Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity (original) (raw)
Abstract
RNA interference is carried out by the small double-stranded RNA-induced silencing complex (RISC). The RISC-bound small RNA guides the RISC complex to identify and cleave mRNAs with complementary sequences. The proteins that make up the RISC complex and cleave mRNA have not been unequivocally defined. Here, we report the biochemical purification of RISC activity to homogeneity from Drosophila Schnieder 2 cell extracts. Argonaute 2 (Ago-2) is the sole protein component present in the purified, functional RISC. By using a bioinformatics method that combines sequence-profile analysis with predicted protein secondary structure, we found homology between the PIWI domain of Ago-2 and endonuclease V and identified potential active-site amino acid residues within the PIWI domain of Ago-2.
RNA interference is the phenomenon in which long double-stranded (ds)RNA is able to silence cognate gene expression (1). First, the dsRNA is processed into small interfering RNA (siRNA) by a Dicer enzyme (2-5). In Drosophila cells, siRNA is retained by Dicer-2 and an associated protein, called R2D2. The Dicer-2-R2D2 complex is then able to facilitate the loading of siRNA onto a complex known as the RNA-induced silencing complex (RISC) (2). The RISC-bound siRNA strand acts as a guide to identify mRNA targets with complementary sequence for nucleolytic cleavage (6). RISC activity is defined simply as siRNA-guided, site-specific cleavage of an mRNA target.
The guide RNA in RISC is held by the PAZ domain of Argonaute 2 (Ago-2). This protein was recognized in Caenorhabditis elegans when mutants RNA interference-defective were found (7). It was also found to correlate with partially purified RISC activity from Drosophila and human cells (6, 8). Furthermore, siRNAs were demonstrated to copurify with Ago-containing complexes (6). Finally, the PAZ domain of Ago-2 was determined to be an siRNA-binding domain by structural studies (9-13).
Several characteristic features of RISC nucleolytic activity have been determined. RISC cleaves the target mRNA strand between positions 10 and 11, counted from the 5′ to 3′ end of the guide RNA (14). These cleavage products have a 5′ phosphate and a 3′ hydroxyl group (15, 16). Finally, RISC activity is magnesium-dependent (16). Although a long list of proteins have been proposed to be RISC components, none has been identified that accounts for the nuclease activity of RISC.
In this study, we used a biochemical fractionation approach to purify the RISC nuclease activity to homogeneity from Drosophila Schnieder 2 (S2) cells. We found that RISC activity is generated by a single protein, Ago-2. We then used metabasic (18) to predict that the PIWI domain of Ago-2 is similar to the endonuclease V in sequence and secondary structure, suggesting that the PIWI domain provides the missing nuclease activity of RISC. Last, we used our alignment of PIWI domain with several endonucleases to predict three residues, aspartate 965 (GADVT), glutamate 1016 (TLEHL), and aspartate 1037 (YRDGV), as being magnesium-coordinating residues at the catalytic center of Ago-2 nuclease. The corresponding two aspartate residues have also been recognized as part of the nuclease active site in mammalian and archaebacterium Pyrococcus furiosus Ago-2 by structural and mutagenesis studies (19, 20).
Materials and Methods
In Vitro siRNA Loading. ATP (final concentration, 1 mM) (Sigma) was added with 5′ phophorylated let-7 siRNA duplex (final concentration, 25 nM) (5′-P-UGAGGUAGUAGGUUGUAUAUU-3′, 5′-P-UAUACAACCUACUACCUCAUU-3′; or with biotinylated 3′ ends) to S-100 S2 extract prepared from a fresh S2 cell pellet, as described in ref. 2. The extract was incubated for 2 h at room temperature for the large-scale purification. For salt-sensitivity experiments, the loading incubation was performed for 30 min at room temperature.
RISC Assay. The RISC assay was performed as described (2). In brief, a let-7 target site was cloned between the _Xba_I and _Fse_I sites on pGL3 vector (Promega). The vector was used as template for PCR using primers spanning the cloning site. The PCR product was ethanol-precipitated, and an aliquot of 2 μg was used as template for in vitro transcription using a megascript in vitro transcription kit (Ambion, Austin, TX). The RNA was then 5′ radiolabeled by using [α-32P]GTP and a partially purified guanylyl transferase enzyme, which was generously provided by Lance Ford (Ambion). The RNA was then gel-purified and stored at a dilution of ≈100,000 cpm/μl. RISC reactions were carried out with 100 mM KOAc and 50,000 cpm of mRNA target for 30 min at 30°C. A typical reaction contained 5 μl of sample in a total volume of 10 μl. The reactions were stopped by addition of 200 μl of 0.3 M NaOAc (pH 5.2) and 100 μl of phenol/chloroform/isoamyl alcohol. The RNA was ethanol-precipitated in the presence of 2.5 μg/ml glycogen. The pellet was resolubilized by boiling in formamide loading dye (Ambion) and subjected to electrophoresis on 6% acrylamide/7 M urea gels for 20 min at 300 V.
Preloaded RISC Purification. We loaded 200 ml of S2 extract (≈5 mg/ml) with siRNA (see above). The extract was then spun at 200,000 × g for 3 h and resuspended in 400 mM KOAc as described in ref. 8. Solid ammonium sulfate was added to 20% saturation. The extract was respun at 100,000 × g to pellet the ammonium sulfate pellet. The supernatant was then loaded onto a 20-ml phenyl-Sepharose column equilibrated in buffer PS. Buffer PS is buffer A (10 mM KOAc/2 mM MgOAc/10 mM Hepes, pH 7.4) plus 400 mM KOAc (pH readjusted to 7.4) and 20% ammonium sulfate. The column was eluted by using gradient of buffer PS and buffer A over 10 column volumes.
The column fractions were dialyzed overnight in buffer A plus 200 mM KOAc (pH 7.4) and assayed for activity, and the peak was pooled for loading onto a 5-ml HiTrap Q-Sepharose column. The column was run with the 200 mM KOAc in buffer A to match the salt in the dialyzed samples. The Q flow-through was collected, diluted to a final concentration of 130 mM KOAc by using buffer A, and loaded onto a 5-ml HiTrap heparin agarose column using buffer A plus 130 mM KOAc as the base buffer. The elution was done over two steps: first to 15% for four column volumes and then to 100% buffer B (buffer A/1 M KOAc, pH 7.4). The 15-100% step peak was dialyzed into PD buffer (buffer A/100 mM NaCl). For the pull-down step, 1% Triton X-100 was added before adding 80 μl of Dynabeads M-280 streptavidin-coated beads (Dynal, Great Neck, NY). The mixture was rotated at 4°C overnight and collected by using the MPC-S magnet (Dynal). This pellet was then washed five times with PD buffer containing 1% Triton X-100, followed by five washes with buffer PS.
Assaying Beads for RISC Activity. The RISC assay was carried out on Dynabeads M-280 streptavidin-conjugated beads by first coating the beads by incubating them on ice in 5× Denhardt's reagent (0.02% polyvinylpyrrolidone/0.02% Ficoll/0.02% molecular-biology-grade BSA) for 1 h to keep the mRNA from nonspecifically sticking to the beads. The Denhardt's reagent was replaced with PD buffer, and the reaction proceeded as described above.
MS Peptide Analysis. Proteins in the beads were digested in 50 mM NH4HCO3/trypsin at 37°C overnight. HPLC and tandem MS (MS/MS) analyses were performed in an LCQ Deca XP Plus ion-trap mass spectrometer (ThermoFinnigan, San Jose, CA) coupled online to a nano-HPLC system (1100 Nano Pump, Agilent Technologies, San Jose, CA) and nanospray source. We manually injected 2 μl of the peptide solution in buffer C (5% acetonitrile/94.9% water/0.1% acetic acid, vol/vol/vol) and separated it in a nano-HPLC column (length, 50 mm; i.d., 75 μm; particle size, 5 μm; pore diameter, 100 Å) packed in-house with Luna C18 resin (Phenomenex, Belmont, CA). The peptides were eluted from the column with a linear gradient of 25-80% buffer D (90% acetonitrile/9.9% water/0.1% acetic acid, vol/vol/vol) in buffer C over 30 min.
The eluted peptides were electrosprayed directly into the LCQ mass spectrometer. The tandem MS (MS/MS) spectra were acquired in a data-dependent mode. The four strongest ions in each MS spectrum were automatically selected for fragmentation. The resulting spectra were used to identify protein candidates in The National Center for Biotechnology Information (NCBI) nonredundant protein sequence database with the MAS-COT search engine (Matrix Science, London).
metabasic Homology Search. metabasic is a sensitive approach to find distant homology between protein families (18). metabasic detects similarity between two alignments by a combination of a standard position-specific-iterative blast-style sequence profile with predicted secondary structure. Several scoring systems and alignment algorithms are used in the process. Found hits are ordered according to the Z score, and Z scores of >12 correspond to a <5% probability of being incorrect.
Results
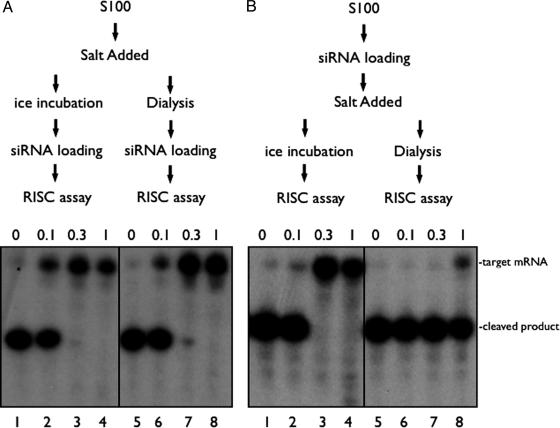
Differential Salt Sensitivity for RISC Activity Before and After siRNA Loading. To study RISC activity, we added let-7 siRNA to S-100 extracts prepared from Drosophila S2 cells and generated siRNA-dependent cleavage of a complementary mRNA (Fig. 1). We analyzed the sensitivity of RISC activity to high salt (up to 1 M) before or after siRNA addition. As shown in Fig. 1_B_, high salt exposure after siRNA addition reversibly masked the ability of RISC to cleave target substrate (Fig. 1 A, lanes 1-4). After the salt was removed by dialysis, full RISC activity was recovered (Fig. 1 A, lanes 5-8). However, upon exposure of S2 cell extracts to high salt prior to siRNA loading, RISC activity was irreversibly lost; it could not be recovered even after the salt was completely dialyzed away (Fig. 1 A, lanes 7-8). The difference in salt sensitivity suggested that the assembly of RISC (by addition of siRNA to naive extracts) involves a component (or components) with a molecular conformation that is irreversibly damaged upon salt addition, and it also suggested that the salt-labile, functioning conformation is upstream of the siRNA-loaded RISC. The salt sensitivity made it difficult to purify the components needed for de novo RISC assembly. However, resistance of preloaded RISC to high salt exposure makes it a better subject for protein-purification procedures. Therefore, we developed a procedure for the purification of preloaded RISC nuclease activity to homogeneity.
Fig. 1.
RISC assembly is sensitive, but preassembled RISC is resistant to high salt. S2 S100 was loaded in vitro with siRNA to program RISC to cleave a G-cap radiolabeled mRNA target (see Materials and Methods). (A) We added 0, 0.1, 0.3, or 1 M NaCl to extract, and the samples were either incubated on ice (lanes 1-4) or dialyzed back to starting buffer conditions at 4°C for 4 h (lanes 5-8) before siRNA and 1 mM ATP was introduced. After a 30-min incubation at room temperature, radiolabeled target mRNA was added, and the reactions were incubated for an additional 30 min at 30°C to measure RISC activity. (B) The indicated concentration of salt was added after siRNA loading. The samples were placed on ice (lanes 1-4) or dialyzed (in buffer A) (lanes 5-8) for 4 h at 4°C. All samples were then tested for RISC (as described above).
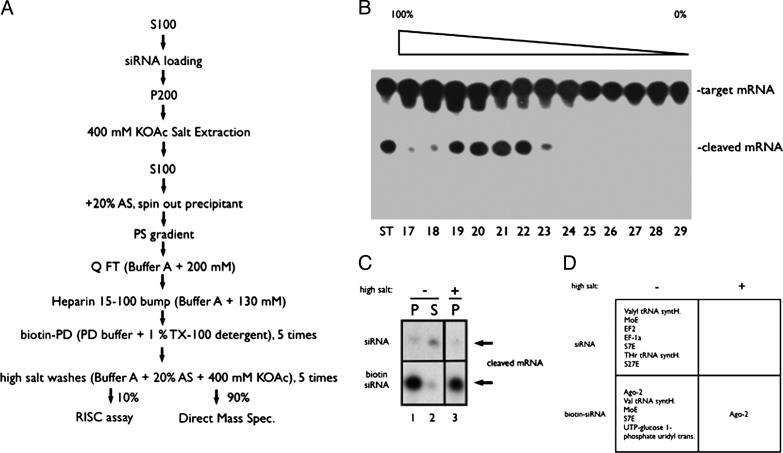
Ago-2 Alone Is Sufficient for siRNA-Guided Cleavage Activity. The purification procedure is shown in Fig. 2_A_. The procedure began with the addition of a 3′-biotinylated siRNA to 200 ml of S-100 extract (1 g of total protein) from S2 cells. After incubation at room temperature for 120 min to load the siRNA into RISC, the RISC activity was collected by centrifugation at 200,000 × g. The pelleted RISC activity was then solubilized by extraction with 400 mM potassium acetate. After addition of ammonium sulfate to 20% saturation, the 100,000 × g supernatant, containing RISC activity, was loaded onto a phenyl-Sepharose column and eluted with a decreasing ammonium sulfate/potassium acetate gradient over 10 column volumes. As shown in Fig. 2_B_, the RISC activity was eluted in a sharp peak around fractions 19-22. The active fractions were pooled, dialyzed to 200 mM KOAc, and loaded onto a Q-Sepharose column. The RISC activity that flowed through this column (data not shown) was subsequently loaded onto a heparin agarose column, and the RISC activity was eluted by a 15-100% step of 1 M KOAc (data not shown). The eluate was dialyzed into PD buffer (buffer A/100 mM NaCl), and the RISC activity was pulled down by using streptavidin-conjugated magnetic beads by overnight incubation with rotation at 4°C in the presence of 1% Triton X-100. The RISC activity pulled down by the beads was then split. An aliquot of 10% of the beads was used for the RISC assay, and the remaining 90% was used for protein identification. Before assaying, the beads were coated with Denhardt's reagent (0.02% polyvinylpyrrolidone/0.02% Ficoll/0.02% molecular-biology-grade BSA) to keep the mRNA substrate from sticking to the beads, a step critical for assaying RISC activity directly on beads. The RISC activity was enriched and pulled down by the beads when biotin-labeled, but not normal, siRNA was used (Fig. 2_C_, lanes 1-3). Because the RISC activity was resistant to high salt (Fig. 1), another five washes of 400 mM KOAc/20% ammonium sulfate were added. The final high-salt wash did not affect the RISC activity on the beads (Fig. 2_C_ Bottom, compare lanes 1 and 3), and the target RNA was completely processed.
Fig. 2.
Purification of preassembled RISC activity. (A) Schematic representation of the purification procedure. (B) RISC assay with individual fractions from the phenyl-Sepharose column (17-29). The first lane shows starting material activity. All samples were dialyzed to 200 mM KOAc before being assayed for activity (because high salt masks activity). (C) The cleaved mRNA product after incubation with the streptavidin-coated magnetic beads after detergent and salt washes (see Materials and Methods for details). Upper is from a parallel purification in which the loaded siRNA was not biotinylated. Lower shows the biotinylated siRNA pull-down activity. Lanes 1 and 3 show the activity before and after the high-salt wash, respectively. Lane 2 shows the activity that is left in the supernatant after the pull down. (D) The corresponding proteins identified from MS of the trypsinized beads. Under the most stringent conditions (biotin tag, detergent wash, and salt wash), only Ago-2 was detected (see Fig. 3 for peptide list).
Protein purifications are normally finished by running an SDS gel electrophoresis to identify correlating bands. However, proteins with unusual sizes, extremely positive charges, low abundance, or that are poor substrates for staining (for instance with silver stain) may be lost during this step. We decided to avoid all of these potential pitfalls by directly digesting all of the proteins on the beads with trypsin and subjecting all resulting peptides directly to MS. Interestingly, if the beads were not subjected to the final high-salt wash, many proteins (including previously identified RISC component Ago-2) were identified (Fig. 2_D_ Lower Left). Most of other proteins were ribosomal and other RNA-binding proteins. However, when the beads were subjected to high-salt wash before digestion with trypsin, all of the 33 peptides recovered were from Ago-2 (see Fig. 3). Importantly, not a single peptide from any other protein was detected under this condition. Also, all of the nonspecific proteins that were pulled down by the bead when normal siRNA was used for RISC assembly (Fig. 2_D_ Upper Left) were washed away by high salt (Fig. 2_D_ Upper Right). Therefore, we conclude that one protein, Ago-2, composes the core RISC nuclease activity.
Fig. 3.
Peptides recovered from the RISC purification. The 33 peptides recovered all map to Ago-2 (GenBank accession no. 24664664; full-length Ago-2) and cover a significant portion of the entire protein. Three nonredundant peptides had amino acids that were different from GenBank entry 24664664 but that agree with the sequence in GenBank entry 20151489 (likely an Ago-2 allelic variant fragment). The amino acid detected in our data (taken from S2 cells) is placed above the 24664664 sequence for each case. Bars indicate the best match sequence for each collected MS signal.
PIWI Domain Is Predicted to Have an Endonuclease V-Like Structure. Our RISC purification suggested that Ago-2 must have an unidentified nuclease activity. Therefore, we took a bioinformatics approach to help us find the region that is responsible for this putative activity. Similarity search engines based purely on sequence (e.g., position-specific-iterative blast; ref. 18) have been unable to detect any homology between Ago-2 and known nucleases. Instead, we turned to an algorithm that identifies relationships based on a combination of sequence profiles with secondary-structure prediction, metabasic (18). Found homology should imply similarity in fold and possibly function. The PIWI domain of Drosophila melanogaster Ago-2 (GenBank accession no. 24664664) found the endonuclease V family as the first metabasic hit with the score of 15.2. metabasic scores of >12 correspond to <5% incorrect matches (18). Unfortunately, no mutants or structures from the endonuclease V protein family have been reported that allow for identification of the active site. However, we were able to use a region of amino acid conservation between the endonuclease V family and the better characterized UVRC DNA-repair endonuclease families active site to predict where the endonuclease V active site should be. In turn, this alignment allowed us to make a prediction of the residues that might form the active site of PIWI based on obvious conservation between the active sites (see Fig. 4). We predicted sites aspartate 965 (GADVT), glutamate 1016 (TLEHL), and aspartate 1037 (YRDGV) of Drosophila Ago-2 (see Fig. 4) as likely active-site residues for the Ago-2 nuclease activity. Coordination by these three residues with water coordinated in position to make the attack on the phosphodiester backbone would account for the normal tetrahedral coordination of a Mg2+-dependent nuclease.
Fig. 4.
Alignment of PIWI domains with selected endonucleases. Ago-2 PIWI domains from D. melanogaster (Dm) and Mus musculus (Mm) are aligned with PIWI domain from P. furiosus (Pf) and endonucleases V, UVRC and RNaseHI from E. coli (Ec). GenBank accession nos. are given at the end of sequence names. Secondary-structure prediction for Ago-2_Dm and secondary structure of RNaseHI (PDB ID 2RN2) are given above and below the alignment, respectively. The β-strands and α-helices are indicated by E and H, respectively. Functionally important conserved residues are shown in white letters boxed in black, uncharged residues at mainly hydrophobic positions are highlighted in yellow, and conserved small residues are highlighted in gray. The first and last residue numbers of the shown sequences are indicated before and after each sequence. Some nonconserved residues are omitted, and the number of omitted residues is shown in parentheses.
Discussion
Previous attempts to purify RISC nuclease from human and Drosophila systems have identified Ago-2 as well as several other proteins, including dFXMR, VIG (21), and tudor-SN (22), that are associated with RISC. However, the roles of these proteins in the fundamental activity of RISC (siRNA-guided, site-specific endonucleolytic cleavage) are still unclear. Martinez et al. (6) also purified RISC activity to near homogeneity from HeLa cell extract. They saw a correlation only between two proteins, eIF2C2 and eIF2C1 (two Ago family members), and RISC activity. This work is especially important because it ruled out many of the previously identified RISC-associated proteins as being required for RISC activity, and it suggested that RISC activity might be provided by an Ago family protein alone. However, this possibility was not demonstrated conclusively because there were still other proteins present in the active fraction (6). Furthermore, because no protein was identified with recognizable nuclease function, it remained possible that an associated nuclease had just evaded detection. We reasoned that purification of RISC activity to homogeneity judged by the most rigorous standard would provide answers to what the actual composition of the RISC complex was and, therefore, lead us to identification of the RISC nuclease. Therefore, we derived a purification procedure that combined conventional chromatographic steps with affinity purification after forming the RISC with a 3′ biotin-labeled siRNA in vitro before purification. The final purified product was measured for its RISC activity in situ and subjected to proteolysis, followed by MS analysis directly. To our surprise, Ago-2 is the only detected protein with the recovered peptides covering most of the protein. This result provides strong evidence that Ago-2 alone is sufficient for siRNA-loaded RISC activity.
This finding was surprising because there had been no previous indication that Ago-2 had any nuclease domain. After a careful bioinformatics analysis based on sequence and predicted secondary structure, we realized that the PIWI domain of Ago-2 shows similarity to endonuclease V. This alignment predicts three active-site residues. Two recently published articles (19, 20) reported a nearly identical conclusion from two different experimental systems.
In the first article, the structure for the Ago protein from archaebacterium P. furiosus was solved (19). The structure revealed that the PIWI domain fold is similar to RNase H, with two conserved active-site residues that fit perfectly with our bioinformatics-based prediction of the active site of PIWI to endonuclease V (GenBank accession no. 16131828), but disagrees on the third site (explanation given below). Although no structure is available for endonuclease V, our sequence analysis strongly suggests that it should possess an RNase H-like fold. RNase H hydrolyzes RNA from an RNA-DNA hybrid ds molecule, whereas endonuclease V nicks DNA near chemically damaged sites on dsDNA substrates (23). Endonuclease V also has the ability to cleave undamaged single-stranded DNA. Both proteins are identical to the RISC endonuclease in terms of magnesium dependence and the molecular ends produced after cleavage.
In the second article, site-directed mutagenesis on two of the structurally predicted and alignment-predicted active-site residues in mammalian Ago-2 protein abolished RISC activity when the mutant proteins were expressed in the Ago-2 knockout mouse embryonic fibroblasts (20). The third Mg2+ coordination site has not yet been experimentally determined. We predict that it resides near the middle of helix 1 of our alignment (light-green helix in figure 3 of ref. 19) rather than at site 635 of the P. furiosus sequence. For our prediction to be correct, helix 1 would have to take on a more “upright” position, in other words, a position that is more closely parallel to the β-pleated sheets. This position would more closely mimic the structure of RNase HI, and RNase HII. Perhaps Mg2+, absent from this structure but present in active RISC, would be sufficient to cause this shift. Alternatively, “migration” of the third ligand could have occurred, and it is contributed by different sites in distant homologs (namely, by a site in the middle of the α-helix in RNase H and by a site after the β-strand in Ago-2). The answer to this question would guide our understanding of the PIWI family as a whole and help us to predict which other members might function as nucleases. Mutagenesis of the sites in question could possibly resolve this issue. When considering our biochemical purification and the published structure analysis of Ago-2 together, it is clear the siRNA-Ago-2 complex alone is sufficient for RISC activity (siRNA-guided, site-specific cleavage of mRNA targets), with the PIWI domain of Ago-2 functioning as the nuclease.
Acknowledgments
We thank Dr. Qinghua Liu for helpful suggestions and encouragement, Zhengzheng Li for S2 cell-culture work, and Dr. Sung Won Kwon for MS analysis. The work is supported by National Institutes of Health Grant GMRO157158 and Welch Foundation Grant I-1412.
Abbreviations: siRNA, small interfering RNA; RISC, RNA-induced silencing complex; ds, double stranded; Ago-2, Argonaute 2; S2, Schnieder 2.
References
- 1.Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391**,** 806-811. [DOI] [PubMed] [Google Scholar]
- 2.Liu, Q., Rand, T. A., Kalidas, S., Du, F., Kim, H. E., Smith, D. P. & Wang, X. (2003) Science 301**,** 1921-1925. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. (2001) Nature 409**,** 363-366. [DOI] [PubMed] [Google Scholar]
- 4.Provost, P., Dishart, D., Doucet, J., Frendewey, D., Samuelsson, B. & Radmark, O. (2002) EMBO J. 21**,** 5864-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang, H., Kolb, F. A., Brondani, V., Billy, E. & Filipowicz, W. (2002) EMBO J. 21**,** 5875-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R. & Tuschl, T. (2002) Cell 110**,** 563-574. [DOI] [PubMed] [Google Scholar]
- 7.Tabara, H., Sarkissian, M., Kelly, W. G., Fleenor, J., Grishok, A., Timmons, L., Fire, A. & Mello, C. C. (1999) Cell 99**,** 123-132. [DOI] [PubMed] [Google Scholar]
- 8.Hammond, S. M., Boettcher, S., Caudy, A. A., Kobayashi, R. & Hannon, G. J. (2001) Science 293**,** 1146-1150. [DOI] [PubMed] [Google Scholar]
- 9.Song, J. J., Liu, J., Tolia, N. H., Schneiderman, J., Smith, S. K., Martienssen, R. A., Hannon, G. J. & Joshua-Tor, L. (2003) Nat. Struct. Biol. 10**,** 1026-1032. [DOI] [PubMed] [Google Scholar]
- 10.Yan, K. S., Yan, S., Farooq, A., Han, A., Zeng, L. & Zhou, M. M. (2003) Nature 426**,** 468-474. [DOI] [PubMed] [Google Scholar]
- 11.Lingel, A., Simon, B., Izaurralde, E. & Sattler, M. (2003) Nature 426**,** 465-469. [DOI] [PubMed] [Google Scholar]
- 12.Lingel, A., Simon, B., Izaurralde, E. & Sattler, M. (2004) J. Biomol. NMR 29**,** 421-422. [DOI] [PubMed] [Google Scholar]
- 13.Lingel, A., Simon, B., Izaurralde, E. & Sattler, M. (2004) Nat. Struct. Mol. Biol. 11**,** 576-577. [DOI] [PubMed] [Google Scholar]
- 14.Elbashir, S. M., Martinez, J., Patkaniowska, A., Lendeckel, W. & Tuschl, T. (2001) EMBO J. 20**,** 6877-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez, J. & Tuschl, T. (2004) Genes Dev. 18**,** 975-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarz, D. S., Tomari, Y. & Zamore, P. D. (2004) Curr. Biol. 14**,** 787-791. [DOI] [PubMed] [Google Scholar]
- 17.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25**,** 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginalski, K., von Grotthuss, M., Grishin, N. V. & Rychlewski, L. (2004) Nucleic Acids Res. 32**,** W576-W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song, J. J., Smith, S. K., Hannon, G. J. & Joshua-Tor, L. (2004) Science 305**,** 1434-1437. [DOI] [PubMed] [Google Scholar]
- 20.Liu, J., Carmell, M. A., Rivas, F. V., Marsden, C. G., Thomson, J. M., Song, J. J., Hammond, S. M., Joshua-Tor, L. & Hannon, G. J. (2004) Science 305**,** 1437-1441. [DOI] [PubMed] [Google Scholar]
- 21.Caudy, A. A., Myers, M., Hannon, G. J. & Hammond, S. M. (2002) Genes Dev. 16**,** 2491-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caudy, A. A., Ketting, R. F., Hammond, S. M., Denli, A. M., Bathoorn, A. M., Tops, B. B., Silva, J. M., Myers, M. M., Hannon, G. J. & Plasterk, R. H. (2003) Nature 425**,** 411-414. [DOI] [PubMed] [Google Scholar]
- 23.Guo, G., Ding, Y. & Weiss, B. (1997) J. Bacteriol. 179**,** 310-316. [DOI] [PMC free article] [PubMed] [Google Scholar]