a particularly critical role in feeding TCA metabolism (Fig. 1C).
Loss of autophagy as a metabolic input would be expected to also impact metabolite concentrations. However, concentration changes could be offset by decreased metabolite consumption. For most essential amino acids for which the contribution of autophagy was significant but modest using our pulse-chase approach, we did not detect significant changes in concentrations between Atg7 wild-type and _Atg7-_null cells in starvation (Fig. 1D). For UDP-glucose, serine, pentose phosphate pathway compounds, TCA intermediates, and TCA-associated amino acids, we detected significant depletion in the starved autophagy-deficient cells compared with the starved wild-type cells (Fig. 1D). Thus, autophagy is an important contributor to pentose phosphate pathway and TCA cycle metabolism.
Autophagy does not influence metabolic flux in nutrient-rich conditions
Both glucose and glutamine can contribute carbon to the TCA cycle. We first examined whether autophagy deficiency alters glucose metabolism in nutrient-replete conditions by tracing with [U13C6]-Glc in Atg7 wild-type and _Atg7-_deficient TDCLs. Although glucose uptake and lactate secretion rates in _Atg7_-deficient TDCLs were slightly higher than in wild-type TDCLs (Supplemental Fig. S1A), there was no discernable difference in the kinetic incorporation rate of [U13C6]-Glc carbon into glycolytic intermediates (Supplemental Fig. S1B) or subsequently into TCA cycle intermediates (Supplemental Fig. S1C). As expected, the majority of glycolytic intermediates, including glucose-6-phosphate (G6P), fructose-1,6-biphosphate (FBP), dihydroxyacetone phosphate (DHAP), and 3-phosphoglyceric acid (3PG), were fully labeled within 5 min in both Atg7 wild-type and _Atg7-_deficient TDCLs (Supplemental Fig. S1B). However, <20% of the carbon of TCA cycle intermediates was derived from glucose in both Atg7 wild-type and _Atg7-_deficient TDCLs (Supplemental Fig. S1C,D). Similarly, we found that the contribution of glutamine, which is a major TCA cycle substrate in almost all cultured cancer cells, did not differ depending on autophagy status (Supplemental Fig. S2A–C). Thus, the utilization of glucose and glutamine in _Kras_-driven lung cancer cells is independent of autophagy in nutrient-replete conditions.
Glutamine and glutamate enable survival in starvation
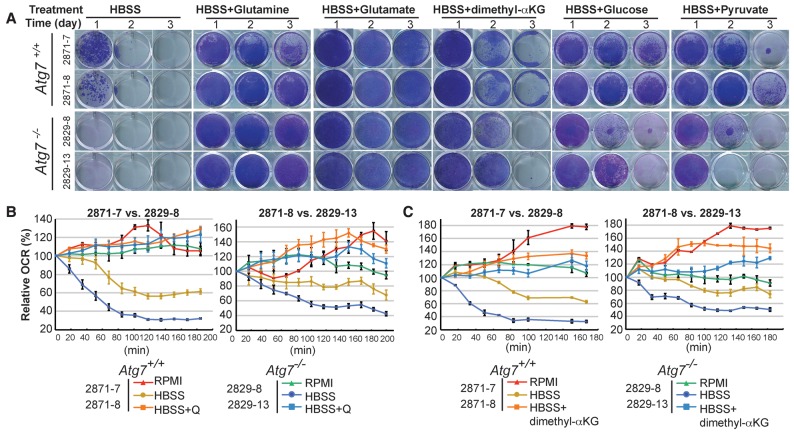
Autophagy deficiency impairs survival of _Kras_- and _Braf_-driven lung tumor cells in starvation, and initial studies pointed to the importance of glutamine in rescuing this survival (Guo et al. 2013; Strohecker and White 2014). To determine which nutrients were essential for _Kras_-driven _Atg7_-deficient tumor cells to survive starvation, we supplemented HBSS with individual amino acids, glucose, membrane-permeable α-KG analog (dimethyl-α-KG), or pyruvate and assessed clonogenic survival. We found that _Atg7_-deficient tumor cells were more sensitive to starvation than wild type, glutamine or glutamate supplementation of HBSS fully rescued the survival of _Atg7_-deficient tumor cells (Fig. 2A), and proline supplementation partially rescued the survival of _Atg7_-null cells, whereas supplementation with other amino acids had no effect on survival (Supplemental Fig. S3A,B). In contrast, glucose and pyruvate only partially rescued the survival of the autophagy-deficient cells (Fig. 2A). Glutamine and glutamate provide nitrogen in addition to TCA carbon. Dimethyl-α-KG provides the carbon skeleton of glutamine and glutamate without any nitrogen. Similar to glucose or pyruvate, which are pure carbon sources, dimethyl-α-KG only partially rescued starved _Atg7_-deficient cells (Fig. 2A).
Figure 2.
Glutamine and its derivatives sustain mitochondria metabolism to support _Kras_-driven TDCL survival in starvation. (A) Clonogenic survival assay of Atg7 wild-type and _Atg7-_deficient cells in HBSS, HBSS supplemented with 2 mM glutamine, 2 mM glutamate, 1 mM dimethyl-α-KG, 2 g/L glucose, or 1 mM sodium pyruvate. Treated cells were recovered for 3 d in RPMI medium at days 1–3 after starvation. (B) Oxygen consumption rate (OCR) of Atg7+/+ and _Atg7_−/− TDCLs in RPMI, HBSS, and HBSS with 2 mM glutamine (Q) supplementation conditions. The error bar indicates ±SEM. n = 4. (C) OCR of Atg7+/+ or _Atg7_−/− TDCLs in RPMI, HBSS, and HBSS with 1 mM dimethyl-α-KG supplementation conditions. The error bar indicates ±SEM. n = 4.
Consistent with impaired substrate supply during starvation, compared with wild type, most autophagy-deficient cells showed less mitochondrial respiration in HBSS (Fig. 2B,C; Supplemental Fig. S4). The decrease in respiration in HBSS was fully rescued by supplementation with either glutamine (Fig. 2B; Supplemental Fig. S4) or dimethyl-α-KG (Fig. 2C), indicating that both glutamine and dimethyl-α-KG provide adequate TCA carbon substrates. Nevertheless, glutamine and glutamate better supported starvation survival. Thus, for survival in starvation, _Atg7_-deficient cells appear to be dependent on both carbon and nitrogen from glutamine or glutamate.
Autophagy sustains redox balance in starvation
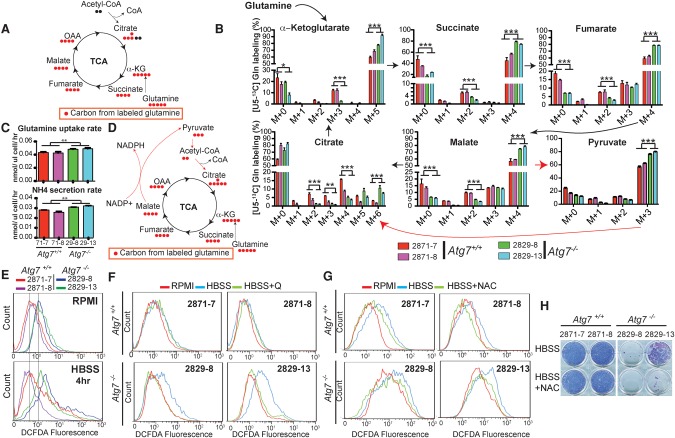
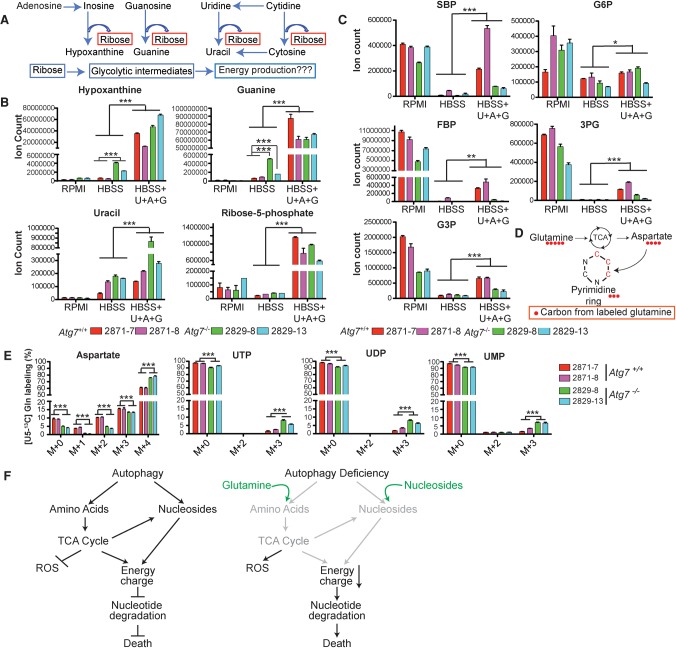
To identify the metabolic pathways maintained by glutamine supplementation during starvation in _Kras_-driven tumor cells, we performed isotope tracer studies using [U13C5]-Gln. As expected (Fig. 3A), glutamine was used to replenish TCA cycle intermediates, as evidenced by fully labeled α-KG (M+5), succinate (M+4), fumarate (M+4), and malate (M+4) (Fig. 3B), with the glutamine uptake rate, associated ammonia excretion rate, and fractional carbon contribution to TCA intermediates significantly higher in _Atg7_-deficient tumor cells compared with wild-type tumor cells (Fig. 3B,C).
Figure 3.
Autophagy maintains the redox state of _Kras_-driven TDCLs in starvation. (A) Schematic of carbon atom (circles) transitions and tracers used to detect glutamine flux to TCA cycle intermediates. (B) [U13C5]-Gln flux to TCA cycle intermediates of _Kras_-driven TDCLs in HBSS. The error bar indicates ±SEM. n = 3. (**) P < 0.01; (***) P < 0.001, _t_-test. (C) Glutamine uptake and ammonium secretion rates of TDCLs in HBSS. The error bar indicates ±SEM. n = 3. (**) P < 0.01, _t_-test. (D) The schematic of carbon atom (circles) transitions and tracers shows the metabolic pathway for citrate (M+6) generation from [U13C5]-Gln. (E) ROS measured by 2′-7′-dichlorodihydrofluorescene diacetate (DCFDA) fluorescence in Atg7 wild-type and _Atg7-_deficient cells in RPMI and 4-h HBSS conditions. (F) ROS measured by DCFDA fluorescence shows that 2 mM glutamine supplementation reduced starvation-induced ROS production in _Atg7-_deficient tumor cells. (G) ROS measured by DCFDA fluorescence shows that 0.5 mM N-acetyl-L-cysteine (NAC) supplementation reduced starvation-induced ROS production in _Atg7−/−-_deficient tumor cells. (H) Clonogenic survival assays show that 0.5 mM NAC supplementation did not rescue _Atg7_-deficient cell death in starvation.
There was a particularly striking increase in M+6 citrate labeling from glutamine (Le et al. 2012) in autophagy-deficient cells (Fig. 3B). Citrate (M+6) can be generated from [U13C5]-glutamine by flux from malate (M+4) to pyruvate (M+3) to acetyl-CoA (M+2), which condenses with oxaloacetic acid (OAA) (M+4) to generate citrate (M+6) (Fig. 3D; Le et al. 2012). Pyruvate (M+3) was significantly higher in _At_g7-deficient tumor cells than in wild type (Fig. 3B). Malic enzyme converts malate to pyruvate and, in so doing, generates NADPH (Fig. 3D; Le et al. 2012). _Kras_-driven pancreatic cancer cells reprogram glutamine metabolism by converting malate to pyruvate to generate NADPH to maintain the cellular redox state (Son et al. 2013). This prompted us to hypothesize that autophagy is required to maintain redox balance in starvation. To test this hypothesis, we first examined the ROS production in _Kras_-driven tumor cells. Compared with Atg7 wild-type tumor cells, _Atg7-_deficient tumor cells had higher basal ROS levels in nutrient-rich conditions, which was further elevated by starvation (Fig. 3E), demonstrating that autophagy suppressed ROS production. Importantly, glutamine supplementation attenuated ROS production in _Atg7_-deficient tumor cells (Fig. 3F). This starvation-induced ROS production was prevented by addition of the ROS scavenger N-acetyl-L-cysteine (NAC) (Fig. 3G). However, NAC supplementation did not rescue the survival of _Atg7_-deficient tumor cells in starvation (Fig. 3H). Thus, while autophagy-derived substrates appear to contribute to antioxidant defense in starving tumor cells, other mechanisms also underlie starvation survival.
Autophagy maintains energy homeostasis
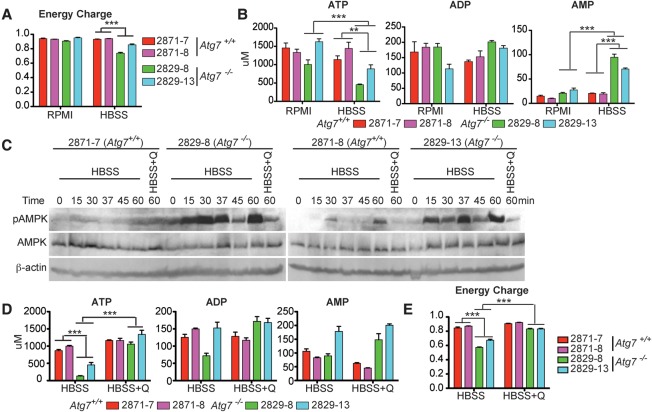
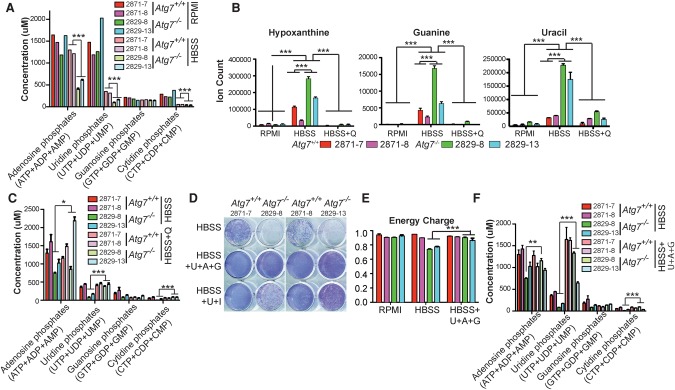
The primary source of ATP in mammals is TCA cycle-driven oxidative phosphorylation. Based on our observations that TCA metabolites and oxidative phosphorylation are both decreased in starving autophagy-deficient cells (Figs. 1D, 2B,C; Supplemental Fig. S4), we hypothesized that autophagy makes a key contribution to energy homeostasis. Adenylate energy charge, defined as (ATP + 0.5 ADP)/(ATP + ADP + AMP), was not influenced by Atg7 in nutrient-rich conditions but was significantly reduced in starved _Atg7_-deficient tumor cells (Fig. 4A). This decreased energy charge in starved _Atg7_-deficient tumor cells was caused by decreased ATP and elevated AMP and was associated with AMPK activation (Fig. 4B,C). Similarly, we observed decreases in GTP, UTP, and CTP and increases in GMP, UMP, and CMP in autophagy-deficient starving cells (Supplemental Fig. S5). Glutamine supplementation restored ATP, reduced AMP, maintained energy charge, and prevented AMPK activation (Fig. 4C–E). These findings suggest that substrates derived from autophagy are required to maintain energy charge in starvation.
Figure 4.
Autophagy sustains energy homeostasis of _Kras_-driven tumor cells in starvation. (A) Energy charge of Atg7+/+ or _Atg7_−/− TDCLs in RPMI and HBSS. The error bar indicates ±SEM (n = 3). (***) P < 0.001, _t_-test. (B) Concentration of adenosine phosphates (ATP, ADP, and AMP) of Atg7 wild-type and _Atg7-_deficient tumor cells in RPMI and HBSS. The error bar indicates ±SEM. n = 3. (**) P < 0.01; (***) P < 0.001, _t_-test. (C) Western blot for pAMPK, total AMPK, and β-actin of Atg7 wild-type and _Atg7-_deficient tumor cells. (D) The level of adenosine phosphates (ATP, ADP, and AMP) in Atg7 wild-type and _Atg7-_deficient tumor cells in HBSS or HBSS supplemented with 2 mM glutamine. The error bar indicates ±SEM. n = 3. (***) P < 0.001, _t_-test. (E) The energy charge of Atg7+/+ and _Atg7_−/− TDCLs in HBSS without or with 2 mM glutamine supplementation. The error bar indicates ±SEM. n = 3. (***) P < 0.001, _t_-test.
Autophagy prevents fatal total nucleotide pool depletion in starvation
In healthy cells, nucleotides are present predominantly (>90%) in the energy-rich triphosphate forms. Based on the substantial decrease in ATP and other triphosphates in starving autophagy-null cells, we hypothesized that total nucleotide pools were depleted. Consistent with prior literature (Park et al. 2016), we found that the total pools of adenosine and uridine nucleotides were substantially larger than the pools of cytidine and guanosine nucleotides (Fig. 5A). The total adenosine, uridine, and cytidine nucleotide pools decreased in starvation in both wild-type and _Atg7_-null cells (intriguingly, the relatively small guanosine nucleotide pools were spared such depletion). The remaining total nucleotide pool in the autophagy-deficient cells was approximately twofold lower than that in wild-type cells (Fig. 5A).
Figure 5.
Autophagy sustains nucleotide pools in starvation. (A) The concentration of nucleoside phosphates in Atg7 wild-type and _Atg7-_deficient tumor cells in RPMI and HBSS (4 h). The error bar indicates ±SEM. n = 3. (***) P < 0.001, _t_-test. (B) The level of nucleotide degradation products in Atg7+/+ and _Atg7_−/− TDCLs in RPMI and HBSS (4 h) without or with glutamine supplementation. The error bar indicates ±SEM. n = 3. (***) P < 0.001, _t_-test. (C) The concentration of nucleoside phosphates in Atg7 wild-type and _Atg7-_deficient tumor cells in HBSS (4 h) without or with 2 mM glutamine supplementation. (D) The clonogenic survival assay shows that nucleoside supplementation (2 mM each) rescued starvation-induced _Atg7_-deficient cell death. Treated cells were recovered for 3 d by replacing HBSS with normal RPMI medium after 1 d of starvation. (U) Uridine; (A) adenosine; (G) guanosine; (I) inosine. (E) The energy charge of Atg7+/+ and _Atg7_−/− TDCLs after HBSS (4 h) without or with nucleoside supplementation (2 mM each). (F) The concentration of nucleoside phosphates in _Kras_-driven tumor cells after HBSS (4 h) without or with nucleoside supplementation (2 mM each).
Because profound nucleotide depletion occurred over a relatively brief starvation interval (4 h), we considered it likely that the pool depletion was driven by nucleotide catabolism. Indeed, there was substantial accumulation of both purine and pyrimidine bases (hypoxanthine, guanine, and uracil) in starved _Atg7_-deficient tumor cells (Fig. 5B). Glutamine supplementation prevented this nucleic acid base accumulation (Fig. 5B) and sustained the pool of nucleotides (Fig. 5C). Because glutamine has a myriad of effects, including on redox state (Fig. 3F), energy charge (Fig. 4A), and nitrogen status (Fig. 3C), we were curious whether substrates that more directly support total nucleotide pools would similarly rescue starving _Atg7_-null tumor cells. We found that a combination of the purine and pyrimidine nucleosides inosine and uridine partially rescued starvation survival, and, more impressively, adenosine, guanosine, and uridine together fully rescued survival in starvation (Fig. 5D). This combination of nucleosides also rescued energy charge (Fig. 5E) and total nucleotide pools (Fig. 5F). Thus, in starvation, autophagy deficiency results in total nucleotide pool depletion, and restoration of nucleotide pools by supplementation with nucleosides rescues survival.
Mechanisms underlying rescue by glutamine or nucleosides
In yeast, adenosine deaminase actively catabolizes AMP during times of energy stress. This transiently buffers energy charge but in the long term can result in fatal nucleotide depletion (Walther et al. 2010). Subsequent catabolism of the resulting IMP can yield ribose phosphate, a potential energy substrate. Other nucleotide monophosphates can be similarly degraded to release ribose phosphate (Fig. 6A; Walther et al. 2010; Xu et al. 2013).
Figure 6.
Mechanisms underlying rescue of starved _Atg7_-deficient tumor cells by glutamine or nucleosides. (A) Schematic of nucleotide degradation and ribose salvage for energy generation. (B) The level of substrates from nucleotide degradation in HBSS (4 h) in the absence or presence of nucleoside supplementation (2 mM each). The error bar indicates ±SEM. n = 3. (***) P < 0.001, _t_-test. (C) Nucleoside addition in HBSS partially rescues levels of glycolytic and PPP intermediates. The error bar indicates ±SEM. n = 3. (*) P < 0.05; (**) P < 0.01; (***) P < 0.0001, _t_-test. (D) The schematic of carbon atom transitions and tracers shows that glutamine contributes three carbons for de novo pyrimidine synthesis. (E) The [U13C5]-Gln tracer study shows increased glutamine flux to aspartate and uridine phosphates (M+3) in _Atg7_-deficient tumor cells compared with wild type in HBSS (4 h). (F) Model depicting how autophagy sustains energy charge and nucleotide pools to enable the survival of _Kras_-driven tumor cells in starvation. Glutamine and nucleoside supplementation (green) is sufficient to maintain energy and nucleotide pools to prevent the death of _Atg7_−/− TDCLs in starvation.
Upon supplementation of the starving cells with nucleosides, in addition to restoration of energy charge and nucleotide pools (Fig. 5E,F), we observed accumulation of nucleic acid bases and partial rescue of pentose phosphate and glycolytic intermediate levels (Fig. 6B,C). While the extent of the rescue was more complete in wild-type than in autophagy-deficient cells, it was significant also in autophagy deficiency (Fig. 6C). However, the supplementation of nucleosides in starvation did not rescue the TCA cycle intermediates (Supplemental Fig. S6A) and the decreased mitochondria respiration (Supplemental Fig. S6B). Thus, in starvation, falling energy charge results in accumulation of nucleotide monophosphates, which are substrates for catabolism into bases plus pentose phosphate intermediates (Fig. 6B,C). Such catabolism can help maintain energy charge by both clearing AMP and providing central carbon intermediates. If the supply of nucleosides is copious (e.g., due to their supplementation), it enables starvation survival even in the absence of autophagy. However, without a persistent nucleoside supply, it results in nucleotide pool depletion and cell death.
A common feature of nucleosides, glutamine, and glutamate—the supplements that rescued starvation survival in autophagy-deficient tumor cells—is that each provides both carbon and nitrogen and can support nucleotide synthesis (Fig. 6D). Indeed, upon glutamine supplementation of starving cells, we observed flux into the pyrimidine precursor aspartate and pyrimidine end product uridine nucleotides (Fig. 6E). These observations further reinforce the critical role of maintenance of nucleotide pools in the survival of autophagy-deficient cells.
Discussion
It has been generally thought that autophagy may maintain cancer cell metabolism by protein and organelle quality control and intracellular recycling functions, although data to support this are lacking. By sequencing the mitochondrial genomes from Atg7 wild-type and _Atg7-_deficient _Kras_-driven lung tumors derived from a GEMM for NSCLC (Guo et al. 2013), we demonstrated for the first time that autophagy suppresses allelic variation in the mitochondrial genome. Since the error rate of sequencing is 0.1% and there are hundreds of mtDNA molecules per cell, we cannot differentiate de novo mutations from allelic drift. The fact that two mitochondrial allele variants were found in two samples at a high frequency in comparison with normal lungs suggests that the variants may be pre-existing at very low allele frequency or that they are mutational hot spots.
Elimination of entire mitochondria by a specialized form of autophagy, mitophagy, is a major mechanism of mitochondrial quality control (Youle and Narendra 2011). Depolarization of the mitochondrial outer membrane is thought to trigger the PINK1/Parkin system to eliminate dysfunctional mitochondria. All of the mitochondrial genome variants that we observed at increased levels in _Atg7_-deficient tumors are not pathogenic and thus are not expected to produce mitochondrial membrane depolarization and trigger mitophagy.
In contrast, Parkin deficiency and the resulting impairment of mitophagy in mice cause mitochondrial dysfunction and pathogenicity but do not increase the levels of mtDNA somatic mutations on the background of the high mutational burden in the mitochondrial genome in Mutator mice (Pickrell et al. 2015). This suggests that failure to cull mitochondrial genome mutations is not the cause of pathogenicity due to loss of Parkin-mediated mitochondrial clearance, similar to what we report here for _Atg7_-deficient tumors. As we did detect increased allelic variation in the mitochondrial genome in _Atg7_-deficient tumors, this could be caused by a more complete block of autophagy with Atg7 compared with Parkin deficiency or by an increased rate of variant detection. Note also that each lung tumor arises from amplification of a clone derived from a single cell, which may increase the sensitivity of mtDNA variant detection. Regardless of the origins of these mitochondrial genome variants in _Atg7_-deficient tumors, they do not represent the underlying cause of metabolic and growth impairment.
Another proposed critical function of autophagy is the degradation and recycling of intracellular components into metabolic pathways to sustain survival in starvation. By stably labeling cellular macromolecules of Atg7 wild-type and _Atg7-_deficient tumor cells followed by a chase with unlabeled medium and then starvation, we were able to demonstrate for the first time that autophagy does recycle macromolecules into metabolism. Interestingly, autophagy-mediated recycling induced by starvation was not uniform and ubiquitous. Isotope tracing demonstrated that autophagy supplies substrates to sustain levels of amino acids, pentose phosphate intermediates, UDP-glucose, glycerol phosphate, and α-KG through intracellular recycling. Among these substrates, glutamine and glutamate were critical to feed the TCA cycle to maintain mitochondrial function, and their exogenous supplementation enables survival of _Atg7_-deficient _Kras_-driven tumor cells in starvation. In addition, Atg7-null cell death was recused by proline supplementation in starvation, likely because proline is metabolized to glutamate. Another interesting finding was the observation of some recycling even in _Atg7_-deleted tumor cells, suggesting that other pathways also contribute to recycling. These could include the proteasome pathway and other autophagy-independent mechanisms to deliver macromolecules to lysosomes for degradation.
Further 13C isotope tracing studies comparing Atg7 wild-type and _Atg7-_deficient tumor cells identified three metabolic pathways distinctly altered by the absence of autophagy. First, glutamine was the major carbon source replenishing mitochondrial TCA cycle intermediates in starvation that was indispensable for mitochondrial respiration and energy homeostasis, eventually preventing nucleotide degradation and cell death. Second, glutamine metabolism was required to sustain redox balance and prevent ROS production. Third, glutamine maintained functional mitochondrial respiration for aspartate synthesis, which was indispensable for de novo nucleotide synthesis. However, defective autophagy limits substrate supply to mitochondria, resulting in increased ROS production, reduced energy charge, and depletion of nucleotides (Fig. 6F).
In proliferating cells, maintaining functional mitochondrial respiration is essential for aspartate synthesis, which is used for protein and nucleotide synthesis (Birsoy et al. 2015; Sullivan et al. 2015). In starvation, _Kras_-driven tumor cells depend on glutamine to maintain aspartate levels. Aspartate was one of the substrates significantly supplied by _Atg7_-dependent recycling in starvation. Although aspartate supplementation did not rescue survival in starvation, the lack of aspartate transport across the plasma membrane prevented aspartate uptake (Birsoy et al. 2015). Thus, cells are generally dependent on de novo aspartate synthesis (Birsoy et al. 2015; Sullivan et al. 2015) and, evidently, aspartate generated by autophagy and recycling of intracellular components (Fig. 1C).
Metabolic reprogramming is a characteristic of cancer cells that enables proliferation and survival. _Kras_-driven tumor cells depend on the autophagy substrate glutamine and its derivatives to sustain mitochondria metabolism to survive starvation. To compensate, _Atg7_-deficient tumor cells significantly increase glutamine uptake and flux to the TCA cycle, de novo nucleotide synthesis, and NADPH generation by up-regulating flux from malate, pyruvate, acetyl-CoA, and citrate for antioxidant defense. _Kras_-driven pancreatic tumor cells are similarly dependent on NADPH generation for redox balance and survival (Son et al. 2013),
In response to energy crisis, high cellular AMP activates AMPK (Faubert et al. 2015). Upon starvation, we found significantly reduced ATP and increased AMP levels correlating with accelerated and more robust AMPK activation in _Atg7_-deficient tumor cells compared with wild type. This is consistent with autophagy sustaining energy homeostasis during starvation.
Total nucleotide pools were dramatically depleted in _Atg7_-deficient tumor cells in starvation, likely due to insufficient substrates for de novo nucleotide synthesis or increased nucleotide degradation. The increased glutamine flux to nucleotide synthesis in _Atg7_-deficient tumor cells suggests that substrate limitation is one reason for nucleotide pool depletion. We also observed that autophagy-deficient cells up-regulated nucleotide degradation. This could be a strategy for Ras-driven cancer cells to remove excess AMP to maintain energy charge, which is critical for metabolic homeostasis (Walther et al. 2010). As a result, increased consumption and defective restoration of nucleotides in autophagy-deficient cells lead to nucleotide pool depletion and lethality. Nucleoside supplementation was sufficient to sustain energy charge and nucleotide pools and rescue starvation-induced _Atg7_-deficient tumor cell death. Therefore, autophagy plays a critical role in preventing fatal nucleotide pool depletion.
Nucleotides are nitrogen-rich molecules, and nucleotide triphosphate levels are maintained through carbon burning. Therefore, long-term support of total nucleotide pools requires both carbon and nitrogen, consistent with the superiority of glutamine and glutamate, as compared with pure carbon substrates, in enabling autophagy-deficient starvation survival.
We observed that nucleoside supplementation partially rescued levels of pentose phosphate intermediates, which feed into the pentose phosphate pathway downstream from the NADPH-producing steps. Thus, the ability of nucleosides to support starvation survival indicates that NADPH production is not limiting in this context.
The autophagy dependency of some tumors has elevated the concept of autophagy inhibition as a cancer therapy (White et al. 2015). Here, we identified how autophagy promotes the survival of _Kras_-driven lung tumor cells during starvation: Autophagy provides substrates for mitochondrial metabolism to avert energy crisis and fatal nucleotide pool depletion (Fig. 6F). Although human cancer cell lines can be selected to tolerate autophagy deficiency (Eng et al. 2016), our findings reveal that up-regulation of glutamine flux to aspartate and nucleotide synthesis is one compensatory adaptation to loss of autophagy. Therefore, inhibiting autophagy, glutamine-mediated metabolic pathways, mitochondrial respiration, or nucleotide synthesis to deplete nucleotide pools is a potential therapeutic strategy for _Kras_-driven lung cancers. As with most other approaches to cancer therapy, however, inherent resistance mechanisms are prevalent. Autophagy has been reported to support the growth of different tumors driven by distinct oncogenic events and originated from different tissues (White et al. 2015). Future work should clarify whether the autophagy-mediated metabolic pathways identified in _Kras_-driven lung cancer can be generalized to other cancers. As autophagy-related proteins are also involved in nonautophagic functions (Subramani and Malhotra 2013), it will be interesting to assess the role of other ATGs in metabolic recycling and cancer metabolism.
Materials and methods
mtDNA isolation, enrichment, ultradeep sequencing, and bioinformatics data analysis
mtDNA isolated from p53_−/−;Kras_ G12D/+;Atg7+/+ or _p53−/−;KrasG12D/+;Atg7_−/− lung tumor samples from GEMMs for NSCLC were used for mitochondria genome sequencing (N = 8 for each genotype) (Guo et al. 2013). Two normal lung tissues obtained from p53 F/F;Kras G12D/+;Atg7+/+ or p53 F/F;Kras G12D/+;Atg7 F/F mice without adenovirus-Cre infection were controls (Guo et al. 2013).
Library preparation was carried out using the Agilent SureSelectXT mouse mitochondrial custom enrichment protocol (Agilent Technologies). The barcoded libraries were assessed on an Agilent Bioanalyzer for proper sizing and then quantified using the KAPA library quantification kit for Illumina sequencing platforms (KAPA BioSystems). Libraries were individually diluted to a 10 nM concentration and then symmetrically pooled for sequencing. Each pool of 16 samples was clustered and sequenced on an Illumina MiSeq instrument using two 150-base-pair (bp) paired-end reads, and ∼2.6 million reads per sample were obtained (Supplemental Table S1). Raw sequencing data were processed using the standard Illumina pipeline. Detailed bioinformatics data analysis is in the Supplemental Material.
Cell culture and reagents
Kras_-driven TDCLs were generated from p53_−/−;Kras G12D/+;Atg7+/+ or p53_−/−;Kras_ G12D/+_;Atg7_−/− lung tumors and cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and 1% sodium bicarbonate at 38.5°C with 8.5% CO2 (Guo et al. 2013).
For starvation assays, HBSS without glucose was used to maintain pH and osmotic balance as well as provide cells with water and essential inorganic ions.
Glutamine, pyruvate, dimethyl-α-KG, uridine, adenosine, guanosine, inosine, and NAC were purchased from Sigma-Aldrich. [U13C5]-Gln, [U13C6]-Glc, and other 13C and 15N uniformly labeled amino acids were purchased from Cambridge Isotope Laboratories.
pAMPK and AMPK antibodies were purchased from Cell Signaling. β-Actin antibody was purchased from Sigma-Aldrich.
Clonogenic survival assays
TDCLs were seeded in 12-well plates in normal growth medium (RPMI). The following day, when TDCLs were at 80% confluence in the well, RPMI was removed and replaced with HBSS for 20 h or for the indicated time. TDCLs were then returned to normal medium for 3 d, fixed with cold methanol for 10 min, and stained with Giemsa (Sigma-Aldrich).
Assessment of oxygen consumption rate (OCR)
OCR of TDCLs was measured using a Seahorse Biosciences extracellular flux analyzer (XF24) as described previously (Guo et al. 2011). Cells were seeded at 3 × 104 cells for Atg7 wild-type TDCLs and 2.5 × 104 cells per well for _Atg7_-deficient TDCLs to accommodate cell volume differences in the XF24 plates overnight prior to XF assay. Real-time OCR measurements were performed in RPMI, HBSS, HBSS with 2 mM glutamine, or 1 mM dimethyl-α-KG for 3 h, and measurements were taken every 15 min. Relative OCR (percentage) was normalized to the 0-min time point.
Glutamine and glucose consumption rate assay
To measure the glutamine and glucose consumption rate in nutrient-rich conditions, TDCLs were plated at 0.5 × 106 cells per 10-cm dish. The following day, fresh RPMI medium was replaced, and medium was collected at 0, 5, 10, 15, 20, and 24 h. The concentration of glucose, glutamine, lactate, and ammonium was determined using a Nova BioProfile flex analyzer. Cell number and volume were measured from each time point.
The glucose/glutamine consumption rate r was obtained by fitting data to the following equation:
where c is the glucose/glutamine concentration in medium, d is doubling time, and _V_0 is the cell volume at time 0.
To measure the glutamine consumption rate in HBSS, TDCLs cells were plated at 2 × 106 cells per 10-cm dish overnight to reach 80% confluence. The following day, medium was replaced with HBSS + 2 mM glutamine and collected at 0, 5, 10, and 15 h. Concentrations of glutamine and ammonium were determined using a Nova BioProfile flex analyzer, and the glutamine consumption rate was calculated as described above.
Assessment of ROS levels
TDCLs were seeded in 12-well plates (0.07 × 106 cells per well) in normal growth medium (RPMI) overnight. The following day, cells were treated with normal growth medium (RPMI), HBSS, HBSS with glutamine supplementation, or HBSS with NAC supplementation for 4 h. Cells were then stained with 10 µM 2′-7′-dichlorodihydrofluorescene diacetate (DCFDA; Molecular Probes) for 20 min and analyzed by flow cytometry (BD influx cell sorter, BD Biosciences).
Metabolomic analysis by LC-MS
Soluble metabolites from Atg7+/+ and _Atg7_−/− TDCLs (two clones each analyzed in triplicate) in RPMI, HBSS, HBSS + 2 mM glutamine, HBSS + 2 g/L glucose, or HBSS + 2 mM nucleosides (for each U, A, and G) for 4 h were quickly washed twice with PBS to exclude floating dead cells and then extracted in 2 mL of −80°C 80:20 methanol:water. Insoluble pellets were re-extracted twice with 0.5 mL each of 80:20 methanol:water for 5 min on dry ice. The supernatants from the three rounds of extraction were combined, dried under N2, and reconstituted in LC-MS-grade water. The samples were analyzed using reversed-phase ion-pairing chromatography coupled by negative mode electrospray ionization to a stand-alone Orbitrap mass spectrometer (Thermo Scientific) scanning from m/z 85−1000 at 1 Hz at 100,000 resolution with LC separation on a Synergy Hydro-RP column (100 mm × 2 mm, 2.5-µm particle size; Phenomenex) using a gradient of solvent A (97:3 H2O/MeOH with 10 mM tributylamine, 15 mM acetic acid) and solvent B (100% MeOH) (Lu et al. 2010). Data were analyzed using Maven software (Melamud et al. 2010). The results are adjusted for natural 13C abundance and enrichment impurity of labeled substrate supplied to cells.
To analyze glucose and glutamine flux into metabolic pathways in steady state, TDCLs were cultured in RPMI medium with [U13C5]-Gln or [U13C6]-Glc for 3 d and then changed to fresh labeled medium 2 h before extraction. Water-soluble metabolites were then extracted and analyzed as described above.
For kinetic incorporation rates of glucose or glutamine into metabolic pathways in normal growth conditions, TDCLs were cultured in RPMI medium with [U13C5]-Gln or [U13C6]-Glc, and soluble metabolites were extracted at the indicated time points and analyzed by a LC-MS system as described above.
To analyze glutamine flux into metabolic pathways in starvation, TDCLs were cultured in HBSS with 2 mM isotope [U13C5]-Gln for 4 h, and soluble metabolites were then extracted and analyzed as described above.
All the pool size data were normalized to the cell volume measured using TPP cell-counting PVC tubes (Midwest Scientific, Inc.). Statistical significance was calculated by Student's _t_-test with false discovery rate (FDR) correction (Atg7 wild-type vs. _Atg7-_deficient TDCLs). In the figures, error bars indicate ±SEM, n = 3, one asterisk indicates P < 0.05, two asterisks indicate P < 0.01, and three asterisks indicate P < 0.001.
Assessment of nucleoside phosphate concentration
Absolute concentrations of nucleotides were determined following a protocol modified from a previous report (Bennett et al. 2008). Specifically, TDCLs were cultured in [U13C6]-Glc for 3 d and then changed to fresh medium for 2 h or HBSS for 4 h. Water-soluble metabolites were extracted and measured as described above with spiked-in known amounts of unlabeled nucleotide standards (Sigma-Aldrich) before drying under N2 flow.
Assessment of autophagy-mediated substrate recycling
_Kras_-driven Atg7 wild-type and _Atg7-_deficient TDCLs were cultured in customized RPMI medium with uniformly 13C-labeled and 15N-labeled amino acids and 13C-labeled glucose for 3 d to label endogenous components, including proteins and metabolites. This was followed by a chase with unlabeled RPMI medium for 3 h to replace labeled with unlabeled metabolites. The cells were then subjected to HBSS starvation for 4 h. Water-soluble metabolites were extracted and examined by LC-MS as described above after 3 d of labeling (R0), after 3 h of chase (R3), and after 4 h of starvation (H4). Evidence of recycling was determined by normalizing H4 to R3 (H4/R3). Statistical significance was calculated by Student's _t_-test with FDR correction by comparing labeling percentages in starvation between Atg7 wild-type and _Atg7_-deficient cells.
Supplementary Material
Supplemental Material
Acknowledgments
We thank the Functional Genomics shared resources of Rutgers Cancer Institute New Jersey for mtDNA extraction and DNA sequencing. This work was supported by National Institutes of Health grants R01 CA130893, R01 CA188096, and R01 CA193970 to E.W.; R01 CA163591 to E.W. and J.D.R.; K22 CA190521 to J.Y.G.; and P30 CA72720 to Rutgers Cancer Institute New Jersey.
Footnotes
Supplemental material is available for this article.
References
- Bennett BD, Yuan J, Kimball EH, Rabinowitz JD. 2008. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat Protoc 3: 1299–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. 2015. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162: 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comerford SA, Huang Z, Du X, Wang Y, Cai L, Witkiewicz AK, Walters H, Tantawy MN, Fu A, Manning HC, et al. 2014. Acetate dependence of tumors. Cell 159: 1591–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. 2007. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci 104: 19345–19350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, et al. 2006. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10: 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X, Kang Y, Rabinowitz JD. 2016. Reversal of cytosolic one-carbon flux compensates for loss of the mitochondrial folate pathway. Cell Metab 23: 1140–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng CH, Wang Z, Tkach D, Toral-Barza L, Ugwonali S, Liu S, Fitzgerald SL, George E, Frias E, Cochran N, et al. 2016. Macroautophagy is dispensable for growth of KRAS mutant tumors and chloroquine efficacy. Proc Natl Acad Sci 113: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TW, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, Miller DM. 2009. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM). Mol Cancer 8: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Kamphorst JJ, Mathew R, Chung MK, White E, Shlomi T, Rabinowitz JD. 2013. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol Syst Biol 9: 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert B, Vincent EE, Poffenberger MC, Jones RG. 2015. The AMP-activated protein kinase (AMPK) and cancer: many faces of a metabolic regulator. Cancer Lett 356: 165–170. [DOI] [PubMed] [Google Scholar]
- Gaglio D, Metallo CM, Gameiro PA, Hiller K, Danna LS, Balestrieri C, Alberghina L, Stephanopoulos G, Chiaradonna F. 2011. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol Syst Biol 7: 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, et al. 2011. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 25: 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, Chen G, Price S, Lu W, Teng X, et al. 2013. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev 27: 1447–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Tolkunov D, Aviv H, Hakimi AA, Yao M, Hsieh JJ, Ganesan CS, White E. 2015. The genomic landscape of renal oncocytoma identifies a metabolic Barrier to tumorigenesis. Cell Rep 13: 1895–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju YS, Alexandrov LB, Gerstung M, Martincorena I, Nik-Zainal S, Ramakrishna M, Davies HR, Papaemmanuil E, Gundem G, Shlien A, et al. 2014. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. Elife 3: e02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, Kalaany NY, Jacks T, Chan CS, Rabinowitz JD, et al. 2014. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov 4: 914–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khutornenko AA, Roudko VV, Chernyak BV, Vartapetian AB, Chumakov PM, Evstafieva AG. 2010. Pyrimidine biosynthesis links mitochondrial respiration to the p53 pathway. Proc Natl Acad Sci 107: 12828–12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169: 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. 2004. The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036. [DOI] [PubMed] [Google Scholar]
- Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, et al. 2012. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab 15: 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy J, Cacheux W, Bara MA, L'Hermitte A, Lepage P, Fraudeau M, Trentesaux C, Lemarchand J, Durand A, Crain AM, et al. 2015. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat Cell Biol 17: 1062–1073. [DOI] [PubMed] [Google Scholar]
- Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. 2010. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone Orbitrap mass spectrometer. Anal Chem 82: 3212–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo T, Pichumani K, Vemireddy V, Hatanpaa KJ, Singh DK, Sirasanagandla S, Nannepaga S, Piccirillo SG, Kovacs Z, Foong C, et al. 2014. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell 159: 1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamud E, Vastag L, Rabinowitz JD. 2010. Metabolomic analysis and visualization engine for LC-MS data. Anal Chem 82: 9818–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B. 2010. Autophagy in mammalian development and differentiation. Nat Cell Biol 12: 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimore GE, Poso AR. 1987. Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu Rev Nutr 7: 539–564. [DOI] [PubMed] [Google Scholar]
- Park JO, Rubin SA, Xu YF, Amador-Noguez D, Fan J, Shlomi T, Rabinowitz JD. 2016. Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nat Chem Biol 12: 482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, Lengrand J, Deshpande V, Selig MK, Ferrone CR, et al. 2015. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature 524: 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell AM, Huang CH, Kennedy SR, Ordureau A, Sideris DP, Hoekstra JG, Harper JW, Youle RJ. 2015. Endogenous Parkin preserves dopaminergic substantia nigral neurons following mitochondrial DNA mutagenic stress. Neuron 87: 371–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, White E. 2010. Autophagy and metabolism. Science 330: 1344–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Tortola L, Perlot T, Wirnsberger G, Novatchkova M, Nitsch R, Sykacek P, Frank L, Schramek D, Komnenovic V, et al. 2014. A dual role for autophagy in a murine model of lung cancer. Nat Commun 5: 3056. [DOI] [PubMed] [Google Scholar]
- Rosenfeldt MT, O'Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, Au A, Rai TS, Zheng L, Ridgway R, et al. 2013. p53 status determines the role of autophagy in pancreatic tumour development. Nature 504: 296–300. [DOI] [PubMed] [Google Scholar]
- Santanam U, Banach-Petrosky W, Abate-Shen C, Shen MM, White E, DiPaola RS. 2016. Atg7 cooperates with Pten loss to drive prostate cancer tumor growth. Genes Dev 30: 399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, Perera RM, Ferrone CR, Mullarky E, Shyh-Chang N, et al. 2013. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496: 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohecker AM, White E. 2014. Targeting mitochondrial metabolism by inhibiting autophagy in BRAF-driven cancers. Cancer Discov 4: 766–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S, Malhotra V. 2013. Non-autophagic roles of autophagy-related proteins. EMBO Rep 14: 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. 2015. Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162: 552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan AS, Baty JW, Dong LF, Bezawork-Geleta A, Endaya B, Goodwin J, Bajzikova M, Kovarova J, Peterka M, Yan B, et al. 2015. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab 21: 81–94. [DOI] [PubMed] [Google Scholar]
- Tardito S, Oudin A, Ahmed SU, Fack F, Keunen O, Zheng L, Miletic H, Sakariassen PO, Weinstock A, Wagner A, et al. 2015. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat Cell Biol 17: 1556–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333: 169–174. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324: 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. 2012. Mitochondria and cancer. Nat Rev Cancer 12: 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T, Novo M, Rossger K, Letisse F, Loret MO, Portais JC, Francois JM. 2010. Control of ATP homeostasis during the respiro-fermentative transition in yeast. Mol Syst Biol 6: 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Wang C, Croce CM, Guan JL. 2014. p62/SQSTM1 synergizes with autophagy for tumor growth in vivo. Genes Dev 28: 1204–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS. 2010. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci 107: 8788–8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. 2012. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 12: 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. 2015. The role for autophagy in cancer. J Clin Invest 125: 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E, Mehnert JM, Chan CS. 2015. Autophagy, metabolism, and cancer. Clin Cancer Res 21: 5037–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong PM, Feng Y, Wang J, Shi R, Jiang X. 2015. Regulation of autophagy by coordinated action of mTORC1 and protein phosphatase 2A. Nat Commun 6: 8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Koh JY, Price S, White E, Mehnert JM. 2015. Atg7 overcomes senescence and promotes growth of BrafV600E-driven melanoma. Cancer Discov 5: 410–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YF, Letisse F, Absalan F, Lu W, Kuznetsova E, Brown G, Caudy AA, Yakunin AF, Broach JR, Rabinowitz JD. 2013. Nucleotide degradation and ribose salvage in yeast. Mol Syst Biol 9: 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell'antonio G, et al. 2011. Pancreatic cancers require autophagy for tumor growth. Genes Dev 25: 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Rajeshkumar NV, Wang X, Yabuuchi S, Alexander BM, Chu GC, Von Hoff DD, Maitra A, Kimmelman AC. 2014. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov 4: 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Fan J, Venneti S, Wan YW, Pawel BR, Zhang J, Finley LW, Lu C, Lindsten T, Cross JR, et al. 2014. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov 4: 1406–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. 2011. Mechanisms of mitophagy. Nat Rev Mol Cell Biol 12: 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. 2007. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol 178: 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong WX, Rabinowitz JD, White E. 2016. Mitochondria and cancer. Mol Cell 61: 667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material