The mouse kinome: Discovery and comparative genomics of all mouse protein kinases (original) (raw)
Abstract
We have determined the full protein kinase (PK) complement (kinome) of mouse. This set of 540 genes includes many novel kinases and corrections or extensions to >150 published sequences. The mouse has orthologs for 510 of the 518 human PKs. Nonorthologous kinases arise only by retrotransposition and gene decay. Orthologous kinase pairs vary in sequence conservation along their length, creating a map of functionally important regions for every kinase pair. Many species-specific sequence inserts exist and are frequently alternatively spliced, allowing for the creation of evolutionary lineage-specific functions. Ninety-seven kinase pseudogenes were found, all distinct from the 107 human kinase pseudogenes. Chromosomal mapping links 163 kinases to mutant phenotypes and unlocks the use of mouse genetics to determine functions of orthologous human kinases.
Eukaryotic protein kinases (PKs) constitute one of the largest of mammalian gene families and are key regulators of a wide variety of conserved cellular processes including cell cycle, cell growth and death, metabolism, transcription, morphology and motility, and differentiation. By adding phosphate groups to substrate proteins, kinases alter the activity, location, and lifetime of a large fraction of proteins and coordinate complex cellular functions. Most PKs belong to a single superfamily containing a conserved eukaryotic PK (ePK) catalytic domain. The remaining, atypical PKs (aPKs), for the most part lack sequence similarity to the ePK domain but are known to have catalytic activity. Fifty-one distinct kinase subfamilies are conserved from yeast to human, reflecting the ancient diversity of kinase functions (1). The recent publication of a comprehensive catalog of 518 human kinases (2) includes scores of novel or poorly understood kinases. The draft mouse genome now provides a key to better understand each human kinase, by comparative analysis of protein and regulatory DNA sequences, and by use of mouse genetics and functional assays to probe the shared functions of mouse kinases and their human orthologs. The detailed comparison of such a large superfamily also casts light on the current state and utility of the draft mouse genome.
The ≈70 million years that separate mouse from human have allowed evolution to test the functional effect of mutations throughout the sequence of every gene. This allows a mapping of functionally important conserved regions within most genes. Initial analysis of the mouse genome (3) showed that within protein coding regions, synonymous nucleotide substitutions (those that do not change protein sequence) occur at a rate [synonymous substitution rate (Ks)] of ≈0.6 substitutions per base between mouse and human orthologs, whereas nonsynonymous substitutions are selectively reduced, to a rate [nonsynonymous substitution rate (Ka)] of ≈0.01–0.1 per base, indicating that most protein sequence changes are rejected by evolution. Accordingly, protein sequence conservation ranges from an average of 71% for regions outside of known domains, to 97% within catalytic domains. Thus, alignment of mouse and human sequences can reveal novel conserved domains and motifs that are important for function, within each kinase protein, as well as conserved DNA features such as promoter elements (4). Here, we determine the sequences of all mouse PKs and compare these genes to their human orthologs to find conserved and lineage-specific sequences and functions.
Methods
Mouse loci orthologous to human PKs were identified by blast search of human protein sequences against the draft mouse genome (Mouse Genome Sequencing Consortium, February 2003 Arachne assembly). The surrounding genomic sequence was subjected to genewise homology-based gene prediction, with the orthologous human kinase. genewise predictions were confirmed, corrected, and extended by aligning EST/cDNA sequences to the genomic sequence and by blast followed by manual inspection. EST/cDNA sequences were from the mouse section of dbEST (GenBank release 134), Incyte's ZooSeq EST database (September 2002), in-house EST and cDNA databases, and a mouse cDNA subset of GenBank (March 1, 2003, download). Additional kinase domains were predicted with ePK and aPK kinase domain Hidden Markov Model (HMM) profiles searched against a six-frame translation of the mouse genomic, EST, and cDNA sequences. Novel HMM matches were mapped to a genomic locus and extended as above.
Mouse kinases were mapped to chromosomal bands by using data from Ensembl (www.ensembl.org). These bands were assigned by aligning band sizes to nucleotide positions, giving a rough approximation of the true locus. Detailed methods are found in Supporting Text, which is published as supporting information on the PNAS web site.
Results
Cataloging and Extending the Mouse Kinome. We searched all available mouse sequences (public mouse genomic and expressed sequences and Incyte and in-house mouse ESTs) for orthologs of every human PK. We then searched for additional PKs, using Hidden Markov Model profiles of the ePK domain and for aPK families (2) and used a series of gene prediction tools and expressed sequences to extend initial predictions to full length genes. We identified 540 putative PK genes and 97 pseudogenes (Tables 2–8, which are published as supporting information on the PNAS web site).
Our use of multiple sequence sources, multiple prediction methods, homology to the human kinome, and manual curation enabled the discovery of previously unreported mouse kinase genes and the extension or correction of >150 known kinase sequences. We compared our protein sequences with the closest match in public cloned proteins (GenPept), reference sequences (RefSeq), and three sets of predictions from genomic sequence (Fig. 1). twinscan (5) is an extension of the ab initio methods used in genscan, which adds conservation of mouse–human genomic sequence in its predictions. Accordingly, it has increased prediction accuracy. Ensembl predictions add information from known protein sequences and so greatly improve prediction quality for cloned genes. RefSeq lacks ≈190 kinase genes (35%) but has high quality sequences for those it does contain. GenPept outscores all prediction methods and contains at least fragments for most kinase genes but has at best partial or incorrect sequences for almost half of the kinome (256 sequences).
Fig. 1.
Comparison of our kinase protein sequences with those of public databases. Each line indicates the number of matching sequences, at a given allowed sequence divergence; as the stringency is loosened, more genes are matched. The SUGEN line is set at 540, the total number of kinase genes. (Inset) The number of perfect matches, very similar matches (<2% difference), and unmatched (>98% difference).
Gene prediction methods are hampered by the incompleteness of the genomic sequence. Using orthologous human kinases as a query, we found that 138 kinases (25.6%) could not be fully mapped to the draft mouse genome assembly, missing an average of 7% of their protein sequence. Thirty-eight kinases contained multiple genomic gaps (Table 3). We used mouse ESTs and cDNAs to bridge most of these gaps and are left with 23 incomplete kinases, which still lack an average of 10% of their sequence as compared with their human homologs.
Comparison of Human and Mouse Kinomes. Almost all mouse and human kinases exist as orthologous pairs, with similar functions in both organisms. There are 510 such orthologous pairs. Eight kinases are found only in human and 30 only in mouse, of which 25 are from a single subfamily of microtubule affinity-regulating kinase (MARK) kinases (Tables 1 and 2). This is in broad agreement with recent estimates from automated gene predictions that >96% of mouse and human genes are orthologous (3, 6–8). Excluding the MARKs, the 13 genes found in only one kinome can be traced to gene loss (eight genes), gene duplication by retrotransposition (four genes), and likely incomplete genome sequence (one gene).
Table 1. Lineage-specific kinases in mouse and human.
| Gene | Found only in | Introns | Rat ortholog | Other orthologs | Closest paralog (%) | Notes |
|---|---|---|---|---|---|---|
| Retrotransposed copies | ||||||
| CK1α2 | Human | No | No | Chimp | CK1α (91) | Ka/Ks = 0.0463/0.2656 = 0.0463 |
| TAF1L | Human | No | No | Primate | TAF1 (92) | Ka/Ks = 0.0217/0.0814 = 0.2666 |
| PKACγ | Human | No | No | Primate | PKACα (83) | Ka/Ks = 0.0829/0.4658 = 0.1880 |
| CK2α1-rs | Mouse | No | No | No | CK2α1 (99) | Ka/Ks = 0.0011/0.0037 = 0.2972 |
| A6-rs | Mouse | No | No | No | A6 (96) | Ka/Ks = 0.0150/0.0689 = 0.217 |
| NDR2-rs | Mouse | No | No | No | NDR2 (97) | Ka/Ks = 0.0111/0.0233 = 0.4764 |
| Gene loss | ||||||
| PSKH2 | Human | Yes | No | Chimp | PSKH1 (≈70) | |
| DRAK1 | Human | Yes | No | Dog, rabbit | DRAK2 (54) | |
| GPRK7 | Human | Yes | No | Squirrel, pig, cow, fishes | GPRK4 (47) | |
| CDK3 | Human | Yes | Pseudogene | Chimp | CDK2 (76) | Pseudogene in mouse, rat |
| KSGC | Mouse | Yes | Yes | No | ANPa (35) | Pseudogene in human |
| PLK5 | Mouse | Yes | Yes | No | PLK2 (37) | Pseudogene in human |
| CYGX | Mouse | Yes | Yes | No | CYGF (51) | Pseudogene in human |
| TSSK5 | Mouse | Yes | Yes | No | TSSK2 (36) | Pseudogene in human |
| Other | ||||||
| PRKY | Human | Yes | No | No | PRKX (92) | Mouse Chr Y not yet sequenced |
| MARKs | Mouse | Some | Some | No | Varied | Twenty-five genes; fast evolving |
Human-Specific Kinases. Orthologs of eight human kinases were absent from mouse. The mouse Y chromosome has not been fully sequenced, which explains the lack of an ortholog for the single human Y chromosome kinase, PRKY. The absence of the other seven kinases is probably not due to incomplete genomic sequence, because they are also absent from EST and cDNA databases and from the draft rat genome. Three of these kinases (CK1α2, PKACγ, and TAF1L) are intronless almost identical copies of other kinases expressed selectively in testis and so are probably retrotransposed copies of these genes (Table 1). Unlike most retrotransposed copies that degenerate into pseudogenes, these sequences are expressed, and sequence analysis indicates that they continue to be under functional pressures, with a low ratio of nonsynonymous to synonymous substitutions (Ka/Ks = 0.05–0.27) relative to their parental genes. Intact homologs of both TAF1L and PKACγ are present in some primate genomes, and their origins have been estimated at 25–40 million years ago (9, 10).
The last four kinases (CDK3, GPRK7, DRAK1, and PSKH2) were probably present in the common ancestor of human and mouse and subsequently lost from the mouse lineage (Table 1). CDK3 survives as a transcribed mouse pseudogene with a single stop within the kinase domain and other nonconservative substitutions and is also a pseudogene in rat, although it may exist as a functional gene in other mouse species (11). GPRK7 is present in a variety of organisms, including squirrel, but is not found in mouse or rat genomes, indicating its loss within this rodent sublineage ≈40 million years ago (12). Similarly, DRAK1 is absent from rat but found in rabbit and was likely lost from the rodent lineage. PSKH2 has been seen only in human and chimp, but its degree of divergence from PSKH1 indicates that the duplication that created these genes happened early in vertebrate evolution, and that one copy was later lost.
Mouse-Specific Kinases. Orthologs of four mouse kinases are found only as pseudogenes in the human genome. These are the receptor guanylyl cyclases CYGX and KSGC, the cell cycle kinase PLK5, and the testis-specific kinase TSSK5. Three other mouse-specific kinase sequences (A6-rs, NDR2-rs, and CK2α1-rs) are recently retrotransposed copies of other genes. All three have a slight bias for synonymous substitutions relative to their parents, but their divergence is too low to show whether their sequences are under selective pressure (Table 1). Only CK2α1-rs is known to be expressed. In agreement with the human kinome analysis, A6-rs and NDR2-rs are classified as tentative pseudogenes. The 25 mouse-specific MARK kinases will be dealt with separately below.
Mouse–Human Comparisons Reveal Functionally Important Sequences. Protein sequence alignment of orthologous kinase pairs shows a wide variation in local sequence conservation. Highly conserved regions map to known domains or reveal previously unknown conserved regions of likely functional importance. For instance, the four Wnk kinases are long (≈1,200–2,300 aa) proteins that have little sequence similarity outside their kinase domain. Pairwise alignment of mouse and human Wnk2 and Wnk3 identifies several previously undescribed highly conserved domains, separated by poorly conserved sequences, occurring in similar regions within both ortholog pairs (Fig. 2). Addition of mouse sequence also improves the quality of family sequence alignments, improving the detection of shorter motifs that are conserved among all family members. Similar alignments for all ortholog pairs are available through KinBase (http://kinase.com).
Fig. 2.
Schematic of Wnk kinase protein sequences, with the kinase domain (KD) boxed in red and previously undescribed blocks of sequence highly conserved between mouse and human boxed in black. Percentage ortholog identity is given for each block and interblock region of lesser conservation.
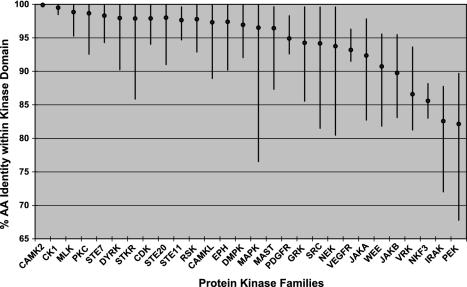
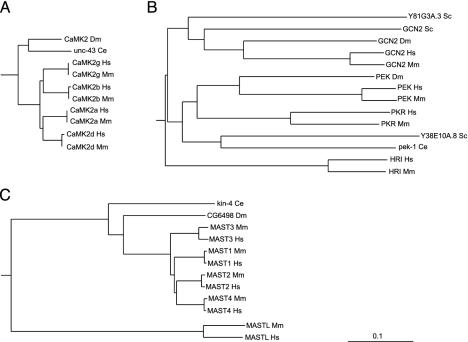
Even within conserved domains, these comparisons are informative. The basic structural constraints of the ePK domain are common across all kinases, yet there are marked differences in the degree of conservation in different kinase families. Orthologous ePK domains are on average 95% identical, similar to that of other known domains, but some are as low as 65%, and 49 pairs are identical across the full 261-residue domain, indicating strong functional pressure throughout the domain. (Table 2). This variability is clearly family-dependent (Fig. 3; Table 4). For instance, of the four calmodulin-dependent kinase 2 (CaMK2) family domain pairs, two are identical, and the other two differ by a single residue, an average difference of only 0.2%. Collectively, this indicates that changes in almost any amino acid within the domain destroy some function and have been eliminated by evolution. CaMK2 orthologs in worm and fly are also highly conserved, indicating that this family has been constrained for hundreds of millions of years (Fig. 4_A_). This conservation may be explained by the unusual CaMK2 structure, which forms tetradecameric multimers in which subunit packing may place strong structural limitations on sequence (13). At the other extreme, PEK/PKR family domain pairs are 68–90% identical, with even lower conservation in invertebrates, indicating that the core functions of this family of eIF2α kinases do not greatly constrain the domain sequence (Fig. 4_B_). Other families, such as microtubule-associated serine/threonine kinase (MAST) (Fig. 4_C_), have variable conservation: the MAST-like (MASTL) kinase domain has diverged greatly from the rest of the family from its birth early in vertebrate evolution, and ortholog comparison shows that this divergence has continued since the mouse–human split. This gene is likely to have changed or lost a function relative to other MAST family members.
Fig. 3.
Conservation within orthologous kinase domains is family-dependent. Diamonds indicate the mean kinase domain identity within selected families; bars indicate the range (data from Table 4).
Fig. 4.
ePK domain conservation is family-dependent. (A) The calmodulin-dependent kinase 2 family is highly conserved, with zero to one amino acid changes between mouse and human orthologs and 82–85% sequence identity between human and invertebrate orthologs. (B) Kinase domains of the PEK family are poorly conserved between human and mouse (68–90% identity) and highly divergent from invertebrate orthologs (29–43% identity). (C) Divergent members of well conserved families may indicate changed or lost function, as in MAST-like, a highly divergent member of the MAST family, which also displays high divergence between mouse and human orthologs. (Bar = 0.1 substitutions per site.) Hs, Homo sapiens; Mm, Mus musculus; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Sc, Saccharomyces cerevisiae.
Although most differences between orthologs are due to amino acid substitutions, many proteins contain substantial inserts or deletions (indels) between orthologs, which may account for much of their functional differences between species. One hundred sixty-two of the 510 ortholog pairs (32%) contain indels of four or more amino acids (92 have novel insertions in human, 40 in mouse and 30 in both; Table 5). These indels are products of unique exons, alternative splice sites, and insertion of unique sequence within exons. They range from 1 to 151 aa in length and are all supported by expressed sequences. Twenty-three more genes were initially thought to have mouse-specific inserts, but analyses of new human cDNA or genomic sequence led to the discovery of equivalent human sequence, leading to the extension of those human kinases.
Indels account for 18% of the sequence difference between the two kinomes, but as blocks of unique sequence, their functional impact may be much greater. For example, human and mouse DCAMKL1 differ by just 11 amino acid substitutions, but the mouse protein also contains a unique 16 amino acid alternatively spliced module in 7 of 22 ESTs. The protein sequence of the module is perfectly conserved in rat, but the equivalent human genomic sequence has greatly degenerated, and the module is absent from all human ESTs. Many other indels are alternatively spliced, allowing evolution to experiment with new protein isoforms, while retaining the function of the original form (14).
Catalytically Inactive Kinases. Several kinases are known to lack catalytic function and instead serve as scaffolds or kinase substrates. We previously reported (2) that 50 human ePK domains might be inactive, due to the lack of one of three highly conserved catalytic residues (Lys-30, Asp-125, or Asp-143). The mouse kinome shows an almost identical set of predicted inactive kinases (Table 6), with only one difference among the orthologous pairs, the EGF receptor family member ErbB3. Human ErbB3 lacks Asp-125 (the catalytic base), has been shown to be catalytically inactive, and instead dimerizes with other EGF receptor family members, acting as a substrate and docking protein (15). Asp-125 is also absent in ErbB3 sequences from dog, pufferfish, and zebrafish but is present in both mouse and rat ErbB3, indicating a secondary reactivation of this residue in rodents after its loss early in vertebrate development. However, mouse ErbB3 is probably still inactive, because it lacks another key conserved residue (Glu-46), and attempts to cure human ErbB3 by replacement of Asp-125 and Glu-46 have failed to restore catalytic activity (16). All of the mouse-specific kinases are predicted to be active, except for the guanylate cyclases (all members of this family are inactive), and three MARK genes that are potential pseudogenes. This strong conservation of inactivity between kinomes supports a conserved noncatalytic function for many kinases.
MARK Kinases. The MARK family is by far the most divergent between mouse and human kinomes. Human, mouse, and rat contain four highly conserved members (MARK1–4), which have a range of functions, including microtubule stability, spermatogenesis, cell polarity, cell cycle control, and Wnt and mitogen-activated protein kinase signaling (17–21). We have identified at least 51 additional mouse-specific MARK genes and pseudogenes. The exact number is unclear, because many are near-identical copies of each other and/or are in poorly assembled regions of the genome, making it difficult to distinguish close paralogs from genome assembly errors or allelic variation. We merged MARK sequences with >99.5% nucleotide identity. Twenty-six MARKs have ORFs interrupted by confirmed frameshifts or stops and are probably pseudogenes. The other 25 generally have long ORFs and selective conservation of family-specific residues and are classified as functional genes, although 22 are intronless and some may be retrotransposed young pseudogenes.
Eleven novel MARKs are seen in EST databases. Their expression is predominantly in testis, although MARKmE1 has been found only in ESTs from two-cell stage embryos, suggesting that it may act in determination of early embryonic polarity, as do the MARK family par-1 genes of fly and worm (22, 23).
Pseudogenes. We found 97 ePK pseudogenes, similar to the human count of 106 (Table 7). All have kinase domains and ORFs that are fragmentary or are interrupted by stops or frameshifts. Thirteen have been found in EST databases, although some may be due to genomic contamination; only seven have multiple ESTs. Expressed pseudogenes may confer some function as RNA or truncated protein, although most probably lack function. Almost all are recent copies of functional mouse kinases; only CDK3ps results from the degeneration of a previously functional gene (Table 1). Most pseudogenes cluster in a few families: MARK (26 pseudogenes), CDK (15 pseudogenes), Aurora (11 pseudogenes), and Erk (6 pseudogenes). This is distinct from the human distribution (2), even within a family: all three human Erk pseudogenes are copies of Erk3, whereas all six mouse Erk pseudogenes are copies of Erk1. No orthology or synteny was seen between human and mouse pseudogenes, because any common pseudogenes would have degenerated beyond recognition within each lineage since their divergence >70 million years ago.
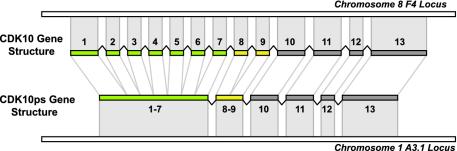
Only two pseudogenes contain introns, and neither is a genomic duplication of functional genes: CDK3ps is a degenerate functional gene, and CDK10ps is a retrotransposed copy of a partially spliced CDK10 mRNA, in which 7 of 12 introns have been spliced out (Fig. 5). Fifteen kinase pseudogenes have gaps in their genomic alignment that are not introns but rather inserts of repetitive DNA; the remaining 80 pseudogenes are intronless and derive from fully processed transcripts or genomic duplications of single exons.
Fig. 5.
CDK10ps is a retrotransposed pseudogene copy of CDK10, which was partially processed, losing the first six and the eighth introns before its reintegration into the genome.
Mouse Kinome Genetics. We mapped the mouse kinome to The Jackson Laboratory's database of mouse mutations (Mouse Genome Database, www.informatics.jax.org, July 2003) (Table 8). One hundred sixty-three kinases (30%) have recorded mutation data, of which 157 have targeted knockouts or transgenic mice, and 21 have mutations due to random mutagenesis. Many other kinases map close to phenotypes whose molecular defect has not yet been determined. These phenotypes cover a wide range of diseases and functions, ranging from kidney, liver, neurological, cardiovascular, circulatory, skeletal, urogenital, respiratory, reproductive, immune system, and vision disorders to tumorigenesis and survival phenotypes. Several ongoing mutagenesis projects promise to saturate the mouse kinome with mutations and shed light on the functions and disease association of almost all human kinases.
Discussion
We have predicted the mouse complement of 540 PKs by using a combination of data sources, prediction methods, and comparison with the human kinome. Careful curation enabled the discovery or extension of >150 kinase sequences. Even though the mouse genome contains fragments of >99% of known mouse genes, we find that the genome is incomplete for ≈25% of kinases, highlighting the importance of finishing the current draft sequence. Ortholog comparison allowed the discovery of longer splice isoforms for 23 human kinases, and comparative genomic analysis is likely to detect additional exons in both species. A recent report (24) gives an alternative count of 561 mouse kinases. This shares 484 sequences with our catalog and also includes 10 pseudogenes, 24 duplicates, and 43 genes that have no meaningful similarity to known kinases or the ePK kinase domain, although some contain a ProSite kinase domain motif (PS00107), which is known to have many false positive hits (see http://kinase.com/mouse for details of comparison). A comparison to UniProt sequences annotated with InterPro kinase (IPR000719) annotations finds 969 matches to 442 ePK sequences and one aPK (Table 2). InterPro also annotates 5 kinase pseudogenes and 29 genes with no similarity either to known kinases or to other mouse sequences.
Five hundred and ten kinases have 1:1 orthology between mouse and human, confirming mouse as a model system for studying human kinases. This sequence and map information coupled with ongoing random and targeted mutagenesis projects will lead to mutations in most mouse kinases within a few years and may allow the development of probes to assay kinase expression, localization, and activation in a wide variety of disease-related and other mutant mice. Sequence conservation between orthologous pairs reveals novel functional domains and motifs; conversely, several kinases have unique regions found only in one species, which may encode species-specific functions. Both genomes have lost four kinases present in their common ancestor. Surprisingly, all new kinases within each lineage are derived from retrotransposition rather than genomic duplication. The functions of lineage-specific genes, where known, often relate to lineage-specific biological functions. CYGX is implicated in olfaction (25), and its loss in human parallels that of other olfactory genes. Similarly, the loss in mouse of GPRK7, a potential cone opsin kinase, correlates with the reduced color vision of rodents (26). Cell cycle-related kinases (CDK3 and PLK5) are less likely candidates for gene loss and may indicate redundancy in cell cycle machinery. Finally, several lineage-specific kinases are expressed in testis or related to reproduction (TSSK5, TAF1L, PKACγ, CK1α2, and MARKs) whose genes are known to evolve quickly (27).
The dramatic expansion of mouse MARK genes implies rapid evolution and possible viral or nongenomic origin. The draft rat genome contains >80 MARK sequences (not shown), but only two are clearly orthologous to novel mouse MARKs (Ks ≈ 0.2–0.25), and most have <75% protein sequence identity to their closest mouse homologs. Many MARKs are flanked by long interspersed nuclear elements (LINE)-like elements in both genomes, suggesting that they derive from mobile genetic elements. Many map to poorly assembled or sequenced regions of the genome; the final completion of these genomes will help explain their evolutionary dynamics. A possible role for the expansion in segregation distortion is suggested by the mouse t haplotype, a region containing a number of distorter loci that adversely affect the motility of all sperm and a responder locus (a duplicated MARK gene called SmokTcr), which protects t haplotype sperm from this effect, providing a selective advantage for the t haplotype in the progeny of heterozygous mice (28). If MARKs can provide a competitive advantage for sperm, this could provide a rationale for the expansion of this family. There are 29 human MARK pseudogenes, suggesting that a separate MARK expansion once occurred in the human lineage but is no longer functional.
The central role of kinases in cellular regulation places them at critical points in many disease pathways, and their conserved catalytic cleft has made them attractive targets for drug therapy (29). The mouse kinome will facilitate the generation of mutants to study kinase functions and the effects of ablating kinase activity and will enhance the exploration of the roles of all kinases in mouse models of human diseases. Current large-scale random and targeted mutagenesis projects (30) promise to eventually provide mutations in almost all PKs and establish models for many human disorders. The kinome catalog will also further large-scale experimental analyses, such as the kinase cloning and localization efforts of the Alliance for Cell Signaling (31). A detailed analysis of the ortholog pairs, sequences, chromosome mapping, and other aspects of the kinome is available online through KinBase at http://kinase.com.
Acknowledgments
We thank Paul Szauter of The Jackson Laboratory for help with their mouse mutation database, the mouse and human genome projects for sequences and annotation, the Rat Genome Sequencing Consortium for access to unpublished genomic sequence (Rnor3.1), and SUGEN colleagues for support and critical review. This work was supported entirely by SUGEN, Inc. T.H. is a Frank and Else Schilling American Cancer Society Research Professor.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ePK, eukaryotic PK; aPK, atypical PK; MARK, microtubule affinity-regulating kinase; Ka, nonsynonomous substitution rate; Ks, synonomous substitution rate; MAST, microtubule-associated serine/threonine kinase.
Data deposition: The sequences reported in this paper have been deposited at kinase.com.
References
- 1.Manning, G., Plowman, G. D., Hunter, T. & Sudarsanam, S. (2002) Trends Biochem. Sci. 27**,** 514–520. [DOI] [PubMed] [Google Scholar]
- 2.Manning, G., Whyte, D. B., Martinez, R., Hunter, T. & Sudarsanam, S. (2002) Science 298**,** 1912–1934. [DOI] [PubMed] [Google Scholar]
- 3.Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. (2002) Nature 420**,** 520–562. [DOI] [PubMed] [Google Scholar]
- 4.Cheremushkin, E. & Kel, A. (2003) Pac. Symp. Biocomput. 291–302. [DOI] [PubMed]
- 5.Korf, I., Flicek, P., Duan, D. & Brent, M. R. (2001) Bioinformatics 17**,** Suppl. 1, S140–S148. [DOI] [PubMed] [Google Scholar]
- 6.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409**,** 860–921. [DOI] [PubMed] [Google Scholar]
- 7.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291**,** 1304–1351. [DOI] [PubMed] [Google Scholar]
- 8.Mural, R. J., Adams, M. D., Myers, E. W., Smith, H. O., Miklos, G. L., Wides, R., Halpern, A., Li, P. W., Sutton, G. G., Nadeau, J., et al. (2002) Science 296**,** 1661–1671. [DOI] [PubMed] [Google Scholar]
- 9.Reinton, N., Haugen, T. B., Orstavik, S., Skalhegg, B. S., Hansson, V., Jahnsen, T. & Tasken, K. (1998) Genomics 49**,** 290–297. [DOI] [PubMed] [Google Scholar]
- 10.Wang, P. J. & Page, D. C. (2002) Hum. Mol. Genet. 11**,** 2341–2346. [DOI] [PubMed] [Google Scholar]
- 11.Ye, X., Zhu, C. & Harper, J. W. (2001) Proc. Natl. Acad. Sci. USA 98**,** 1682–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar, S. & Hedges, S. B. (1998) Nature 392**,** 917–920. [DOI] [PubMed] [Google Scholar]
- 13.Hoelz, A., Nairn, A. C. & Kuriyan, J. (2003) Mol. Cell 11**,** 1241–1251. [DOI] [PubMed] [Google Scholar]
- 14.Modrek, B. & Lee, C. J. (2003) Nat. Genet. 34**,** 177–180. [DOI] [PubMed] [Google Scholar]
- 15.Riese, D. J., II, van Raaij, T. M., Plowman, G. D., Andrews, G. C. & Stern, D. F. (1995) Mol. Cell. Biol. 15**,** 5770–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prigent, S. A. & Gullick, W. J. (1994) EMBO J. 13**,** 2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller, J., Ory, S., Copeland, T., Piwnica-Worms, H. & Morrison, D. K. (2001) Mol. Cell 8**,** 983–993. [DOI] [PubMed] [Google Scholar]
- 18.Peng, C. Y., Graves, P. R., Ogg, S., Thoma, R. S., Byrnes, M. J., III, Wu, Z., Stephenson, M. T. & Piwnica-Worms, H. (1998) Cell Growth Differ. 9**,** 197–208. [PubMed] [Google Scholar]
- 19.Biernat, J., Wu, Y. Z., Timm, T., Zheng-Fischhofer, Q., Mandelkow, E., Meijer, L. & Mandelkow, E. M. (2002) Mol. Biol. Cell 13**,** 4013–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drewes, G., Ebneth, A. & Mandelkow, E. M. (1998) Trends Biochem. Sci. 23**,** 307–311. [DOI] [PubMed] [Google Scholar]
- 21.Sun, T. Q., Lu, B., Feng, J. J., Reinhard, C., Jan, Y. N., Fantl, W. J. & Williams, L. T. (2001) Nat. Cell Biol. 3**,** 628–636. [DOI] [PubMed] [Google Scholar]
- 22.Pellettieri, J. & Seydoux, G. (2002) Science 298**,** 1946–1950. [DOI] [PubMed] [Google Scholar]
- 23.Zernicka-Goetz, M. (2002) Development (Cambridge, U.K.) 129**,** 815–829. [DOI] [PubMed] [Google Scholar]
- 24.Forrest, A. R., Ravasi, T., Taylor, D., Huber, T., Hume, D. A. & Grimmond, S. (2003) Genome Res. 13**,** 1443–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fulle, H. J., Vassar, R., Foster, D. C., Yang, R. B., Axel, R. & Garbers, D. L. (1995) Proc. Natl. Acad. Sci. USA 92**,** 3571–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiss, E. R., Ducceschi, M. H., Horner, T. J., Li, A., Craft, C. M. & Osawa, S. (2001) J. Neurosci. 21**,** 9175–9184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torgerson, D. G., Kulathinal, R. J. & Singh, R. S. (2002) Mol. Biol. Evol. 19**,** 1973–1980. [DOI] [PubMed] [Google Scholar]
- 28.Herrmann, B. G., Koschorz, B., Wertz, K., McLaughlin, K. J. & Kispert, A. (1999) Nature 402**,** 141–146. [DOI] [PubMed] [Google Scholar]
- 29.Cohen, P. (2002) Nat. Rev. Drug Discov. 1**,** 309–315. [DOI] [PubMed] [Google Scholar]
- 30.Rossant, J. & McKerlie, C. (2001) Trends Mol. Med. 7**,** 502–507. [DOI] [PubMed] [Google Scholar]
- 31.Gilman, A. G., Simon, M. I., Bourne, H. R., Harris, B. A., Long, R., Ross, E. M., Stull, J. T., Taussig, R., Arkin, A. P., Cobb, M. H., et al. (2002) Nature 420**,** 703–706. [DOI] [PubMed] [Google Scholar]