5-Formylcytosine can be a stable DNA modification in mammals (original) (raw)
. Author manuscript; available in PMC: 2017 Jun 27.
Published in final edited form as: Nat Chem Biol. 2015 Jun 22;11(8):555–557. doi: 10.1038/nchembio.1848
Abstract
5-Formylcytosine (5fC) is a rare base found in mammalian DNA and thought to be involved in active DNA demethylation. Here, we show that developmental dynamics of 5fC levels in mouse DNA differ from those of 5-hydroxymethylcytosine (5hmC), and using stable isotope labeling in vivo, we show that 5fC can be a stable DNA modification. These results suggest that 5fC has functional roles in DNA that go beyond being a demethylation intermediate.
DNA of all mammalian cells and tissues is methylated at specific loci, mainly in the 5′-cytosine-phosphate-guanine-3′ (CpG) context, to modulate the expression of genes1. 5-Methylcytosine (5mC) is produced from cytosine (C) by dedicated DNA methyltransferases using _S_-adenosylmethionine (SAM) as a source of the methyl group2. In 2009, two independent laboratories found that 5-hydroxymethylcytosine (5hmC) is present in mammalian DNA and is the product of ten-eleven translocation (TET) enzyme–mediated oxidation of 5mC3,4. This oxidized base occurs in all mammalian cells and tissues with global levels ranging between 0.005% and 0.7% of all Cs5,6. The iron(II)- and 2-oxoglutarate-dependent TET enzymes can also oxidize 5hmC further to 5fC and 5-carboxycytosine (5caC), which were found at levels below 0.002% (or 20 p.p.m.) of all Cs in the genomic DNA of mouse embryonic stem (mES) cells and several adult mouse tissues7–9.
One proposed role for these oxidized cytosine bases is to serve as intermediates of enzyme-mediated DNA demethylation initiated by oxidation of 5mC10,11. Indeed, thymine-DNA glycosylase (TDG) can selectively recognize and excise 5fC and 5caC from the genome and trigger a repair process, which can lead to restoration of unmodified C7,12. Moreover, mES cells lacking TDG show increased levels of 5fC and 5caC, suggesting that a proportion of these modifications is constantly being removed from the genome of mES cells7,12. On the other hand, we recently demonstrated that 5hmC is a predominantly stable modification in mammalian DNA, especially in the adult mouse brain where 5hmC is most abundant6. Herein we investigate the temporal dynamics of 5fC in genomic DNA in vivo to consider whether this rare modification may be stable rather than an active demethylation intermediate (Fig. 1a).
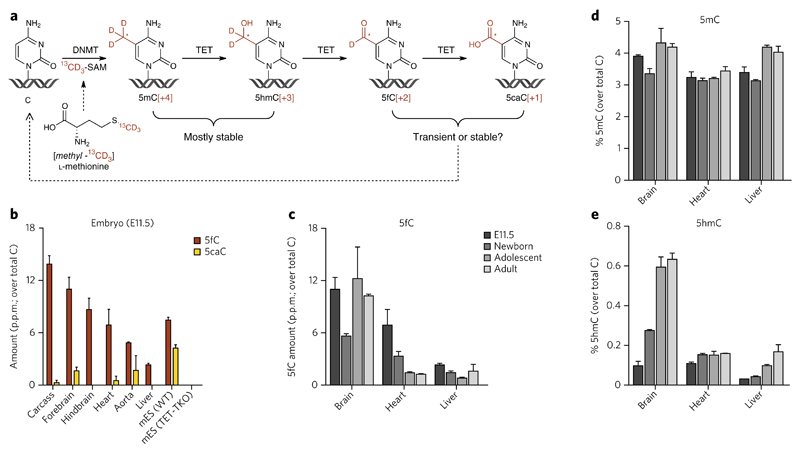
Figure 1. Dynamics of global levels of 5fC during mouse development are distinct from those of 5hmC.
(a) Metabolism of cytosine modifications in DNA. Although the majority of 5mC and 5hmC was known to persist in the genomic DNa, the stability of 5fC and 5caC in vivo was unknown. Feeding [_methyl_-13CD3]l-methionine can be used as a means to measure the lifetime of cytosine modifications in cells and in vivo. Labeling pattern is indicated in red. See also Figure 2. (b) Global levels of 5fC and 5caC in genomic DNA from mouse embryos (embryonic day 11.5, E11.5). Shown are mean ± s.e.m. of three animals. Each sample was analyzed in technical duplicate and the mean value was used. WT, wild type. (c–e) Changes of global 5fC (c), 5mC (d) and 5hmC (e) levels during development in selected C57Bl/6 mouse tissues (further data in Supplementary Fig. 6). shown are mean ± s.e.m. of three embryos (E11.5) (data from Fig. 1b), three newborns (1 d old), and two adolescent (21 d old) and two adult (15 week old) mice. see also Supplementary Figures 7 and 8.
We first analyzed global levels of all cytosine modifications in the genomic DNA of C57BL/6 mouse tissues to see whether we could detect and quantify 5fC and identify a relationship between its levels and those of its precursors 5mC and 5hmC or its metabolite 5caC. We included a range of postnatal tissues from newborn (1 d old), adolescent (21-d-old) and adult (15-week-old) mice, as their genomic DNA is known to have different levels of 5hmC depending on the overall proliferation rate (and therefore the age) of the tissue6. We also included C57BL/6 embryos at 11.5 d post-fertilization as this is the lethal age for mice lacking TDG13,14, and mES cells derived from the same strain were added for comparison. To achieve quantification of the rare modifications (5fC and 5caC) with the highest possible sensitivity and accuracy, we employed a nano high-performance liquid chromatography–tandem high-resolution mass spectrometry (nanoHPLC-MS/HRMS) method, which is able to resolve genuine rare modified bases (5fC and 5caC) from potential impurities of the same nominal mass and retention time, and can detect down to 0.1 p.p.m. of total Cs in as little as 100 ng of digested genomic DNA. In addition, the use of isotopically labeled internal standards (IS) of C, 5mC and 5hmC substantially improved the quality of the measurements, ensured excellent reproducibility between technical replicates and excluded spontaneous oxidation of 5hmC as the source of 5fC or 5caC. Example mass spectra, extracted ion chromatograms and calibration curves are shown in Supplementary Results, Supplementary Figures 1–5. 5mC and 5hmC were present in all tissues (Supplementary Fig. 6), and the levels that we detected were in good agreement with available published data5,6,8,9. While 5mC levels show a relatively uniform distribution between tissues, global 5hmC content is highly correlated to the proportion of proliferating cells in the tissue, as we have shown previously6. We found 5fC to be also present in all studied tissues at levels ranging between 0.2 p.p.m. and 15 p.p.m. of all Cs (Fig. 1b and Supplementary Fig. 6). Notably, 5caC was not detected in any postnatal tissues from C57BL/6 mice, even in those with high 5fC content, but several tissues from C57BL/6 embryos (Fig. 1b) and adult (12 week old) CD1 mice (Supplementary Fig. 7) contained up to 2 p.p.m. of this rare DNA base modification. Overall, we found no correlation between the levels of 5fC and the levels of its precursors 5mC or 5hmC (Supplementary Fig. 8), nor did we find any clear pattern of DNA modification changes as the tissues age. They can retain the levels of 5fC while gaining 5hmC (e.g., brain), lose 5fC while retaining the levels of 5hmC (e.g., heart) or even lose 5fC while gaining 5hmC (e.g., liver) (Fig. 1c–e). We found that DNA from mES cells lacking all three TET enzymes (TET triple-knockout (TET-TKO))15 contained no detectable 5hmC, 5fC or 5caC (Fig. 1a and Supplementary Fig. 6), confirming that 5hmC is the only source of 5fC and 5caC in mES cell DNA. Although we have no measure of tissue-specific susceptibility to oxidation (such as the quantity of the oxidative lesion 8-oxoguanine), the lack of correlation between global levels of 5hmC and 5fC (Supplementary Fig. 8) together with the lack of positional overlap between 5hmC and 5fC in mES cells16,17 strongly suggest that 5fC and 5caC are not generated by spontaneous oxidation of 5hmC and 5fC.
To elucidate the stability of 5fC in genomic DNA toward turnover in vivo, we applied a stable isotope tracing method consisting of feeding cultured cells and mice with [_methyl_-13CD3]l-methionine, as we have done previously to study the lifetime of 5hmC6. The _methyl_-13CD3 group enters the intracellular pool of SAM and is transferred into newly methylated Cs by the action of DNA methyltransferase (DNMT) enzymes. The TET enzymes can then convert labeled [_methyl_-13CD3]5mC (5mC[+4]) into [_hydroxymethyl_-13CD2]5hmC (5hmC[+3]) and [_formyl_-13CD]5fC (5fC[+2]) (Fig. 1a). The labeling ratios (e.g., % 5fC[+2] over total 5fC) change according to the dynamics and half-life of the given modification in the genomic DNA. For example, a modification that is quickly turning over in DNA would show a high labeling ratio, whereas a very stable modification would show no labeling in nonproliferating cells or tissues. The maximum obtainable ratio also depends on the activity of other biosynthetic pathways feeding into the one-carbon metabolism. The labeling ratios can be determined very accurately for each modified C using LC-MS/HRMS due to unique masses of the labeled base fragments (Supplementary Fig. 9 and Supplementary Table 1).
We first cultured mES cells in the labeled ([_methyl_-13CD3] l-methionine) medium for 8 d, and found that the labeling ratio of 5fC increased much more slowly than that of 5mC and 5hmC. This indicates either a substantial time lag in making 5fC from newly formed 5hmC or the presence of a population of more slowly dividing or nondividing (unlabeled) cells with higher global levels of 5fC than the quickly dividing (labeled) population of mES cells (Fig. 2a).
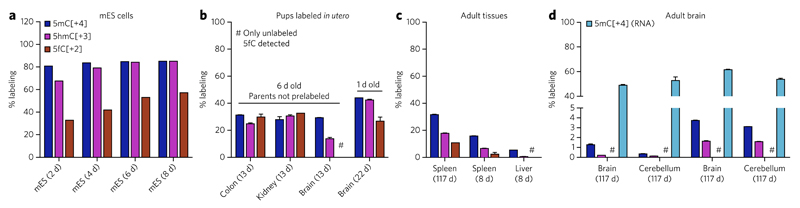
Figure 2. 5fC can be a stable DNA modification in vivo.
(a) Labeling ratios of 5mC, 5hmC and 5fC in the genomic DNA of mEs cells cultured in the presence of [_methyl_-13CD3]l-methionine. Shown are single measurements; total labeling time is given in brackets. (b–d) Labeling ratios of 5mC, 5hmC and 5fC in the genomic DNA of C57Bl/6 mice fed with the [_methyl_-13CD3]l-methionine diet. (b) 6-d-old pups labeled from 1 week before birth (total labeling time of 13 d) or 1-d-old newborns (total labeling time of 22 d, parents on labeled diet for 52 d before conception). Shown are mean ± s.e.m. of two animals (6 d old pups) or two technical replicates (1 d old pup). (c,d) Mice labeled in adulthood. Shown are mean ± s.e.m. of at least two technical replicates from individual mice, and total labeling time is shown in brackets. The absence of 5fC[+2] in the brain (d), where 5fC is most abundant (see Fig. 1 and Supplementary Fig. 6), indicates minimal or no further generation of 5fC once placed in postmitotic tissues. Moreover, if 5fC was involved in cycles of methylation and demethylation, its labeling ratio would be similar to that of 5mC in RNA (d). See also Supplementary Figure 9 and Supplementary Table 1.
We then analyzed genomic DNA from C57BL/6 mice fed with a diet in which all l-methionine was replaced with [_methyl_-13CD3] l-methionine. To gain information about 5fC in developing tissues, we fed a pregnant female the labeled diet starting from 7 d before birth, and kept the family on the diet for 6 more days (so that the 6-d-old pups had been labeled for 13 d when their tissues were harvested). The genomic DNA in tissues such as kidney or colon showed uniform labeling of around 30% for all detectable modifications (5mC, 5hmC and 5fC) (Fig. 2b). However, brain tissue from the same pups showed much less 5hmC[+3] and no detectable 5fC[+2]. This indicates that 5fC was formed in these tissues before the start of labeling and remained there for 13 d until the DNA was harvested. 1-d-old newborns labeled from conception and with prelabeled parents (starting 52 d before conception, so that total labeling time of pup was 22 d) already showed higher 5fC labeling in the brain, but the ratio was still lower than those of 5mC and 5hmC (27% vs. 44% and 42.5%, respectively) (Fig. 2b). During the gestation period, the labeling ratio of the methionine pool in the pregnant female was still increasing, and therefore this observation is consistent with 5fC being more abundant in the older or more slowly proliferating DNA, as we concluded above. Proliferating tissues (e.g., spleen) from adult mice showed a similar trend in which the 5fC labeling ratio was always smaller than that of 5mC or 5hmC, even in animals labeled for as long as 4 months (117 d) (Fig. 2c). This effect is best explained by 5fC being mostly stable, and again by the presence of nondividing (unlabeled) cells alongside a population of proliferating (labeled) cells that have lower global 5fC levels than the nondividing cells.
Finally, in the mostly nondividing adult brain, where 5fC is most abundant (see Fig. 1 and Supplementary Fig. 6) and only 1.3% and 3.7% of 5mC becomes labeled during the 117-d feeding period, there was no detectable labeled 5fC. Again, this is not due to the lack of an intracellular pool of [_methyl_-13CD3]SAM, as we could measure more than 50% [_methyl_-13CD3]5mC in RNA in adult brain and cerebellum (Fig. 2d). If 5fC was a short-lived DNA modification and was constantly being turned over, its labeling ratio would be close to the labeling ratio of intracellular SAM and 5mC in RNA. If 5fC was short-lived and produced only from preexisting unlabeled 5hmC in the adult brain, the levels of 5hmC would be depleted over time, which is inconsistent with the high levels of 5hmC in this tissue and is the opposite of what has been described for aging brain18. Therefore, the lack of 5fC labeling in the adult brain means that this modified base must be stable in the genome as opposed to generally acting as a dynamic intermediate of active DNA demethylation.
In summary, we present the first direct evidence that 5fC (derived from mC by TET-mediated oxidation) can be a stable DNA modification in vivo, and provide quantitative measurements of the levels of all modified Cs in mouse tissues across several developmental stages. 5fC levels do not correlate with those of its precursors 5mC and 5hmC or its metabolite 5caC or with the age of the individual. Although there is precedent for removal of 5fC and 5caC from the genome (for example, in mES cells), probably in the process of active DNA demethylation7,12, our findings suggest that the bulk of 5fC can be stable. 5fC has been identified as having more protein binders than 5mC or 5hmC19,20 and having a genomic profile distinct from those of 5mC, 5hmC or 5caC at single-base resolution12,16,17,21–23. Moreover, 5fC has recently been shown to alter the structure of the DNA double helix24. We therefore conclude that such stably 5fC-modified DNA could have profound consequences for the regulation of gene expression that may be distinct to those caused by 5mC and 5hmC. However, direct evidence regarding the biological function of 5fC remains to be demonstrated.
Online Methods
Animals
All in vivo experiments were performed under the terms of a UK Home Office license. C57BL/6 and CD1 mice were bred and housed according to UK Home Office guidelines. Custom l-methionine-free mouse diet supplemented with [_methyl_-13CD3]l-methionine (Sigma) was manufactured by TestDiet.
Cell culture
mES cells were derived by X. Zou at the CRUK Cambridge Institute from a C57BL/6 mouse (Charles River) and cultured on a gelatin-coated plate in a DMEM-KO medium (Invitrogen) supplemented with 10% FCS, MEM non-essential amino acids, glutamine, sodium pyruvate, penicillin, streptomycin, mouse leukemia inhibitory factor (mLIF) and 2i (mitogen-activated protein kinase kinase (MEK) and glycogen synthase kinase (GSK-3β) inhibitors) as described by Ying et al.25. TET-TKO mES cells were obtained from G. Xu15 and cultured in the 2i conditions as above. All cells were regularly tested for mycoplasma contamination. For isotopic labeling experiments, cells were maintained in a custom l-methionine-free DMEM-KO medium (Invitrogen) supplemented with 30 mg/L of [_methyl_-13CD3]l-methionine (Cambridge Isotope) and the respective components above.
Genomic DNA extraction
Tissues and cells were resuspended in lysis buffer (100 mM Tris, pH 5.5, 5 mM EDTA, 200 mM NaCl, 0.2% SDS) supplemented with 400 μg/ml proteinase K (Invitrogen), and were incubated at 55 °C overnight. DNA was purified using phenol:chloroform:isoamyl alcohol (25:24:1, Sigma) and Phase Lock Gel (5 Prime), precipitated from 70% ethanol and resuspended in ultrapure HPLC-grade water.
DNA degradation to 2′-deoxynucleosides and LC-MS analysis
1–2 μg of DNA was incubated with 5 U of DNA Degradase Plus (Zymo Research) in a total volume of 30 μl for 4 h at 37 °C. Samples were filtered through a pre-washed Amicon 10 kDa centrifugal filter unit (Millipore) before LC-MS analysis.
LC-MS analysis of global 5mC, 5hmC, 5fC and 5caC levels
Analysis of global levels of 5mC, 5hmC, 5fC and 5caC was performed on a Q Exactive mass spectrometer (Thermo) fitted with an UltiMate 3000 RSLCnano HPLC (Dionex) and a self-packed hypercarb column (20 mm × 75 μm, 3 μm particle size) at a flow rate of 0.75 μl/min and a gradient of 0.1% formic acid in water and acetonitrile. Calibration curves were generated using a mixture of synthetic standards 2′-deoxycytidine (Sigma), 5-methyl-, 5-hydroxymethyl-, 5-formyl- and 5-carboxy-2′-deoxycytidine (Berry & Associates), in the ranges of 0.5 nM–5 μM for C, 0.025–50 nM for 5mC, and 0.005–50 nM for 5hmC, 5fC and 5caC. Samples and synthetic standards were spiked with an isotopically labeled mix containing 100 nM of 2′-deoxycytidine-(15N,d2) (synthesis and characterization in Bachman et al.6), 5-methyl-2′-deoxycytidine-(d3) and 5-hydroxymethyl-2′-deoxycytidine-(d3) (both from Toronto Research Chemicals). Target ions were fragmented in a positive ion mode at 10% normalized collision energy, and full scans (50–300 Da) were acquired. The inclusion list contained the following masses: C (228.1), C_IS (231.1), 5mC (242.1), 5mC_IS (245.1), 5hmC (258.1), 5hmC_IS (261.1), 5fC (256.1), 5caC (272.1). Extracted ion chromatograms of base fragments (see Supplementary Fig. 1) were used for quantification. Results are expressed as a % or p.p.m. of total cytosines.
LC-MS analysis of isotope incorporation into genomic DNA
Analysis of isotope incorporation into DNA was performed using the same instrumental setup as above, targeting ions of masses 242.1 (mC), 246.1 (mC[+4]), 258.1 (5hmC and 5fC[+2]), 261.1 (5hmC[+3]) and 256.1 (5fC). Extracted ion chromatograms of base fragments were used for quantification of labeling ratios (see also Supplementary Fig. 8). Results are expressed as % labeling (for example, % 5fC[+2] represents the percentage of labeled 5fC[+2] in total 5fCs).
LC-MS analysis of isotope incorporation into 5mC in RNA
Total RNA was carried through during genomic DNA extraction (no RNase treatment), during hydrolysis to nucleosides and during LC-MS/HRMS analysis. An additional mass of 262.1 was targeted for RNA 5mC[+4] (unlabeled 5mC was present in the 258.1 channel), and base fragments 126.0662 (5mC) and 130.0884 (5mC[+4]) were used for quantification of % labeling as above.
Supplementary Material
Supplementary information is available in the online version of the paper. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html. Correspondence and requests for materials should be addressed to S.B.
Supplementary Information
Acknowledgments
We thank C. d’Santos and D. Oxley for their support with mass spectrometry and G. Xu for kindly providing TET-TKO mES cells. This work was supported by Cancer Research UK (C14303/A17197, S.B.), The Wellcome Trust (WT099232, S.B.; WT095645/Z/11/Z, W.R.) and the Biotechnology and Biological Sciences Research Council UK (BB/K010867/1, W.R.).
Footnotes
Author contributions
M.B., S.U.-L. and S.B. conceived the study; S.U.-L., M.B., H.E.B. and M.I. performed experiments; M.B. and X.Y. carried out mass spectrometry and data analysis; S.B., A.M. and W.R. supervised the project; M.B. and S.B. wrote the manuscript with contributions from all authors.
References
- 1.Ooi SK, O’Donnell AH, Bestor TH. J Cell Sci. 2009;122:2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goll MG, Bestor TH. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 3.Kriaucionis S, Heintz N. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tahiliani M, et al. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Globisch D, et al. PLoS ONE. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachman M, et al. Nat Chem. 2014;6:1049–1055. doi: 10.1038/nchem.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He YF, et al. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito S, et al. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfaffeneder T, et al. Nat Chem Biol. 2014;10:574–581. doi: 10.1038/nchembio.1532. [DOI] [PubMed] [Google Scholar]
- 10.Song CX, He C. Trends Biochem Sci. 2013;38:480–484. doi: 10.1016/j.tibs.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schiesser S, et al. J Am Chem Soc. 2013;135:14593–14599. doi: 10.1021/ja403229y. [DOI] [PubMed] [Google Scholar]
- 12.Neri F, et al. Cell Reports. 2015;10:674–683. doi: 10.1016/j.celrep.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Cortázar D, et al. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 14.Cortellino S, et al. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu X, et al. Cell Stem Cell. 2014;14:512–522. doi: 10.1016/j.stem.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Booth MJ, Marsico G, Bachman M, Beraldi D, Balasubramanian S. Nat Chem. 2014;6:435–440. doi: 10.1038/nchem.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song CX, et al. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraus TF, Guibourt V, Kretzschmar HA. J Neural Transm. 2014 Dec 4; doi: 10.1007/s00702-014-1346-4. [DOI] [PubMed] [Google Scholar]
- 19.Iurlaro M, et al. Genome Biol. 2013;14:R119. doi: 10.1186/gb-2013-14-10-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spruijt CG, et al. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Booth MJ, et al. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 22.Yu M, et al. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu X, et al. Cell Res. 2015;25:386–389. doi: 10.1038/cr.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raiber EA, et al. Nat Struct Mol Biol. 2015;22:44–49. doi: 10.1038/nsmb.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ying QL, et al. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information