Functional Dissection of a Conserved Motif within the Pilus Retraction Protein PilT (original) (raw)
Abstract
PilT is a hexameric ATPase required for type IV pilus retraction in gram-negative bacteria. Retraction of type IV pili mediates intimate attachment to and signaling in host cells, surface motility, biofilm formation, natural transformation, and phage sensitivity. We investigated the in vivo and in vitro roles of each amino acid of the distinct, highly conserved C-terminal AIRNLIRE motif in PilT. Substitution of amino acids A288, I289, L292, and I293 as well as a double substitution of R290 and R294 abolished Pseudomonas aeruginosa PilT function in vivo, as measured by a loss of surface motility and phage sensitivity. When introduced into purified Aquifex aeolicus PilT, substitutions in the AIRNLIRE motif did not disrupt ATPase activity or oligomerization. In contrast, a K136Q substitution in the broadly conserved nucleotide binding motif prevented PilT function in vivo as well as in vitro. We propose that the AIRNLIRE motif forms an amphipathic α helix which transmits signals between a surface-exposed protein interaction site and the ATPase core of PilT, and we recognize a potential functional homology in other type II secretion ATPases.
Through their roles in cell adhesion and surface motility, type IV pili contribute significantly to the pathogenicity and symbiotic nature of many gram-negative bacteria (reviewed in references 10 and 14). Pili, which extend up to five times the length of the bacterium, are perhaps most renowned for their role in the binding of bacteria to host cells or abiotic surfaces. Subsequent to initial binding, pilus retraction drives a form of surface motility, originally referred to as twitching motility (reviewed in references 14 and 34). In the context of pathogenic bacteria, this activity leads to intimate attachment and host cell invasion (18, 26). Pilus retraction is also critical for biofilm formation in some species (4, 22) and can mediate phage and DNA uptake, conferring, for example, phage sensitivity to Pseudomonas aeruginosa and natural competence to Neisseria gonorrhoeae (3, 39).
PilT is required for the retraction of type IV pilus filaments, as pilT mutants are hyperpiliated with nonretractile pili (14, 19, 36). Consistent with a role for pilus retraction in motility and intimate cell adhesion, a P. aeruginosa pilT mutant strain with nonretractile pili is significantly less infectious than the wild-type strain in mouse models of corneal infection and acute pneumonia (6, 41).
PilT is a member of the diverse group of bacterial type II secretion nucleoside triphosphatases (NTPases) (36). This large family shares a core domain of four signature motifs: Walker box A and B nucleotide binding motifs and aspartate and histidine boxes (24, 33). Several members of the type II secretion ATPase family, including Actinobacillus actinomycetemcomitans TadA, Legionella pneumophila DotB, and Aquifex aeolicus PilT, form hexameric rings with demonstrated ATPase activity in vitro (1, 9, 11, 30). Despite a conserved NTPase core, the type II secretion NTPase subfamilies differ substantially in their biological roles and in the amino acid sequences of their N- and C-terminal domains. These data suggest that it is these domains which confer specific functions (protein secretion, type IV pilus assembly, type IV pilus retraction, and natural competence in gram-positive bacteria) to the subfamilies.
Although in vivo ATPase activity has not been experimentally demonstrated for PilT, disruption of the ATP binding site in enteropathogenic Escherichia coli BfpF, a presumed functional homolog of PilT, leads to a marked decrease in infectivity (2). An analogous change in DotB, a PilT family member based on sequence similarity, prevents the survival of L. pneumophila within macrophages (30).
Several models have been proposed for the role of PilT in type IV pilus function (14, 17, 34). A hexameric PilT ATPase could actively dissociate pilin monomers from the base of the pilus filament, contributing to the pool observed within the cytoplasmic membrane (20). PilT could remove or inactivate a capping protein that prevents an energetically favorable retraction reaction. Alternatively, PilT could reverse the direction of the PilB motor, whose ATPase activity is required in vivo for the assembly of pilus filaments from pilin monomers (32). However, recent evidence that functional homologs of PilB and PilT do not interact in vivo in enteropathogenic E. coli (7) as well as the observation that the control of pilus biogenesis and the control of retraction are separable in Neisseria meningitidis (20) makes this third model unlikely.
To further the molecular understanding of the role of PilT in pilus retraction, we chose to study a well-conserved, PilT-specific sequence motif. Pairwise alignment of the P. aeruginosa PilT sequence with the sequences of other members of the type II secretion ATPase family reveals that the C-terminal domain (defined here as the final 72 amino acids) is 30 to 90% identical to those of the functionally confirmed PilT and PilU pilus retraction proteins but is less similar to those of members of other subgroups, including P. aeruginosa PilB and Vibrio cholerae EpsE (Fig. 1). The PilT and PilU C-terminal domains, typically shorter than those of other members of the family, are characterized by a GMQTXXXXLXXLXXXXXI motif and the lack of a zinc-binding tetracysteine motif found in most type II secretion ATPases (23, 27). In PilT proteins, there is an additional AIRNLIRE motif whose high degree of sequence conservation suggests a functional role which has been maintained by selective pressure (Fig. 1) (11). Thus, we chose to investigate the requirement for each amino acid of the C-terminal AIRNLIRE motif for in vivo and in vitro PilT functions.
FIG. 1.
Alignment of the C-terminal regions of in vivo functionally confirmed PilT and PilU proteins. The following sequences (NCBI reference number) are shown: Pa T, Pseudomonas aeruginosa PilT (AAG03784); Ps T, Pseudomonas stutzeri PilT (CAB56295); Rs T, Ralstonia solanacearum PilT (CAD16389); Ss T, Synechocystis sp. PilT (BAA18564); Nm T, Neisseria menigitidis PilT (AAF41181); Ng T, Neisseria gonorrhoeae PilT (AAB30824); Tt T, Thermus thermophilus PilT (AAL37755); Mx T, Myxococcus xanthus PilT (40); Aa T, Aquifex aeolicus PilT (AAC06903); Ps U, P. stutzeri PilU ((CAB56296); Pa U, P. aeruginosa PilU (S54702); Ng U, N. gonorrhoeae PilU (23); Ss T2, Synechocystis sp. PilT2 ((BAA18443); Pa B, P. aeruginosa PilB (AAG07914); and Vc E, Vibrio cholerae EpsE ((AAA58786). The alignment, created with Megalign Clustal W and manual optimization, begins with the last residues of the core nucleotide binding domain and the short glycine- and proline-rich linker region and is truncated at the end of the similarity, 6 to 20 amino acids from the C termini of the proteins. Residues that are identical or similar in 7 out of the 13 PilT and PilU sequences are shaded in black or grey, respectively; AIRNLIRE residues are indicated by asterisks. PilB and EpsE residues are shaded in black or grey if they are identical or similar, respectively, to conserved PilT and PilU residues. The amino acids of the tetracysteine loops of PilB and EpsE are not present in PilT or PilU. Percent identity (%I) was calculated from a pairwise alignment of each sequence with Pa T and from a pairwise alignment of Pa B with Vc E beginning at the caret (^).
MATERIALS AND METHODS
Bacterial strains and plasmids.
Isogenic P. aeruginosa PAK wild-type and ΔpilT strains as well as isogenic PA103 wild-type and ΔpilT strains were gifts from S. Lory and J. Engel, respectively (Table 1). The PAK ΔpilT strain was created by using a previously described _sacB_-based method to replace genomic pilT with a pilT deletion allele carried on pEX18Tc (12; M. Wolfgang, personal communication). The in-frame deletion removed the coding sequence for amino acids 39 to 200. The PA103 ΔpilT strain was created through recombination of a pilT fragment marked with β-lactamase into the pilT locus. The pilT deletion fragment contained the first 180 bases joined out of frame to the last 200 bases of pilT. Selection for recombination resulting in the loss of carbenicillin resistance yielded an unmarked pilT deletion (J. Engel, personal communication).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Properties or genotypea | Reference or source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAK | Natural isolate ATCC 25102 | 31 |
| PAK Δ_pilT_ | pilT in-frame deletion | M. Wolfgang and S. Lory |
| PA103 | Toxigenic (exoT exoU), non- protease producing | 13 |
| PA103 Δ_pilT_ | pilT out-of-frame deletion | J. Engel |
| KA305 | PA103 pilT (A288D) | This study |
| KA306 | PA103 pilT (I289A) | This study |
| KA307 | PA103 pilT (R290A) | This study |
| KA308 | PA103 pilT (N291A) | This study |
| KA309 | PA103 pilT (L292A) | This study |
| KA310 | PA103 pilT (I293N) | This study |
| KA311 | PA103 pilT (I293A) | This study |
| KA312 | PA103 pilT (E295A) | This study |
| KA313 | PA103 pilT (R290A/R294A) | This study |
| E. coli S17.1 | recA pro (RP4-2 Tet::Mu Kan::Tn_7_) | 25 |
| Plasmids | ||
| pUCP20 | Pseudomonas-E. coli shuttle vector | 35 |
| pEX19Ap | Pseudomonas integration vector | 12 |
| pKA22 | pUCP20::pilTPa (wild type) | This study |
| pKA50 | pUCP20::pilTPa (K136Q) | This study |
| pKA56 | pUCP20::pilTPa (A288D) | This study |
| pKA58 | pUCP20::pilTPa (I289A) | This study |
| pKA60 | pUCP20::pilTPa (R290A) | This study |
| pKA62 | pUCP20::pilTPa (N291A) | This study |
| pKA64 | pUCP20::pilTPa (L292A) | This study |
| pKA66 | pUCP20::pilTPa (I293N) | This study |
| pKA68 | pUCP20::pilTPa (I293A) | This study |
| pKA70 | pUCP20::pilTPa (R294A) | This study |
| pKA72 | pUCP20::pilTPa (E295A) | This study |
| pKA92 | pUCP20::pilTPa (R290A/R294A) | This study |
| pKA282 | pUCP20::pilTPa (I293L) | This study |
| pKA284 | pUCP20::pilTPa (I289L) | This study |
| pKA286 | pUCP20::pilTPa (A288V) | This study |
| pKA288 | pUCP20::pilTPa (L292I) | This study |
| pKA290 | pUCP20::pilTPa (E295D) | This study |
| pKA293 | pUCP20::pilTPa (N291Q) | This study |
| pKA297 | pUCP20::pilTPa (R290K/R294K) | This study |
| pKA236 | pEX19Ap::pilTPa (A288D) | This study |
| pKA238 | pEX19Ap::pilTPa (I289A) | This study |
| pKA240 | pEX19Ap::pilTPa (R290A) | This study |
| pKA242 | pEX19Ap::pilTPa (N291A) | This study |
| pKA244 | pEX19Ap::pilTPa (L292A) | This study |
| pKA246 | pEX19Ap::pilTPa (I293N) | This study |
| pKA248 | pEX19Ap::pilTPa (I293A) | This study |
| pKA250 | pEX19Ap::pilTPa (R294A) | This study |
| pKA252 | pEX19Ap::pilTPa (E295A) | This study |
| pKA262 | pEX19Ap::pilTPa (R290A/R294A) | This study |
| pTJH1000 | pET23a(+)::_pilTAa_-His6 | 11 |
| pTJH1009 | pET23a(+)::_pilTAa_-His6 (K149Q) | 11 |
| pTJH1001 | pET23a(+)::_pilTAa_-His6 (A301D) | This study |
| pTJH1002 | pET23a(+)::_pilTAa_-His6 (I302A) | This study |
| pTJH1003 | pET23a(+)::_pilTAa_-His6 (R303A) | This study |
| pTJH1004 | pET23a(+)::_pilTAa_-His6 (N304A) | This study |
| pTJH1006 | pET23a(+)::_pilTAa_-His6 (I306A) | This study |
| pTJH1007 | pET23a(+)::_pilTAa_-His6 (R307A) | This study |
| pTJH1008 | pET23a(+)::_pilTAa_-His6 (E308A) | This study |
To express P. aerugionsa PilT from a plasmid, pilT, including putative promoter (430 bp upstream) and terminator (110 bp downstream) regions, was amplified from genomic DNA (PA103) by PCR with 5′-GGTCGGCCAGTTCGGCCTGCTTGCCGAGGGCC-3′ and 5′-GCGCTCGCCGGCAAGGATAGGTAGGAATGCGCC-3′ as primers. By use of standard molecular biology techniques for DNA manipulation and E. coli transformation (28), the genomic PstI and KpnI sites were used to clone the pilT locus into pUCP20 (35), creating pKA22 (Table 1). P. aeruginosa was transformed as previously described (15), and pUCP20-derived plasmids were maintained with carbenicillin (250 μg/ml). All mutations described were engineered into pKA22 for P. aeruginosa pilT or pTJH1000 for A. aeolicus pilT by using the full-circle PCR method of site-directed mutagenesis (Quick Change; Stratagene) (Table 1). Primer sequences are available upon request.
For gene replacement of the ΔpilT allele with the novel pilT alleles, a previously described _sacB_-based strategy was used with modifications (12). First, to create pilT alleles with sufficient sequence overlap for integration into the PA103 ΔpilT strain, the 3′ end of the pilT sequence on pKA22 was extended by insertion of a fragment amplified from genomic DNA with 5′-GGATTCAAGAGCGGGAGAGCGGTACCGTAGG-3′ (natural KpnI site in bold type) and 5′-CGTCCAGAGCTCGCTCCAGCGCCTGGTTGGCGTTGTTCG-3′ (introduced SacI site in bold type) as primers into the KpnI and SacI sites of each pUCP20-derived plasmid containing a pilT allele with a mutation. The PstI-SacI pilT fragments then were subcloned from the pUCP20-derived plasmids into pEX19Ap (12), creating pKA236-262 (Table 1). Each resulting plasmid was conjugated from E. coli S17.1 to PA103 ΔpilT. Merodiploids were selected on Luria-Bertani (LB) medium containing 250 μg of carbenicillin/ml and subsequently resolved on LB medium containing 5% sucrose. Integration of each full-length pilT gene was confirmed on the basis of the size of a PCR amplification product of the locus, and each sequence was confirmed by automated DNA sequencing.
Motility assays.
Single P. aeruginosa colonies carrying the appropriate plasmids were stabbed through the agar layer of thin LB medium-carbenicillin plates as previously described (8). Zones of motility between the agar and the plastic were examined after incubation at 37°C for 24 h. Each deletion strain expressing a PilT variant was assayed in triplicate.
Phage sensitivity assays.
Pilus-dependent sensitivity to bacteriophage PO4 (a gift from S. Lory) was tested as previously described with minor modifications (36). A total of 0.45 OD600 (optical density at 600 nm) cell equivalents (500 to 1,200 μl) of P. aeruginosa harboring a plasmid, grown to an OD600 of 0.3 to 0.6 in LB medium-carbenicillin, were incubated with 100 μl of a high-titer phage stock (approximately 1,000 PFU/ml) at room temperature for 10 min. Three milliliters of LB medium-carbenicillin top agar was added to the cells before they were spread on LB medium-carbenicillin plates. After incubation at 37°C for approximately 18 h, phage sensitivity was determined by counting PFU in a lawn of cells. Assays were performed in duplicate.
Expression and purification of A. aeolicus PilT variants.
PilT and its variants were purified from E. coli BL21(DE3) carrying the appropriate expression plasmid by Ni2+ affinity chromatography followed by heat shock (Table 1) as described for wild-type PilT (11).
ATPase end-point activity assays.
The ATPase activity of purified PilT variants was determined as previously described for wild-type A. aeolicus PilT (11). PilT (final concentration, 0.7 to 1.5 mg/ml) was incubated in buffer containing 5 mM MgCl2 and 2.5 mM ATP at 80°C for 10, 15, and 20 min. Reactions were stopped by flash-freezing, and the final ADP concentration, determined by a modified coupled-enzyme assay, was used to calculate the specific activity (11). Results presented here are the averages of nine measurements from three independent experiments with three replicates each.
Size exclusion chromatography.
The oligomeric status of PilT variants was determined by size exclusion chromatography with a Superdex 200HR 10/30 prepacked column (Pharmacia) at a flow rate of 0.7 ml/min with elution buffer containing 25 mM Tris (pH 7.2), 100 mM imidazole, 200 mM KCl, and 10% glycerol. The column was calibrated with the following standard mixture (Bio-Rad): thyroglobulin, 670 kDa; gamma globulin, 158 kDa; ovalbumin, 44 kDa; myoglobin, 17 kDa; and cyanocobalamin, 1.35 kDa. For the data presented here, standards and each of the samples were separated individually with the same column on the same day. A total of 100 μl of each purified protein was injected. The protein concentration was approximately 0.1 mg/ml for all samples, except for PilT R303A, which was injected at approximately 0.03 mg/ml.
Cell lysis and protein immunoblotting.
To assay PilT accumulation, P. aeruginosa cells were grown to mid-log phase, and 20 OD600 equivalents were harvested. Cells were resuspended in 1.2 ml of 20 mM Tris (pH 8)-250 mM NaCl and lysed by sonication. Cell lysates from 0.017 OD600 equivalents were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12%) and transferred to nitrocellulose. PilT was detected with precleared anti-N. gonorrhoeae PilT antibodies (a gift from M. Koomey) (20). Protein-antibody complexes were visualized by enhanced chemiluminescence (Pierce SuperSignal Femto detection system).
RESULTS
Amino acids of the AIRNLIRE motif are required for in vivo function.
To determine whether the AIRNLIRE motif is required for PilT function in vivo, alanine substitutions were engineered into each position of the P. aeruginosa PilT AIRNLIRE motif, with aspartic acid substituted for the initial alanine. An asparagine substitution for I293 was inadvertently created as well. These pilT genes with site-directed mutations were cloned behind the native pilT promoter on the pUCP20 shuttle vector (Table 1 shows strains and plasmids), and proteins were expressed in a P. aeruginosa PAK pilT deletion strain (ΔpilT). After demonstration of the in vivo requirement for the AIRNLIRE motif with these variant pilT alleles, a second generation of more conservative substitutions was investigated.
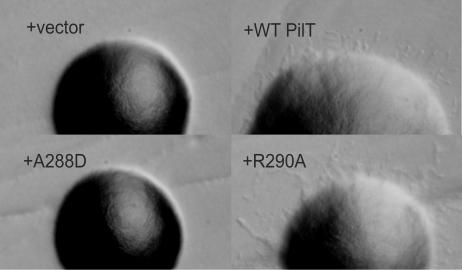
Colony morphology served as an indicator of PilT function. Unlike colonies of the wild-type strain, colonies of the ΔpilT strain are smooth and domed on rich agar plates, lacking finger-like projections (Fig. 2) (29). The smooth morphology of PAK ΔpilT colonies was rescued by complementation with plasmid-derived wild-type P. aeruginosa PilT. Plasmids encoding PilT proteins with individual substitutions A288D, I289A, L292A, I293N, and I293A in the AIRNLIRE motif did not restore normal colony morphology, whereas plasmids encoding the individual substitutions R290A, N291A, R294A, and E295A restored the wild-type phenotype (Fig. 2 and Table 2).
FIG. 2.
Colony morphology of cells harboring wild-type (WT) and mutated pilT genes. Representative colonies are shown for PAK ΔpilT plus the following plasmids: pUCP20 (+vector), pKA22 (+WT PilT), pKA56 (+A288D), and pKA60 (+R290A). Colonies are shown at the same magnifications.
TABLE 2.
Effects of AIRNLIRE substitutions on three PilT-dependent phenotypes
| Plasmid-expressed PilT protein | Morphologya | Motilityb | Bacteriophage PO4 sensitivityc |
|---|---|---|---|
| Wild type (parent) | F | + | S |
| None (Δ_pilT_) | D | − | R |
| None (vector) | D | − | R |
| Wild type | F | + | S |
| K136Q (Walker box A) | D | − | R |
| A288D | D | − | R |
| A288V | D | − | R |
| I289A | D | − | S |
| I289L | F | + | S |
| R290A | F | + | S |
| N291A | F | + | S |
| N291Q | F | + | S |
| L292A | D | − | S |
| L292I | F | + | S |
| I293N | D | − | R |
| I293A | D | − | R |
| I293L | D | − | R |
| R294A | F | Int | S |
| E295A | F | Int | S |
| E295D | F | + | S |
| R290A/R294A | D | − | R |
| R290K/R294K | F | + | S |
Surface motility of cells expressing PilT proteins with substitutions served as a second measure of PilT function. Cells with a pilT deletion lose the ability to spread at an agar-plastic interface and are therefore unable to form large, uniform motility zones (38). The same pilT variants with aberrant colony morphology produced no motility zones (Table 2). Therefore, as determined by two independent assays, certain individual substitutions in A288, I289, L292, and I293 led to the loss of in vivo PilT function. In contrast, alanine substitution in any one of the remaining AIRNLIRE residues did not disrupt surface motility.
As a third test of PilT function in vivo, the effect of amino acid substitutions in the AIRNLIRE region on phage sensitivity was investigated (Table 2). In P. aeruginosa, type IV pili are receptors for some filamentous phage, including PO4, and phage are probably brought in proximity with the bacterium upon pilus retraction (3). While PilT function is required for both surface motility and phage uptake, the latter does not require PilU (38). Phage sensitivity is therefore independent of potential defects in PilU regulation of or interaction with PilT. In addition, phage uptake likely serves as a sensitive measure of PilT function, as plaque formation may require as few as one pilus retraction event per cell. Therefore, a crippled PilT protein may retain enough function to promote phage uptake but not enough to drive coordinated cell motility. Of the PilT variants that disrupted colony morphology and motility, A288D, I293N, and I293A also abolished phage sensitivity, while PilT variants I289A and L292A provided cells with phage sensitivity (Table 1). Therefore, although I289A and L292A PilT function is not adequate for motility, residual PilT function is sufficient for phage uptake.
Because the extents of twitching motility vary among P. aeruginosa strains, we also tested the in vivo requirement for the residues of the PilT AIRNLIRE motif in P. aeruginosa PA103. We examined both plasmid-expressed and chromosomally expressed PilT in PA103 because the regulation of pilT expression is not yet fully understood (37). PA103 carrying the set of PilT-encoding plasmids described above exhibited the same phenotypes as PAK carrying those plasmids, except that the I289A variant led to a loss of phage sensitivity and the L292A variant produced a fully functional protein (Table 3). The phenotypes of the PA103 pilT integrants also paralleled the phenotypes of the PAK pilT transformants, with the exception that the integrated L292A pilT variant caused phage resistance (Table 3). These data indicate that neither strain differences nor plasmid expression of pilT affects the overall in vivo requirement for specific amino acids within the AIRNLIRE motif.
TABLE 3.
Strain and plasmid differences in in vivo phenotypes and accumulation of P. aeruginosa PilT proteins
| PilT protein | Morphologya | Motilitya | Bacteriophage PO4 sensitivitya | PilT accumu- lationb |
|---|---|---|---|---|
| I289A | D | − | S/R/S | </< |
| L292A | D/F/D | −/+/− | S/S/R | +/− |
| R290A/R294A | D | − | R/R/S | +/> |
An amphipathic helix model for the AIRNLIRE motif.
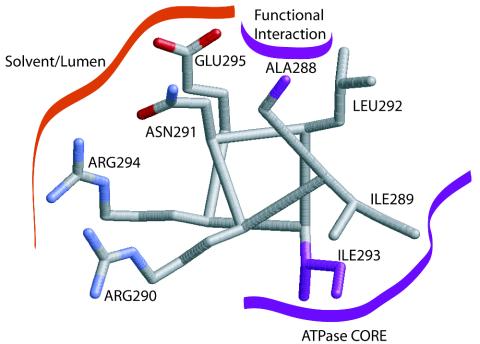
The pattern of hydrophobic and hydrophilic side chains within the AIRNLIRE sequence, bracketed by poor helix-forming residues proline at position 287 and glycine, asparagine, or aspartic acid at position 296, is consistent with a two-turn amphipathic α helix (5). Positioning the AIRNLIRE sequence on a helical wheel clarifies the spatial relationship that these amino acids would have in such a hypothetical α helix (Fig. 3). I289, I293, and L292 fall on one face of this helix, forming a hydrophobic surface, probably buried within the molecule. The charged or polar residues R290, N291, R294, and E295 form the opposite side of the model AIRNLIRE helix and are likely to be solvent exposed (Fig. 3).
FIG. 3.
Structural model of the proposed AIRNLIRE motif amphipathic α helix. Side-chain positions A288 and I293, at which single amino acid substitutions prevent both twitching motility and phage sensitivity in vivo (purple), suggest functional interactions. Additionally, the double mutant R290A/R294A loses PilT function. This stylized helix (grey carbon, red oxygen, and blue nitrogen atoms) was modeled in Xfit (16) by using standard side-chain conformations from the rotamer library and avoiding unfavorable steric clashes.
The in vivo requirement of an AIRNLIRE helix may be its amphipathic nature, and substitutions conserving the pattern of hydrophobic and hydrophilic residues may be sufficient for a functional motif. Therefore, conservative substitutions were introduced into the motif, and the resulting PilT proteins, expressed from plasmids in the P. aeruginosa PAK ΔpilT strain, were assayed for in vivo function. Substitutions in polar or charged residues N291Q and E295D preserved PilT function (Table 2), whereas substitutions in A288 and the three hydrophobic residues yielded two functional (I289L and L292I) and two nonfunctional (A288V and I293L) proteins (Table 2). Because alanine or more charge-conservative substitutions in the hydrophilic residues resulted in fully functional PilT proteins, no specific charged or polar residue in the AIRNLIRE motif is required for PilT function. However, these data demonstrate that while nonspecific hydrophobic residues at positions 289 and 292 are sufficient to retain PilT function, the amino acid identities at positions 288 and 293 are critical. Therefore, the amphipathic nature of the AIRNLIRE motif is necessary but not sufficient for in vivo PilT function.
It is perhaps surprising that the charged and polar residues of the AIRNLIRE motif, which are so highly conserved among PilT proteins and which have the potential to form a distinct, charged surface, did not significantly disrupt PilT function in vivo when substituted individually. To determine whether there was redundancy in the roles of the two adjacent arginine side chains on the modeled helix, we created two double mutants. Double substitutions R290K and R294K, which retained the overall charge and polarity on the presumed solvent-exposed surface, did not result in a loss of PilT function. However, double substitutions R290A and R294A resulted in a complete loss of PilT function in vivo. These results suggest that a single charged residue at either position 290 or position 294 within the conserved motif is necessary and sufficient for in vivo function.
The phenotypes of this collection of substitutions in the AIRNLIRE motif are consistent with the formation of an amphipathic α helix and suggest a multifunctional role: the amphipathic nature must be preserved, the positive charges on one surface are critical, and the specific identities of certain hydrophobic residues are also required for full PilT function in vivo.
Substitutions in the AIRNLIRE region affect but do not prevent PilT accumulation.
To determine whether the loss of PilT function is caused by decreased amounts of protein in vivo, the steady-state expression of PilT was assayed by qualitative protein immunoblot analysis of whole-cell lysates. All of the variant proteins accumulated when encoded by a plasmid in PAK Δ_pilT_ (Fig. 4), although the three I293 variants reproducibly accumulated to a markedly lesser extent than wild-type PilT. Due to the low signal from the available antibody, this assay is not quantitative, and differences in the accumulation of other hydrophobic variants relative to wild-type PilT are potentially significant.
FIG. 4.
Accumulation of PilT proteins for all AIRNLIRE variants. Whole-cell lysates were examined by qualitative protein immunoblotting with an anti-gonococcal PilT antibody to visualize protein accumulation for plasmid-borne pilT variants of PAK ΔpilT. Each PilT variant is indicated by its amino acid substitution(s). Two gels are shown. Levels for I293 variants that were lower than those for the wild type were apparent and reproducible. Differences in accumulation for other variants were not reproducibly observed.
When expressed from the chromosome, the levels of PilT proteins were in general lower and therefore more difficult to accurately assay than when expressed from a plasmid. The only two reproducible differences in expression were consistent with differences in the functions of chromosomally derived PilT and plasmid-derived PilT (Table 3 and data not shown). Specifically, in contrast to the plasmid-expressed version, the integrated L292A pilT variant was not functional and the protein was never detected by immunoblotting. On the other hand, unlike its plasmid-expressed counterpart, integrated R290A/R294A PilT retained residual function sufficient for phage sensitivity, and it was consistently present at significantly higher levels than wild-type PilT.
These immunoblotting data, while qualitative, rule out the diminution of steady-state protein levels as a simple explanation for the phenotypes observed for A288D and the arginine double mutant. These data also suggest that altered levels of PilT protein partially explain the phenotypes caused by other substitutions.
ATPase activity is required for PilT function in vivo.
To serve in addition to the pilT deletion as a negative control, glutamine was substituted for the invariant lysine in the phosphate binding loop of Walker box A (K136Q). This substitution was previously shown to eliminate the ATPase activity of purified PilT (11). Accordingly, in all in vivo assays, cells expressing the Walker box variant PilT protein were indistinguishable from ΔpilT cells carrying the vector only (Table 2). This variant PilT accumulated in cells (Fig. 4); however, the levels were consistently lower than those of wild-type PilT, suggesting that a single amino acid substitution in the nucleotide binding loop disrupts ATP binding and hydrolysis as well as the in vivo stability of PilT. These results are in agreement with published observations for L. pneumophila DotB (30) and provide an important demonstration of the in vivo requirement for an intact nucleoside triphosphate binding site and enzymatic activity of PilT.
Substitutions in the AIRNLIRE region do not disrupt ATPase activity.
Given that the Walker box A variant provides no measurable PilT function, one possible explanation for the phenotypes caused by substitutions in the PilT AIRNLIRE region is that the mutations disrupt the ATPase activity critical to pilus retraction in vivo. Although the AIRNLIRE region is not located in the conserved ATPase domain shared by all type II and type IV secretion ATPases, it is possible that these substitutions alter the structure of the C-terminal domain in a way that affects the catalytic properties of the protein either directly or indirectly.
Purified PilT is required to determine whether the substitutions in the AIRNLIRE region affect ATPase activity or oligomerization. Although mesophilic PilT proteins have been purified, they are poorly soluble (9, 21). Thermostable A. aeolicus PilT, with 51% pairwise sequence identity to P. aeruginosa PilT and N. gonorrhoeae PilT, provides a preferable in vitro system for studying the AIRNLIRE motif, since tens of milligrams of pure protein are easily obtained (11). Therefore, single amino acid substitutions were introduced into thermophilic A. aeolicus PilT to determine whether disruption of the AIRNLIRE motif directly affects enzymatic activity. Wild-type and variant A. aeolicus PilT-His6 proteins were expressed from an E. coli expression system and purified to near homogeneity by standard Ni2+ affinity chromatography followed by heat shock.
PilT proteins with substitutions in the AIRNLIRE motif (residues 301 to 308 in A. aeolicus) were assayed for ATPase activity by a modified coupled-enzyme assay (11). The specific activities of all proteins with substitutions were within 55 to 132% wild-type protein activity, 16.0 nmol of ATP/min/mg of PilT (Table 4). The slight decreases in ATPase activity for variants I302A and I306A were unlikely to be responsible for the complete loss of function of the corresponding P. aeruginosa PilT variants in vivo, as R303A PilT had a similar reduction of in vitro ATPase activity without a disruption of in vivo function. We conclude that the amino acids of the AIRNLIRE region are not directly required for the catalytic activity of PilT.
TABLE 4.
In vitro ATPase activity of A. aeolicus PilT proteins
| PilT variant | Mean ± SD sp act (nmol of ATP hydrolyzed/min/mg of PilT) |
|---|---|
| Wild type | 16.0 ± 1.6 |
| K149Q (Walker box A) | Below detection (11) |
| A301D | 19.7 ± 2 |
| I302A | 8.8 ± 0.9 |
| R303A | 9.3 ± 1.1 |
| N304A | 21.2 ± 1.8 |
| L305A | Not determined |
| I306A | 12.3 ± 1.3 |
| R307A | 20.7 ± 1.5 |
| E308A | 17.6 ± 2.0 |
Oligomerization is not affected by substitutions in the AIRNLIRE region.
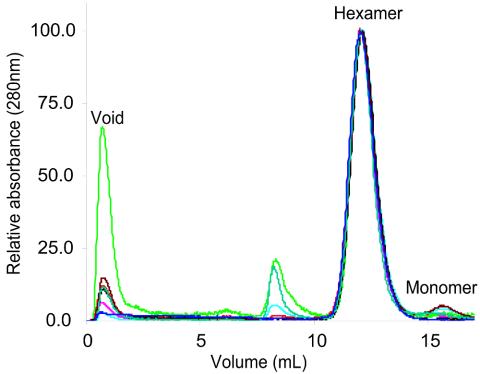
Another potential explanation for the in vivo loss of function of PilT variants A288, I289, L292, I293, R290, and R294 is that these residues influence subunit-subunit interactions required for PilT ring formation. To determine their oligomeric state, all purified A. aeolicus PilT proteins were subjected to size exclusion chromatography (Fig. 5). The retention time for the major elution peak of the variants was identical to that of the wild-type protein, corresponding to a hexameric ring structure (9, 11). This overlapping peak position indicates that under the in vitro conditions tested, the variant proteins are able to form oligomers with the same molecular mass, hydrodynamic radius, and overall shape as the wild-type protein. While these data do not rule out the possibility that the hexamer/monomer ratio has shifted for variant proteins in vivo, they establish that no single amino acid within the AIRNLIRE region is required for hexamerization.
FIG. 5.
Size exclusion chromatography of purified wild-type and variant A. aeolicus PilT proteins. A total of 3 to 10 μg of each protein was injected. The PilT proteins are labeled as follows: A301D, red; I302A, pink; R303A, light green; N304A, light blue; I306A, black; R308A, brown; E308A, dark green; and wild type, dark blue. All chromatograms were normalized to a peak value of 100. The hexamer and monomer peaks are labeled (11). The apparent aggregation of R303A was not seen in all separations.
DISCUSSION
Here we report a detailed investigation of the well-conserved C-terminal AIRNLIRE motif of the pilus retraction protein PilT. We have identified single amino acid substitutions in A288 and the hydrophobic residues I289, L292, and I293 and one double substitution, R290A/R294A, in the AIRNLIRE region that disrupt P. aeruginosa PilT function in vivo by several criteria. Analogous single amino acid substitutions in purified A. aeolicus PilT do not disrupt ATPase activity or overall oligomer size or shape in vitro. Thus, the AIRNLIRE motif is an important if not yet fully understood functional feature of the C-terminal domain of PilT.
There are several models for the in vivo function of the PilT C-terminal domain and the AIRNLIRE motif in particular. One possible role that would explain the strong in vivo but negligible in vitro effects is aberrant membrane localization in the mutants. Our preliminary qualitative results show that PilT proteins with substitutions partition between the membrane and cytosolic fractions, as does wild-type PilT; however, our assay would not have detected changes in the ratio of membrane fractionation to cytosol fractionation in the variant proteins. A second model to explain these results is that the motif serves as a protein interaction domain. Third, the AIRNLIRE motif may regulate the catalytic ATPase activity of PilT in vivo. We favor a scenario in which more than one of these possibilities is correct.
Our data are consistent with a model in which AIRNLIRE adopts an amphipathic α helix whose hydrophobic face (formed by I289, L292, and I293) signals binding events (potentially to pilin subunits, a regulatory protein, or the membrane) at the exposed surface of the helix to the conserved NTPase domain or, conversely, communicates ATP-dependent conformational changes from the core to the surface (Fig. 3). This helix model explains why three classes of mutations led to a null phenotype. First, the defect caused by double substitution of both arginines with alanines may indicate that while no single polar amino acid of the AIRNLIRE motif is required for PilT function, there is a requirement for an exposed charged face of a helix. Second, the position of A288 in the model helix suggests that it may be a surface-exposed yet nonpolar residue. Perhaps this invariant PilT residue (Fig. 1; see also the supplemental material) is the defining nonpolar contribution to the solvent-exposed surface of the AIRNLIRE region and is critical for recognition by an unidentified PilT-interacting protein. Third, alanine substitutions for I289 and L292 and every substitution tested for invariant I293 prevented PilT function. While all of the plasmid-expressed variants with substitutions in hydrophobic residues accumulated protein, our qualitative analysis suggested that the levels of at least the proteins with I293 substitutions were substantially decreased (Fig. 4). We hypothesize that these hydrophobic residues participate in interdomain structural alignment, signaling, and stability and that the combination of a functional defect and a protein stability defect may lead to the phenotypes observed for the substitutions at positions 289, 292, and 293.
The X-ray crystal structure of a V. cholerae type II secretion ATPase, EpsE, was recently reported (27). With a tetracysteine motif and 42% pairwise identity, the C-terminal domain of EpsE more closely resembles PilB than PilT (Fig. 1). However, after the importance of the alanine, leucine, and isoleucine residues in the AIRNLIRE sequence was revealed by this mutagenesis study, we noted that I293 of P. aeruginosa PilT aligns with I458 of EpsE (Fig. 1) (27). Consistent with our model for a functional helix formed by the AIRNLIRE motif, I458 of EpsE is found within the short amphipathic α helix K (27). α helix K has a solvent-exposed face toward the lumen of the EpsE hexameric ring. I458 emanates from the opposite, more hydrophobic face, bridging the C-terminal domain and the ATPase core through several hydrophobic packing interactions. L454 of EpsE, which aligns with I289 of PilT, is located in the same hydrophobic pocket. Therefore, in EpsE these hydrophobic residues maintain proper positioning of the C-terminal domain in relation to the ATPase core and may act as sensors of conformational changes, allowing the transmission of information between the lumenal surface and the core ATPase domain. It is reasonable to assume the same relative structural arrangement in PilT.
In summary, our data verify the in vivo requirement for an intact AIRNLIRE motif for the function of PilT and rule out the direct role of these amino acids in catalytic activity. All evidence is in agreement with a model in which this motif of PilT adopts an amphipathic helical structure and acts as a mediator between the lumenal surface and the NTPase core. Structural, biochemical, and genetic investigations of PilT will provide mechanistic details to confirm or refute this model. Future experiments are required to determine whether the AIRNLIRE region is a protein-protein interaction surface and, if so, with which protein(s) it interacts in vivo.
Supplementary Material
[Supplemental material]
Acknowledgments
The work described in this article was funded by NIH (GM59721) and W. M. Keck Foundation grants to K.T.F.
We thank Joanne Engel, Stephen Lory, and Michael Koomey for gifts of reagents and Amber Pollack for creating our first phenotypically defective P. aeruginosa PilT variant, I293N/E295Q.
Footnotes
REFERENCES
- 1.Bhattacharjee, M. K., S. C. Kachlany, D. H. Fine, and D. H. Figurski. 2001. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J. Bacteriol. 183**:**5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280**:**2114-2118. [DOI] [PubMed] [Google Scholar]
- 3.Bradley, D. E. 1972. Shortening of Pseudomonas aeruginosa pili after RNA-phage adsorption. J. Gen. Microbiol. 72**:**303-319. [DOI] [PubMed] [Google Scholar]
- 4.Chiang, P., and L. L. Burrows. 2003. Biofilm formation by hyperpiliated mutants of Pseudomonas aeruginosa. J. Bacteriol. 185**:**2374-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou, P. Y., and G. D. Fasman. 1974. Prediction of protein conformation. Biochemistry 13**:**222-245. [DOI] [PubMed] [Google Scholar]
- 6.Comolli, J. C., A. R. Hauser, L. Waite, C. B. Whitchurch, J. S. Mattick, and J. N. Engel. 1999. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect. Immun. 67**:**3625-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crowther, L. J., R. P. Anantha, and M. S. Donnenberg. 2004. The inner membrane subassembly of the enteropathogenic Escherichia coli bundle-forming pilus machine. Mol. Microbiol. 52**:**67-79. [DOI] [PubMed] [Google Scholar]
- 8.Darzins, A. 1993. The pilG gene product, required for Pseudomonas aeruginosa pilus production and twitching motility, is homologous to the enteric, single-domain response regulator CheY. J. Bacteriol. 175**:**5934-5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forest, K. T., K. A. Satyshur, G. A. Worzalla, J. K. Hansen, and T. J. Herdendorf. 2004. The pilus-retraction protein PilT: ultrastructure of the biological assembly. Acta Crystallogr. Sect. D 60**:**978-982. [DOI] [PubMed] [Google Scholar]
- 10.Hahn, H. P. 1997. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas _aeruginosa—_a review. Gene 192**:**99-108. [DOI] [PubMed] [Google Scholar]
- 11.Herdendorf, T. J., D. R. McCaslin, and K. T. Forest. 2002. Aquifex aeolicus PilT, homologue of a surface motility protein, is a thermostable oligomeric NTPase. J. Bacteriol. 184**:**6465-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212**:**77-86. [DOI] [PubMed] [Google Scholar]
- 13.Liu, P. V. 1966. The roles of various fractions of Pseudomonas aeruginosa in its pathogenesis. II. Effects of lecithinase and protease. J. Infect. Dis. 116**:**112-116. [DOI] [PubMed] [Google Scholar]
- 14.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56**:**289-314. [DOI] [PubMed] [Google Scholar]
- 15.Mattick, J. S., M. M. Bills, B. J. Anderson, B. Dalrymple, M. R. Mott, and J. R. Egerton. 1987. Morphogenetic expression of Bacteroides nodosus fimbriae in Pseudomonas aeruginosa. J. Bacteriol. 169**:**33-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McRee, D. E. 1999. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125**:**156-165. [DOI] [PubMed] [Google Scholar]
- 17.Merz, A. J., and K. T. Forest. 2002. Bacterial surface motility: slime trails, grappling hooks and nozzles. Curr. Biol. 12**:**R297-R303. [DOI] [PubMed] [Google Scholar]
- 18.Merz, A. J., and M. So. 2000. Interactions of pathogenic Neisseriae with epithelial cell membranes. Annu. Rev. Cell Dev. Biol. 16**:**423-457. [DOI] [PubMed] [Google Scholar]
- 19.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407**:**98-102. [DOI] [PubMed] [Google Scholar]
- 20.Morand, P. C., E. Bille, S. Morelle, E. Eugene, J. L. Beretti, M. Wolfgang, T. F. Meyer, M. Koomey, and X. Nassif. 2004. Type IV pilus retraction in pathogenic Neisseria is regulated by the PilC proteins. EMBO J. 23**:**2009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okamoto, S., and M. Ohmori. 2002. The cyanobacterial PilT protein responsible for cell motility and transformation hydrolyzes ATP. Plant Cell Physiol. 43**:**1127-1136. [DOI] [PubMed] [Google Scholar]
- 22.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30**:**295-304. [DOI] [PubMed] [Google Scholar]
- 23.Park, H. S., M. Wolfgang, and M. Koomey. 2002. Modification of type IV pilus-associated epithelial cell adherence and multicellular behavior by the PilU protein of Neisseria gonorrhoeae. Infect. Immun. 70**:**3891-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Planet, P. J., S. C. Kachlany, R. DeSalle, and D. H. Figurski. 2001. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. USA 98**:**2503-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Priefer, U. B., R. Simon, and A. Puhler. 1985. Extension of the host range of Escherichia coli vectors by incorporation of RSF1010 replication and mobilization functions. J. Bacteriol. 163**:**324-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pujol, C., E. Eugene, M. Marceau, and X. Nassif. 1999. The Meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. Proc. Natl. Acad. Sci. USA 96**:**4017-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robien, M. A., B. E. Krumm, M. Sandkvist, and W. G. Hol. 2003. Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae. J. Mol. Biol. 333**:**657-674. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Semmler, A. B., C. B. Whitchurch, and J. S. Mattick. 1999. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 145**:**2863-2873. [DOI] [PubMed] [Google Scholar]
- 30.Sexton, J. A., J. S. Pinkner, R. Roth, J. E. Heuser, S. J. Hultgren, and J. P. Vogel. 2004. The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J. Bacteriol. 186**:**1658-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeya, K., and K. Amako. 1966. A rod-shaped Pseudomonas phage. Virology 28**:**163-165. [DOI] [PubMed] [Google Scholar]
- 32.Turner, L. R., J. C. Lara, D. N. Nunn, and S. Lory. 1993. Mutations in the consensus ATP-binding sites of XcpR and PilB eliminate extracellular protein secretion and pilus biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 175**:**4962-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1**:**945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32**:**1-10. [DOI] [PubMed] [Google Scholar]
- 35.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148**:**81-86. [DOI] [PubMed] [Google Scholar]
- 36.Whitchurch, C. B., M. Hobbs, S. P. Livingston, V. Krishnapillai, and J. S. Mattick. 1991. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene 101**:**33-44. [DOI] [PubMed] [Google Scholar]
- 37.Whitchurch, C. B., A. J. Leech, M. D. Young, D. Kennedy, J. L. Sargent, J. J. Bertrand, A. B. Semmler, A. S. Mellick, P. R. Martin, R. A. Alm, M. Hobbs, S. A. Beatson, B. Huang, L. Nguyen, J. C. Commolli, J. N. Engel, A. Darzins, and J. S. Mattick. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 52**:**873-893. [DOI] [PubMed] [Google Scholar]
- 38.Whitchurch, C. B., and J. S. Mattick. 1994. Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa. Mol. Microbiol. 13**:**1079-1091. [DOI] [PubMed] [Google Scholar]
- 39.Wolfgang, M., P. Lauer, H. S. Park, L. Brossay, J. Hebert, and M. Koomey. 1998. pilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29**:**321-330. [DOI] [PubMed] [Google Scholar]
- 40.Wu, S. S., J. Wu, and D. Kaiser. 1997. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23**:**109-121. [DOI] [PubMed] [Google Scholar]
- 41.Zolfaghar, I., D. J. Evans, and S. M. Fleiszig. 2003. Twitching motility contributes to the role of pili in corneal infection caused by Pseudomonas aeruginosa. Infect. Immun. 71**:**5389-5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]