Rebuilding Chromosomes After Catastrophe: Emerging Mechanisms of Chromothripsis (original) (raw)
. Author manuscript; available in PMC: 2018 Dec 1.
Published in final edited form as: Trends Cell Biol. 2017 Sep 9;27(12):917–930. doi: 10.1016/j.tcb.2017.08.005
Abstract
Cancer genome sequencing has identified chromothripsis, a complex class of structural genomic rearrangements involving the apparent shattering of an individual chromosome into tens to hundreds of fragments. An initial error during mitosis, producing either chromosome missegregation into a micronucleus or chromatin bridge interconnecting two daughter cells, can trigger the catastrophic pulverization of the spatially isolated chromosome. The resultant chromosomal fragments are re-ligated in random order by DNA double-strand break repair during the subsequent interphase. Chromothripsis scars the cancer genome with localized DNA rearrangements that frequently generate extensive copy number alterations, oncogenic gene fusion products, and/or tumor suppressor gene inactivation. Here we review emerging mechanisms underlying chromothripsis with a focus on the contribution of cell division errors caused by centromere dysfunction.
Keywords: chromothripsis, genomic instability, chromosome rearrangements, mitosis, micronuclei, DNA repair
Hidden in Plain Sight: Chromothripsis in the Cancer Genome
The karyotypes of cancer cells are often remarkably complex – littered not only with mutations but also small- and large-scale changes in both chromosome number and architecture. Copy number alterations in the form of whole-chromosome or segmental aneuploidy are present in the vast majority of tumors, yet its role as a cause or consequence of cancer development remains under debate [1, 2]. Structural aberrations and gross rearrangements alter the linear organization of chromosomes, and in some instances can directly drive tumorigenesis. The Philadelphia chromosome in chronic myelogenous leukemia is the classic example [3] involving a translocation between chromosomes 9 and 22 to generate an oncogenic gene fusion product that can be effectively targeted by clinical therapeutics [4].
Although it appeared firmly established that tumorigenesis develops through the gradual and sequential accumulation of genetic and/or epigenetic changes [5], recent cancer genome sequencing efforts challenged this dogmatic view by identifying several new classes of mutational processes that form simultaneously in a single burst. Adapted from the age-old theory in evolution, the paradigm-shifting concept of punctuated equilibrium (see “Glossary”) has now been applied toward our understanding of cancer progression models, as recently described in the renewed context of pancreatic cancer [6]. These punctuated events currently include alterations such askataegis (regions of clustered hypermutation) [7, 8] and chromoplexy (serial rearrangements linking multiple chromosomes) [9] – the underlying bases of which are not well understood.
The most striking example of punctuated equilibrium ischromothripsis (a Greek neologism for ‘chromosome shattering’), in which tens to hundreds of structural rearrangements are rapidly acquired within a short timeframe [10]. These complex rearrangements are curiously restricted to one or a few chromosome(s) with breakpoints scattered across an entire chromosomal axis or clustered within a specific arm or region. Such alterations were predicted to form de novo from massive DNA damage and repair events, leading to a derivative chromosome that shares little resemblance to its original configuration. The characteristic mutation signature for chromothripsis [11] has now been detected in a broad spectrum of cancers, and of particular interest, at high frequency in specific tumor types – including bone, blood, and brain cancers [6, 12–17]. Although the majority of chromothripsis cases occur somatically, inherited forms of germline chromothripsis have also been documented [18]. Additionally, similar types of complex rearrangements have been reported in individuals with developmental and/or cognitive disorders, which appear to be caused by DNA replication errors [19] in a process termed chromoanasynthesis. The catchall phrasechromoanagenesis (Greek for ‘to be reborn’) has been coined [20] to encompass all the possible types of localized and complex chromosomal rearrangements in human genomes irrespective of their underlying mechanism of formation.
If chromothripsis causes such drastic changes in chromosome structure and occurs frequently in some cancers, why had it not been discovered prior to 2011? Several types of large-scale structural rearrangements, such as gross translocations, can be easily identified by routine cytogenetic methodologies involving chromosome-banding patterns or fluorescent _in situ_hybridization. However, submicroscopic-scale structural variations in the range of kilobase- to even megabase-sized rearrangements or more complex abnormalities, such as chromothripsis, can easily escape recognition. Thus, initially missed by microscopy-based approaches, the recent advent of higher resolution next-generation DNA sequencing technologies enabled Stephens et al. [10] to identify and resolve the complexity of rearrangements characterizing chromothripsis in cancer genomes.
The highly localized and complex nature of chromothripsis initially puzzled both cell biologists and cancer geneticists, leading to a spectrum of proposed hypotheses for the underlying mechanism(s) [10, 20–23]. Considering that the rearrangements are often restricted to a single chromosome, it was strongly suspected that the affected chromosome must have been spatially isolated from the remaining genome, even if only for a transient period. DNA damage in the form of double-strand breaks (DSBs) resulting in chromosome breakage is also likely involved followed by one or more mechanism(s) of error-prone DNA repair to produce the resulting rearrangements [10]. Many key aspects regarding the cellular mechanisms have emerged over the last five years, including evidence supporting a role of cell division errors in the shattering of an initially missegregated chromosome [24–26]. In this review, we cover recent insights into the mechanisms of chromothripsis with a particular focus on the role of mitotic errors driven by centromere dysfunction. We also highlight a number of outstanding questions.
Chromothripsis Driving Tumorigenesis
How does chromothripsis contribute to cancer development? The simultaneous formation of a multiple alterations through chromothripsis can lead to the acquisition of one or more selective advantages (Figure 1A). Because chromothripsis can result in both the loss of DNA segments and the formation of de novo rearrangements, two obvious culprits are the disruption of tumor suppressor genes and the formation of oncogenic fusion products, respectively. Rearrangements formed between two normally distant loci may also juxtapose an active regulatory element (e.g., a promoter) adjacent or in close proximity to an otherwise repressed oncogene. Cancer genome sequencing efforts have indeed documented numerous examples of tumor suppressor loss [10], gene fusion events [14], and perturbed regulatory elements [15, 27] associated with chromothripsis in human malignancies (Box 1).
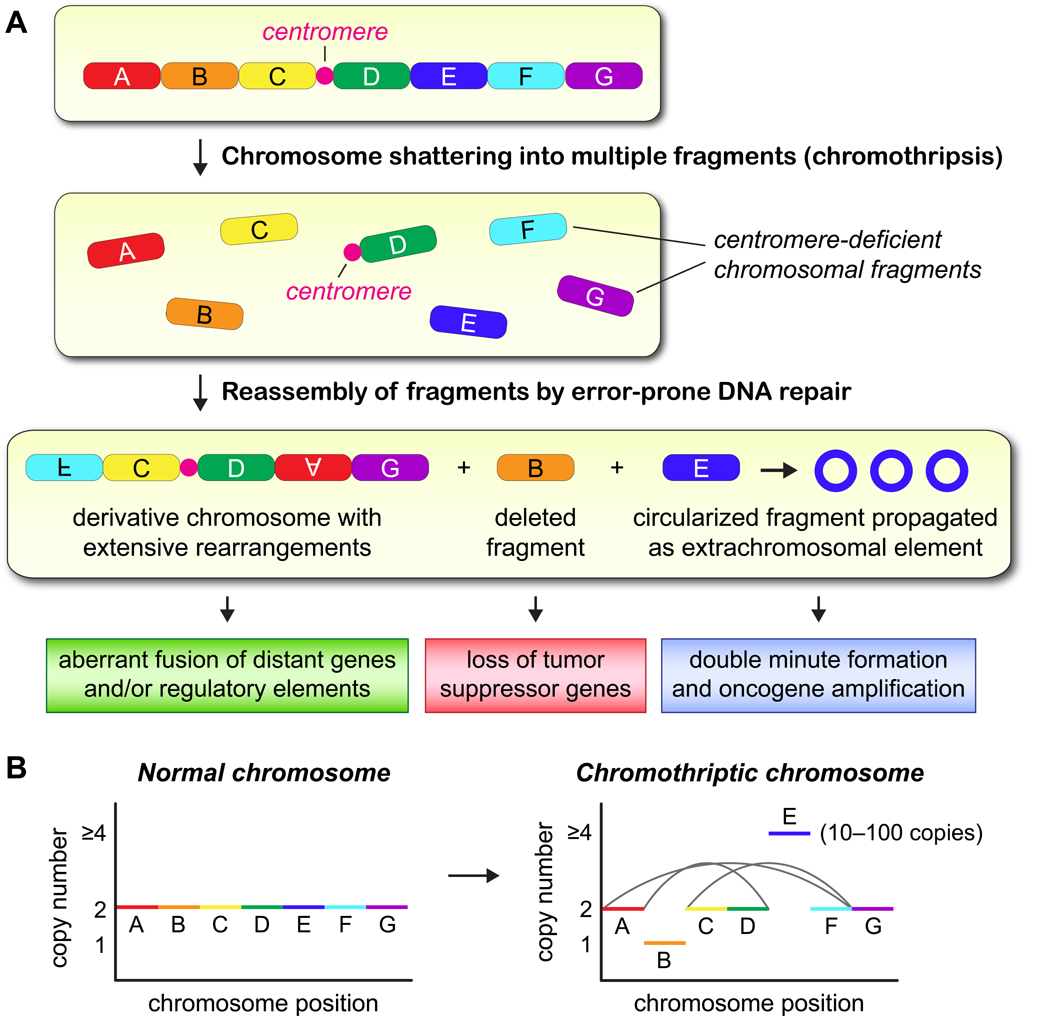
Figure 1. Genomic and tumorigenic consequences of chromothripsis.
(A) The shattering of an individual chromosome can produce tens to hundreds of acentric DNA fragments that persist as intermediates until they are re-ligated and stabilized by intrinsic DNA repair mechanisms. These fragments reassemble to form a scrambled, derivative chromosome containing multiple rearrangements (chromothripsis), become lost, and/or self-ligate into circular DNA structures called double minutes. (B) Chromothriptic events can give rise to a characteristic mutation signature that has been detected in a broad range of cancer genomes, including oscillating copy number states and complex patterns of intrachromosomal rearrangements in apparently random fashion.
Box 1. Three case examples of chromothripsis driving genomic changes associated with cancer and disease.
Pancreatic cancer is thought to develop from an initiating clone that gradually accumulates oncogenic mutations over relatively long timescales to eventually acquire the capacity to disseminate to distant organs and become metastatic. However, the majority of pancreatic cancer patients are asymptomatic and not diagnosed until the tumor has reached the metastatic stage – an endpoint with extremely poor clinical outcome. This observation has challenged the conventional stepwise paradigm of pancreatic cancer progression and instead raises an alternative model: the rapid acceleration of early pancreatic lesions toward metastatic disease occurs through punctuated events that simultaneously drive multiple oncogenic changes. Sequencing of 107 pancreatic cancer genomes revealed that, along with genome duplication events (polyploidy), chromothripsis affecting at least one chromosome was evident in at least two-thirds of examined cases [6]. Multiple chromosomes characterized by the signatures of chromothripsis were also found, which concurrently inactivated several classic pancreatic driver genes, including_CDKN2A_, SMAD4, and TP53. Parallel events involving presumptive breakage-fusion-bridge cycles also triggered further genomic complexity, including the amplification of mutated_KRAS_ alleles.
Whole-genome sequencing analyses of nine supratentorial ependymomas, a type of brain and spinal cord tumor, strikingly revealed chromothripsis affecting chromosome 11 in all nine cases examined [14]. In eight cases, complex rearrangements generated a fusion of the oncogenic_RELA_ gene with an uncharacterized _C11orf95_gene, which are normally located ~2 megabases apart and separated by 73 genes on chromosome 11. Expression of RELA-C11orf95 fusions in neural stem cells implanted into the cerebellum of mice resulted in a marked increase in brain tumors, providing evidence that gene fusion products created through chromothripsis can be highly oncogenic in nature.
In one truly remarkable medical case, a patient with an extremely rare congenital immunodeficiency disorder called WHIM syndrome was serendipitously cured through somatic chromothripsis of chromosome 2 [72]. These rearrangements resulted in the loss of a disease-causative, gain-of-function mutant allele of the CXCR4 gene within a hematopoietic stem cell clone that was capable of repopulating the myeloid lineage and restoring normal neutrophil count.
In certain cases [10, 13, 28], there is also an association between chromothripsis and gene amplification. These can be manifested as extrachromosomal DNA elements in the form of double minute (DM) chromosomes, suggesting that the ends of one or more fragments can ligate and circularize [25] into self-propagating entities. In the initial example from small-cell lung cancer, chromothripsis of chromosome 8 produced a megabase-long DM through the stitching of fifteen fragments that led to the amplification of the_MYC_ oncogene [10]. These regions were lost from the reassembled derivative chromosome 8. Remarkably, such DMs have recently been detected in nearly half of all human cancers and can lead to exceptionally high expression of the corresponding DM-located genes [29] – typically oncogenes and/or genes conferring resistance to therapy [30]. That said, the frequency at which chromothripsis directly contributes to the formation of DMs across varying cancer types is not established.
Loss of the TP53 tumor suppressor gene, which can halt cell cycle progression in response to DNA damage, also appears to be a prerequisite event for chromothripsis [13]. Current experimental models for studying chromothripsis in human cells have required depletion or inactivating mutations of p53 [24–26, 31, 32]. Bypassing the p53 checkpoint is therefore a critical step towards tolerating and overcoming the damage accompanying chromothripsis. Although the vast majority of non-transformed cells experiencing chromothripsis do not survive, the cases exemplified in cancer genomes are likely rare exceptions that underwent clonal selection and expansion towards cancer. Overall, one or a combination of these rearrangements, which collectively produce a hallmark mutation signature [11] for chromothripsis (Figure 1B), could fuel cancer development and tumor evolution through selective processes.
The Micronucleus Revisited
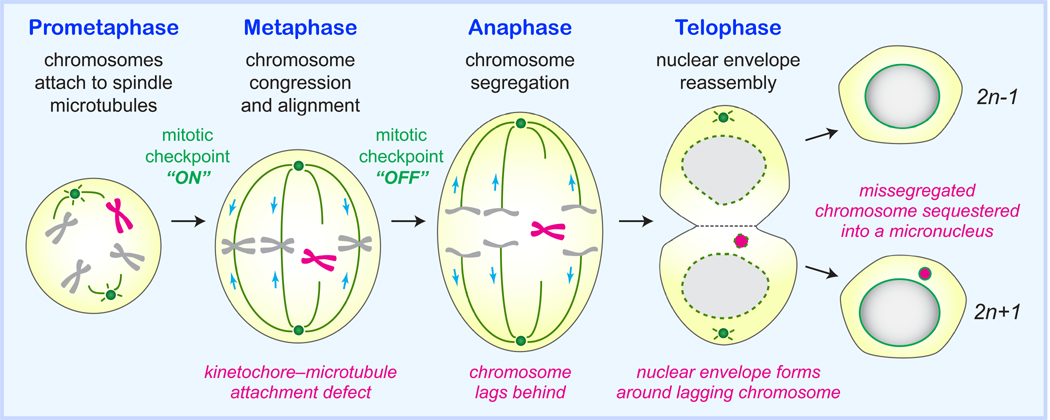
At the exit of mitosis, nuclear lamins and pore complexes redeposit around newly segregated chromosomal masses to encapsulate the genome within the nuclear envelope (NE), ultimately forming the cell nucleus. A chromosome that fails to correctly segregate to either of the two mitotic spindle poles, perhaps due to improper kinetochore–microtubule attachments, will produce a micronuclear envelope (micro-NE) assembled around the lagging chromosome. The resultantmicronucleus spatially isolates one or sometimes few missegregated chromosome(s) into a small nucleus-like structure that is positioned outside of the adjacent primary nucleus (Figure 2). Micronuclei are therefore a consequence of chromosome segregation errors during mitosis.
Figure 2. Chromosome segregation errors during mitotic cell division can entrap DNA within a micronucleus.
During mitosis, a chromosome that fails to congress, align, and/or form proper bipolar spindle microtubule–kinetochore attachments prior to anaphase onset can be left behind during the physical separation of the duplicated genome. A nuclear envelope assembles around the missegregated chromosome, subsequently forming a micronucleus at the exit of mitosis.
Accumulating evidence has established micronuclei as a unique source of genomic instability and DNA damage. The initial proposal for this emerged in 1968 when Kato and Sandberg determined that chromosomes in micronuclei undergo pulverization in mitosis after failing to complete DNA replication prior to mitotic entry [33]. Subsequent cell fusion studies between cells in S-phase and mitosis demonstrated that actively replicating chromosomes become fragmented upon premature chromosome condensation caused by exposure to a mitotic cytoplasm [34].
More than 40-years later, the discovery of chromothripsis in 2011 [10] has reawakened widespread interest in what is now recognized as the distinct biology of micronuclei. Consistent with the proposal by Kato and Sandberg, more recent evidence supports that micronuclear chromosomes do in fact acquire DNA damage and/or exhibit delayed replication kinetics compared to the main nucleus [24, 35–37]. Additionally, elegant use of live-cell imaging combined with single-cell DNA sequencing (a technique called ‘Look-Seq’) detected the presence of multiple, aberrantly ligated DNA fragments in the daughter cell(s) that arise from a previously micronucleated chromosome in a dividing mother cell [25]. These ligation events produced complex patterns of localized rearrangements that are highly reminiscent of cancer-associated chromothripsis, although whether an actual derivative chromosome was produced has not been established. Indeed, the experimental creation of a micronucleus-derived chromothriptic chromosome capable of stable transmission in subsequent cell cycles and that include the defining features of a fully functional chromosome has not yet been accomplished and represents a critical next step forward.
Similar mechanisms of chromosome shattering and re-ligation also appear to be present in genetic plant [38] and fission yeast [39] models of chromosome missegregation. In_Arabidopsis_ models, apparent micronuclei have been reported to produce complex rearrangements [38] that are strikingly similar to chromothripsis observed in human cancer genomes, implicating chromothripsis as a potentially conserved consequence of mitotic errors across evolution. Interestingly and along similar lines, protozoan ciliates such as Tetrahymena carry both a macronucleus and a micronucleus, the latter containing germline chromosomes that undergo deliberate DNA fragmentation and extensive rearrangements for subsequent conversion into the macronucleus [40].
Sources of Micronuclear DNA Damage
In mammalian cells, why might micronuclear chromosomes acquire DNA damage in the form of DSBs? As micronucleated cells progress through interphase (Figure 3), the micro-NE has a substantial tendency to undergo disruption that causes abrupt loss of nuclear contents (as detected by loss of GFP fused to nuclear import sequences) [41]. Correspondingly, the sequestered chromatin becomes exposed to normally cytoplasmic-localized components that diffuse into the micronucleus [41]. Thus, disruption of the micro-NE impairs proper nucleocytoplasmic transport and its exclusion of normally cytoplasmically localized proteins. The frequency of disruption is remarkably high, occurring in approximately half of micronuclei in an asynchronously cycling population, and accumulates with cell cycle progression through interphase [41]. Why micronuclei are prone to envelope disruption has not been solved, but could potentially arise from 1) differences in the stoichiometry of nuclear lamin or pore components assembled into the micro-NE during mitotic exit,2) the more extreme membrane curvature of the micro-NE, and/or_3)_ cytoskeletal or external compressive forces. The main nucleus can also rupture, although transiently as it rapidly reseals [42] through repair by the membrane remodeling ESCRT machinery [43, 44]. In contrast, disruption of the micro-NE is irreversible, partially or completely terminating normal nuclear function [41].
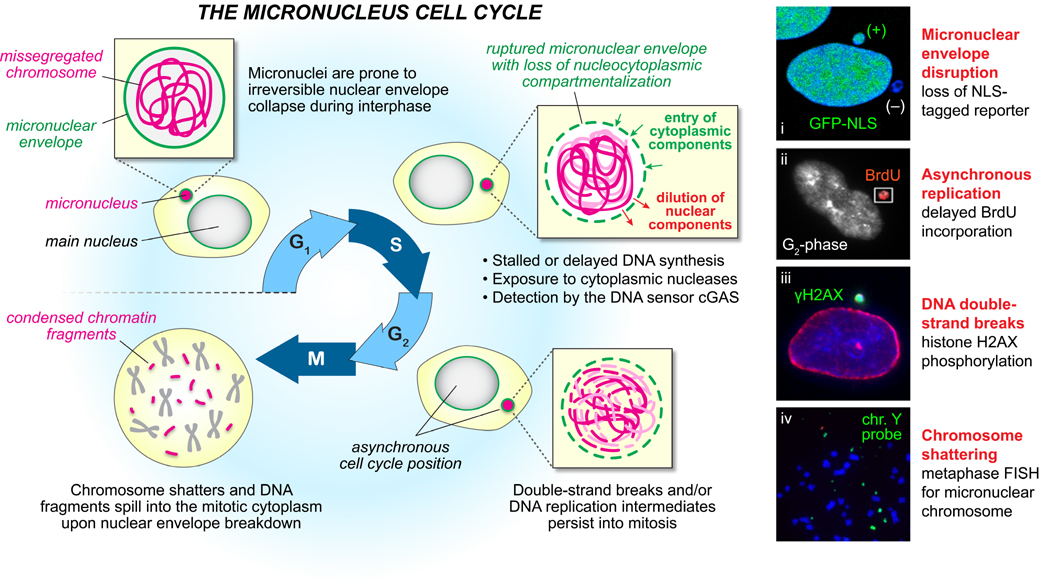
Figure 3. DNA damage in micronuclei triggers in the catastrophic shattering of individual chromosomes.
(A) Chromosomes isolated in micronuclei are sequestered by a highly unstable nuclear envelope, which are susceptible to disruption throughout interphase that interferes with normal nucleocytoplasmic transport and compartmentalization (i) [41]. Disruption can cause DNA replication asynchrony between the main nucleus and micronucleus (ii)[24], as well as permit exposure of micronuclear DNAs to damaging cytoplasmic components such as nucleases. In turn, DNA damage restricted to the micronucleated chromosome(iii) [26] persists throughout the cell cycle and into mitosis. Nuclear envelope breakdown and chromatin condensation initiated by mitotic entry subsequently causes the micronuclear chromosome harboring multiple double-stranded DNA breaks to undergo shattering that is accompanied by the spatial separation of chromosomal fragments (iv) [26]. Data images were modified and reproduced with permission from Elsevier (i) and Nature Publishing Group (ii–vi). NLS, nuclear localization signal.
Two non-mutually exclusive models have been raised to reconcile how micronuclear DSBs are generated after micro-NE disruption. In the first model, disruption during interphase can cause nuclear components of the micronucleus to become diluted through leakage into the cytoplasm. Disruption prior to or during S-phase can delay active replication fork progression, slowing or completely stalling DNA synthesis [24, 33, 37,41], which itself may act as a source of DNA damage owing to replication stress [45]. Disruption has been suggested to be a critical step for chromothriptic-like rearrangement signatures arising from micronuclei [25], which accumulate to peak frequency during S and G2 phases of the cell cycle [41]. The persistence of a large number of unrepaired DNA DSBs and/or unresolved replication intermediates could be catastrophic upon the dramatic remodeling of chromatin organization that occurs during mitosis, in particular the condensation of chromosomes that is required to facilitate the spatial separation of DNA.
The second model involves aberrant exposure of the micronuclear chromosome to one or more potentially damaging component(s) from the cytoplasm. Upon disruption of the micro-NE, influxes of cytosolic-localized proteins into the micronucleus have been observed (as detected by the inclusion of GFP fused to nuclear export sequences). Recent evidence [43,44] suggest that transient NE rupture of the main nucleus triggers the recruitment of the cytosolic DNA sensor cGAS [46], which evolved to detect foreign and pathogenic DNAs in the cytoplasm to activate the innate immune response [47]. During interphase, cGAS can be rapidly detected at sites of NE rupture at the nuclear periphery, which is subsequently followed by the focal acquisition of DNA damage [43, 44]. Similar rupturing events were reported to promote genomic copy number aberrations [48], and cGAS has been shown to associate with condensed chromosomes after NE breakdown during mitosis [49]. Consistent with this, recent work confirmed that cGAS could indeed sense micronuclear DNAs exposed to the cytoplasm following micro-NE disruption as cytosolic self-DNA [50], although the downstream consequences on the micronuclear chromosome itself are unknown.
Harmful cytoplasmic components could include endo- or exonucleases whose localization is tightly regulated or that are activated upon recognition of specific DNA intermediate structures arising from replication stress. MUS81 is one potential nuclease given its role in inducing DSBs at replication stress-induced late-replicating loci, which activates POLD3-dependent mitotic DNA synthesis to safeguard against the formation of ultra-fine anaphase bridges [51]. DNA synthesis errors during mitosis could be one source of chromosomal rearrangements arising from micronuclei that undergo mitotic entry with partially replicated DNA. Altogether, several mechanisms acting in parallel likely converges to promote the massive DNA damage associated with chromothripsis in micronuclei.
One Centromere Too Few: Chromothripsis Driven by Centromere Inactivation
Several experimental approaches have been used to investigate the properties of micronuclei and the eventual fate of the encapsulated chromosome [52]. One widely employed method to generate micronuclei is through prolonged mitotic arrest using microtubule inhibitors, such as nocodazole, followed by release and subsequent missegregation of one or few random chromosomes. An alternative experimental approach was recently developed involving the inactivation of a specific centromere (Box 2) to induce micronuclei containing a defined chromosome-of-interest (the human Y chromosome) – a strategy that has identified the temporal sequence of chromothriptic events over several consecutive cell cycles [26].
Box 2. Epigenetic maintenance and function of the centromere.
Centromeres are unique chromosomal loci that establish assembly of the kinetochore, a large multi-protein complex that directly facilitates chromosome movement and segregation by attachment to spindle microtubules during mitosis. Although the overwhelming majority of human centromeres are found on repetitive alpha-satellite DNA sequences, these sequences are neither sufficient nor necessary for centromere formation, maintenance, or function, as evident by the discovery of mitotically-stable neocentromeres formed at distinct loci located on chromosome arms [73–75]. Instead, the position of each centromere is specified epigenetically by the histone H3 variant CENP-A [76], which self-templates its own propagation every cell cycle (reviewed in detail [77]). These epigenetic mechanisms act to ensure that one – and strictly one – centromere is active per chromosome to safeguard against genomic instability.
Centromere inactivation initiates a series of events beginning with chromosome missegregation into micronuclei at the end of the first cell cycle. DNA damage accumulates within micronuclei, triggering chromosome shattering during mitosis of the second cell cycle. Chromosome fragments are subsequently reassembled throughout interphase of the third cell cycle. Analyses of metaphase spreads by fluorescent in situ hybridization provided direct evidence supporting micronucleus-mediated chromosome shattering, revealing >50 microscopic chromosomal fragments dispersed across the mitotic cytoplasm – all but one or a few of which lacked centromeric DNA sequences [26]. Chromosome shattering is likely an intermediate stage for chromothripsis (Figure 1A) as the vast majority of acentric fragments would be unstable long-term and lost. With exception of circularized extrachromosomal DMs, reassembly would be required to stabilize acentric fragments to at least one fragment containing an active centromere to form a derivative chromosome that can be genetically inherited.
Bringing It All Back Home: Reassembly through DNA Repair
The overwhelming proportion of DNA fragments produced by chromosome shattering are acentric [26], which alone are incapable of attaching to the mitotic spindle. The resulting fragments are therefore – at best – passively distributed, with likely asymmetric partitioning into newly formed daughter cells and reintegration into the main nucleus if they are in close proximity to either of the poleward-segregating chromosome masses (Figure 4A). The ends of these fragments are presumably recognized as DNA DSBs in the subsequent G1, thereby activating the DNA damage response (Figure 4B). Indeed, suppressing classical non-homologous end joining (c-NHEJ) (Box 3) by depleting or inhibiting the activity of LIG4 or DNA-PKcs prevented micronuclei-derived fragments to undergo repair [26], evidence supporting that c-NHEJ is the predominant mechanism for reassembling chromosomal fragments. LIG4 also appears to be required for repairing missegregated chromosomes in genetic plant models [38]. Chromosomal translocations in human cells are mediated primarily through c-NHEJ, although murine cells appear to favor alt-EJ [53,54]. Sequence analyses of the breakpoint junctions from chromothriptic tumors [10] and experimental models of chromothripsis [25, 38] has revealed that a large proportion of breakpoints (but not all, as discussed below) lack significant homology or microhomology – consistent with the repair signature of c-NHEJ.
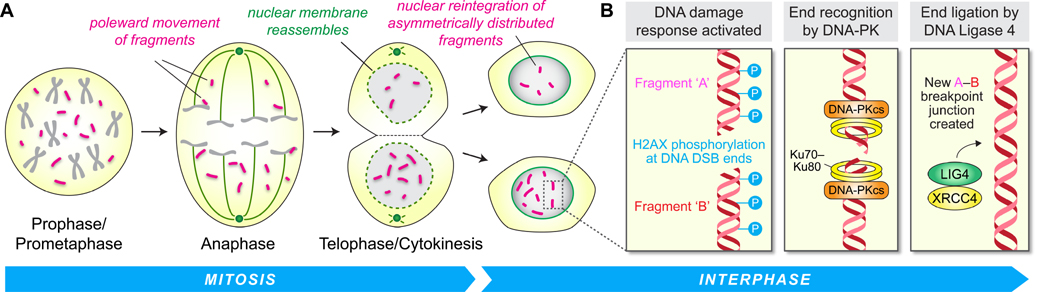
Figure 4. DNA damage repair mechanisms contributing to the reassembly of fragmented chromosomes.
(A) Chromosome fragments produced by chromothripsis spill into the mitotic cytoplasm and are subsequently incorporated into newly formed daughter cell nuclei at the exit of mitosis, possibly through the physical tethering between fragments and/or onto intact, centromere-containing chromosomes.(B) In the next interphase, reintegrated fragments activate the DNA damage response. In the absence of functional p53, DNA double-strand break repair ensues through error-prone non-homologous end joining, which directly links multiple fragments together in a haphazard manner by ligation. The reassembled chromosome is characterized by extensive DNA rearrangements harboring de novo breakpoint junctions that carry the signatures of the underlying DNA repair mechanism.
Box 3. Repair mechanisms for DNA double-stranded breaks.
DNA DSBs are repaired by two primary mechanisms in mammalian cells. In the first, homologous recombination (HR) utilizes long tracts of homologous sequence as a template to repair DSBs and is most active during S and G2 phases of the cell cycle. In the second, non-homologous end joining (NHEJ) directly joins two DSBs together without use of long sequence homology and is therefore a more error-prone repair pathway.
There are at least two recognized subtypes of NHEJ that mechanistically operates through distinct components and pathways: classical NHEJ (c-NHEJ) and the less characterized alternative end joining (alt-EJ) pathway (recently reviewed [78, 79]). c-NHEJ functions through a signaling cascade involving DNA-PK, a heterotrimeric complex composed of DNA-PKcs, Ku70, and Ku80 subunits, with direct end joining activity mediated by DNA Ligase 4 (LIG4)–XRCC4. Because end processing is minimal, repair by c-NHEJ can produce between 0–4 bp of microhomology at junction sequences, the majority of which occur by chance. In contrast, repair by alt-EJ does not require the components involved in c-NHEJ.
Microhomology-mediated end joining (MMEJ) is the major form of alt-EJ (reviewed in detail [80, 81]). MMEJ requires an initial resection step mediated by the MRN complex and CtIP (a step shared with HR) followed by a search for microhomologous sequences between the two resected ends. Once aligned, the ends are subjected to fill-in synthesis by the DNA polymerase Polθ and ligation by DNA Ligase 3. Breakpoint junctions repaired by MMEJ contain scars of 3–8 bp of microhomology (and up to 20 bp) that are usually accompanied by small deletions.
Although DNA DSBs accumulate in micronuclei following rupture in interphase, the resultant breaks are likely not subjected to DNA repair as micro-NE disruption would cause dilution of components involved in the DNA damage response [41]. Micronuclei with phosphorylated histone γH2AX, a marker of DNA DSBs, often failed to recruit or retain detectable levels of the DNA repair factor 53BP1 [35]. As micronucleated cells enter mitosis, a fraction of micronuclei also seemingly fail to disassemble the micro-NE and persist into the next interphase [24, 55]. However, high-throughput sequencing of purified micronuclear DNAs demonstrated that extensive rearrangements do not accumulate to detectable levels within micronuclei [26], further indicating that most reassembly events occur in the main nucleus following fragment reintegration.
Several lines of evidence suggest that nuclear ligation of fragmented DNA ensues rapidly with efficient repair kinetics. Analysis by Look-Seq of daughter cell pairs revealed that a high proportion of the fragments were ligated within ~4 hours after mitotic exit [25], most likely during G1. In 3/8 examples, fragments that were apparently lost from one daughter cell were found ligated within the paired daughter cell [25], which reflects the oscillating regions of copy number loss characteristic of chromothripsis [10, 11]. Additionally, use of the Y centromere-specific inactivation strategy demonstrated that inhibiting c-NHEJ produced chromosome fragments that persists into the next mitosis [26], indicating that the duration of a single cell cycle is sufficient for complete (or nearly complete) fragment reassembly.
How chromosomal fragments are inherited between daughter cells during mitosis remains a key question. Spindle forces and motor proteins drive chromosome movement during mitosis, but these do not engage chromosome fragments lacking a centromere. In Drosophila neuroblasts, acentric chromosomal fragments have been reported to partially segregate poleward [56] through kinetochore-independent microtubules and the chromokinesin Klp3a [57], which shares similarity to human KIF4A. Alternative models include the topological linkage or “tethering” of chromosome fragments to each other, as suggested by 5/8 Look-Seq examples in which the majority of fragments were unequally distributed to a single daughter [25], or perhaps onto other chromosomes, a mechanism analogous to the proposal that extrachromosomal DNAs (including DMs or viral episomes) could tether and segregate in trans with centromere-containing chromosomes [58–60]. Recent evidence suggests that the c-NHEJ components XRCC4-XLF can physically bridge two DSB ends prior to ligation [61]. Although DNA repair is normally suppressed throughout mitosis to preventtelomere fusions [62, 63], whether this or other tethering mechanisms are involved in maintaining chromosome fragments in close spatial proximity until segregation and/or ligation remains unsolved.
One Centromere Too Many: Chromothripsis Driven by Dicentric Chromosomes
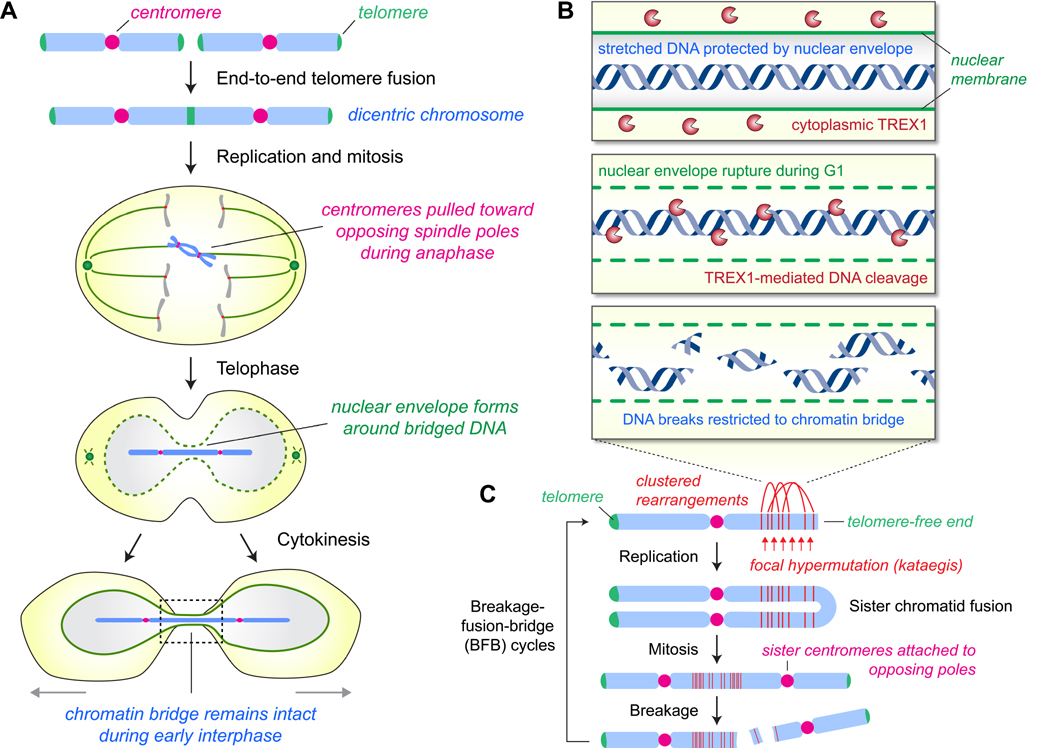
In certain instances, a single chromosome can harbor two active centromeres, both of which are capable of attaching to the mitotic spindle. Thesedicentric chromosomes can be formed through several mechanisms. A neocentromere can spontaneously form at a non-centromeric region on the chromosome arm, an event that naturally occurs at a rare frequency through poorly defined mechanisms (Box 2). Most often and perhaps by telomere shortening, a dicentric can be produced by an end-to-end fusion event between the telomeres of two non-homologous chromosomes (chromosome-type fusion) or between sister chromatids (chromatid-type fusion). In the latter, a pseudodicentric chromosome is formed in which the sister centromeres are properly attached to the opposing spindle poles in mitosis but remain linked at the fused end during chromatid separation at anaphase. Lastly, dicentric chromosomes can also result from the fusion of two chromosome fragments that each contains an active centromere [64]. Regardless of the mechanism of dicentric chromosome formation, a chromatin bridge is usually formed during mitosis that can persist until, or even long after, cytokinesis (Figure 5A).
Figure 5. Chromatin bridges act as a source of focal chromothripsis.
(A) Telomere fusion events can create dicentric chromosomes that harbor two active centromeres, both of which are capable of forming kinetochore–microtubule attachments during mitosis and segregation toward opposite spindle poles. Nuclear envelope reassembly at the exit of mitosis produces a chromatin bridge (dotted box, magnified in B) that persists into interphase connecting two nascent daughter cells.(B) Rupture of the nuclear envelope surrounding the chromatin bridge enables access of the normally cytoplasmic-localized TREX1 exonuclease to the underlying DNA, causing chromosome breaks restricted to the bridge that are likely repaired during the same or subsequent interphase. **(C)**Clustered rearrangements and hypermutation localized to a specific chromosome arm or region are common outcomes for fragmented bridges. Subsequent fusion events between telomere-free ends can facilitate further genomic instability through repeated cycles of breakage-fusion-bridge
Recent efforts identified that dicentric chromosomes developing into chromatin bridges during late mitosis could act as another source of chromothripsis [32]. Anaphase chromatin bridges were created using an established method to induce chromosome-type telomere fusions [65] through expression of a dominant-negative mutant of the telomere-associated shelterin component TRF2 [66]. Following the completion of an apparently normal cytokinesis, the chromatin bridge remains intact and interconnected between the two newly formed daughter cells throughout early interphase. As the daughters migrate away from one another, the NE surrounding the bridged DNA undergoes rupturing during G1 that is accompanied by the acquisition of the single-stranded DNA binding protein RPA. This rupture mediates access of the cytoplasmic 3′ exonuclease TREX1 to the exposed and stretched DNA, driving cleavage and ultimate resolution of the chromatin bridge (Figure 5B). Homozygous deletions of TREX1, however, partially delayed but did not completely prevent eventual bridge resolution, suggesting possible roles of other nucleases and/or physical mechanisms responsible for the breakage of chromatin bridges. Sequencing of clones revealed chromothriptic-like rearrangements involving the fused chromosomes [32] through an unidentified DNA repair mechanism. Consistent with a role for telomere fusions in chromothripsis, depletion of TRF2 to uncap telomeric ends in non-cancerous cells followed by selection for partially transformed cells also generated chromothriptic-like signatures [31]. Combinations of chromothripsis with breakage–fusion– bridge cycles (Figure 5C) can add another layer of complexity to the mutation signatures associated with cancer genomes [6, 64, 67].
How do chromothripsis and rearrangement patterns generated by chromatin bridges differ from those arising through micronuclei? Whereas replication defects have been proposed to be an important component of the DNA damage associated with the micronucleus model, chromothripsis from telomere fusions likely do not require ongoing DNA synthesis. Rather, the fragmentation of the bridged DNA is dependent on its resolution by TREX1, which frequently occurs prior to S-phase entry [32]. Additionally, multiple examples from cancer genomes have been documented in which chromothriptic rearrangements were focally restricted to a single arm or localized to a terminal region of a given chromosome rather than the entire chromosome [10]. Therefore, in contrast to the rearrangements produced from micronuclei that are distributed across an entire chromosome, dicentric bridge formation could explain how focal chromothripsis occurs. Analyses of chromothriptic examples generated from dicentrics revealed that many of the rearrangements were indeed clustered within a focal chromosomal region [32] that is probably the site of TREX1-mediated bridge resolution. Unexpectedly, these regions were often associated with hotspots of local hypermutation preferentially affecting cytosine nucleotides (kataegis) [7], implicating a potential mechanistic linkage between chromothripsis and hypermutation.
Alternative Mechanisms and Forms of Chromothripsis
A fraction of chromothriptic breakpoint junctions contain microhomology [10], suggesting potential repair by alternative end joining (Box 3) or the involvement of other mechanisms for localized rearrangements that are independent of chromosome shattering events, such as chromoanasynthesis. Mechanisms that have been proposed to contribute to the complex structural rearrangements defined by chromoanasynthesis include errors in DNA replication, most notably aberrant DNA template switching at stalled forks (called fork stalling and template switching, or FoSTeS) or collapsed/broken forks (called microhomology-mediated break-induced replication, or MMBIR) [68, 69]. In individuals with inherited genomic disorders, iterative rounds of such replication-based events could produce catastrophic rearrangements that are accompanied by templated insertions, duplications, and/or microhomology at the sequence junctions [19]. Such complex rearrangements can be reminiscent of, but distinct from, chromothripsis through chromosome shattering and fragment re-ligation [70, 71]. Interestingly, a few examples of micronuclei-derived rearrangements also exhibited sequence features at junctions that share similarities with chromoanasynthesis, in particular small insertions and the presence of microhomology [25], suggesting possible overlapping mechanisms between chromothripsis and chromoanasynthesis.
Concluding Remarks
The discovery of chromothripsis has advanced our understanding of the complexities associated with cancer genomes, as well as opened exciting new avenues for research. The development of novel cell biological tools combined with computational methods to examine the fate and sequence characteristics of missegregated chromosomes has recently contributed to the mechanisms underlying chromothripsis. Much remains to be determined (see ‘Outstanding Questions’), in particular the exact causes of NE disruption and DNA damage in micronuclei. Key among these questions is what does it take to assemble a fully functional and heritable human chromosome, and how is this achieved following catastrophic processes such as the shattering of an individual chromosome? Whether other unidentified types of punctuated equilibrium-driven chromosomal alterations are present in cancer or disease remains to be seen, but advancements in DNA sequencing technologies will likely enable further discovery of more unexpected and hidden features of the human genome.
Outstanding Questions.
- ■
What are the contributing factors that predispose micronuclei to undergo nuclear envelope disruption during interphase? - ■
What are the underlying sources of DNA damage in a micronucleus following the disruption of its nuclear envelope? - ■
How are chromosomal fragments segregated between nascent daughter cells during mitosis, and are these fragments topologically or physically tethered to facilitate segregation en masse? - ■
Does the nuclear reintegration of chromosomal fragments activate the DNA damage response to engage DNA repair, and if so, how are these fragments spatially positioned within the interphase nucleus to promote efficient reassembly? - ■
At what frequency do reassembled fragments form a fully functional chromosome that is capable of long-term inheritance? - ■
Do chromothripsis and chromoanasynthesis share similar underlying mechanisms? - ■
What other types of complex DNA alterations caused by “punctuated” events exist in the genomes of individuals suffering from diseases or disorders?
Trends.
- ■
Chromothripsis is a catastrophic event in which one or a few chromosome(s) are shattered and stitched back together in random order, producing a derivative chromosome with complex rearrangements within a few cell cycles. - ■
Chromosome missegregation during cell division frequently produces small nuclear structures called micronuclei, which are prone to irreversible nuclear envelope disruption during interphase and impaired nucleocytoplasmic compartmentalization. - ■
Micronucleated chromosomes accumulate extensive DNA damage and are susceptible to shattering during the next mitosis, generating multiple, distinct DNA fragments. - ■
Chromosome fragments are reassembled by DNA double-strand break repair to form a derivative chromosome. - ■
Chromatin bridges trapped between daughter cells are attacked by a cytoplasmic nuclease (TREX1) during interphase to generate DNA breaks and focal chromothripsis.
Acknowledgments
We thank M. Hetzer, E. Hatch, and D. Pellman for sharing original data images. This work was supported by grants from the National Institutes of Health (R35 GM122476 to D.W.C. and K99 CA218871 to P.L.) and the Hope Funds for Cancer Research (HFCR-14-06-06 to P.L.). D.W.C. receives salary support from the Ludwig Institute for Cancer Research.
Glossary
Acentric
a chromosome, or fragment of a chromosome, that lacks an active centromere
Chromothripsis
complex rearrangements arising from the catastrophic shattering of a single or few chromosome(s)
Chromoanasynthesis
complex rearrangements arising from the defective replication of a single or few chromosome(s)
Chromoanagenesis
a catchall term that encompasses catastrophic mutational processes involving one or a few chromosomes, independent of the precise mechanism(s); included here are chromothripsis and chromoanasynthesis, which arise through distinct mechanisms
Chromoplexy
a series of chained, complex rearrangements frequently involving five or more chromosomes
Centromere
a specialized region on each chromosome designated for assembly of the kinetochore and whose unique position is identified and maintained epigenetically
Dicentric
a chromosome harboring two active centromeres
Double minute (DM) chromosomes
circular and replication-competent extrachromosomal DNA elements that accumulate and amplify multiple gene copies that drive high levels of expression
Kataegis
clusters of focal hypermutation preferentially favoring cytosine substitutions
Micronucleus
small, nuclear structures that encapsulate missegregated chromosomes and are spatially isolated from the main nucleus
Micronuclear envelope disruption
irreversible rupture of the nuclear envelope surrounding a micronucleus that causes abrupt loss of nucleocytoplasmic partitioning and terminates micronuclear function
Non-homologous end joining
a major form of DNA double-strand break repair whereby two damaged ends undergoes direct ligation
Punctuated equilibrium
a long-standing theory in evolutionary biology with recent implications for cancer development, in particular the rapid acquisition of a large number of oncogenic alterations over a short timescale
Telomeres
repetitive sequences that protect each of the terminal ends of chromosomes from shortening during replication and detection by the DNA damage response
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Weaver BA, Cleveland DW. The aneuploidy paradox in cell growth and tumorigenesis. Cancer Cell. 2008;14:431–433. doi: 10.1016/j.ccr.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheltzer JM, Amon A. The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends Genet. 2011;27:446–453. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nowell PC, Hungerford DA. Minute Chromosome in Human Chronic Granulocytic Leukemia. Science. 1960;132:1497–1497. [Google Scholar]
- 4.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 5.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 6.Notta F, Chan-Seng-Yue M, Lemire M, Li Y, Wilson GW, Connor AA, Denroche RE, Liang SB, Brown AM, Kim JC, et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature. 2016;538:378–382. doi: 10.1038/nature19823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 12.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 13.Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, Jager N, Remke M, Shih D, Northcott PA, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker M, Mohankumar KM, Punchihewa C, Weinlich R, Dalton JD, Li Y, Lee R, Tatevossian RG, Phoenix TN, Thiruvenkatam R, et al. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature. 2014;506:451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, Leenders F, Lu X, Fernandez-Cuesta L, Bosco G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scarpa A, Chang DK, Nones K, Corbo V, Patch AM, Bailey P, Lawlor RT, Johns AL, Miller DK, Mafficini A, et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature. 2017;543:65–71. doi: 10.1038/nature21063. [DOI] [PubMed] [Google Scholar]
- 17.Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, Shiah YJ, Yousif F, Lin X, Masella AP, et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature. 2017;541:359–364. doi: 10.1038/nature20788. [DOI] [PubMed] [Google Scholar]
- 18.Kloosterman WP, Guryev V, van Roosmalen M, Duran KJ, de Bruijn E, Bakker SC, Letteboer T, van Nesselrooij B, Hochstenbach R, Poot M, et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet. 2011;20:1916–1924. doi: 10.1093/hmg/ddr073. [DOI] [PubMed] [Google Scholar]
- 19.Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holland AJ, Cleveland DW. Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat Med. 2012;18:1630–1638. doi: 10.1038/nm.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maher CA, Wilson RK. Chromothripsis and human disease: piecing together the shattering process. Cell. 2012;148:29–32. doi: 10.1016/j.cell.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer. 2012;12:663–670. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
- 23.Jones MJ, Jallepalli PV. Chromothripsis: chromosomes in crisis. Dev Cell. 2012;23:908–917. doi: 10.1016/j.devcel.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ly P, Teitz LS, Kim DH, Shoshani O, Skaletsky H, Fachinetti D, Page DC, Cleveland DW. Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat Cell Biol. 2017;19:68–75. doi: 10.1038/ncb3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, Kramer A, Roncaioli JL, Sand F, Heuckmann JM, et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526:700–704. doi: 10.1038/nature14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng C, Zhou Y, Li H, Xiong T, Li S, Bi Y, Kong P, Wang F, Cui H, Li Y, et al. Whole-Genome Sequencing Reveals Diverse Models of Structural Variations in Esophageal Squamous Cell Carcinoma. Am J Hum Genet. 2016;98:256–274. doi: 10.1016/j.ajhg.2015.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, Li B, Arden K, Ren B, Nathanson DA, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543:122–125. doi: 10.1038/nature21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G, Eskin A, Hwang K, Wang J, Masui K, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–76. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mardin BR, Drainas AP, Waszak SM, Weischenfeldt J, Isokane M, Stutz AM, Raeder B, Efthymiopoulos T, Buccitelli C, Segura-Wang M, et al. A cell-based model system links chromothripsis with hyperploidy. Mol Syst Biol. 2015;11:828. doi: 10.15252/msb.20156505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maciejowski J, Li Y, Bosco N, Campbell PJ, de Lange T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell. 2015;163:1641–1654. doi: 10.1016/j.cell.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato H, Sandberg AA. Chromosome pulverization in human cells with micronuclei. J Natl Cancer Inst. 1968;40:165–179. [PubMed] [Google Scholar]
- 34.Johnson RT, Rao PN. Mammalian cell fusion: induction of premature chromosome condensation in interphase nuclei. Nature. 1970;226:717–722. doi: 10.1038/226717a0. [DOI] [PubMed] [Google Scholar]
- 35.Terradas M, Martin M, Tusell L, Genesca A. DNA lesions sequestered in micronuclei induce a local defective-damage response. DNA Repair (Amst) 2009;8:1225–1234. doi: 10.1016/j.dnarep.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Xu B, Sun Z, Liu Z, Guo H, Liu Q, Jiang H, Zou Y, Gong Y, Tischfield JA, Shao C. Replication stress induces micronuclei comprising of aggregated DNA double-strand breaks. PLoS One. 2011;6:e18618. doi: 10.1371/journal.pone.0018618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okamoto A, Utani K, Shimizu N. DNA replication occurs in all lamina positive micronuclei, but never in lamina negative micronuclei. Mutagenesis. 2012;27:323–327. doi: 10.1093/mutage/ger082. [DOI] [PubMed] [Google Scholar]
- 38.Tan EH, Henry IM, Ravi M, Bradnam KR, Mandakova T, Marimuthu MP, Korf I, Lysak MA, Comai L, Chan SW. Catastrophic chromosomal restructuring during genome elimination in plants. Elife. 2015;4 doi: 10.7554/eLife.06516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabatinos SA, Ranatunga NS, Yuan JP, Green MD, Forsburg SL. Replication stress in early S phase generates apparent micronuclei and chromosome rearrangement in fission yeast. Mol Biol Cell. 2015;26:3439–3450. doi: 10.1091/mbc.E15-05-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60. doi: 10.1016/j.cell.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vargas JD, Hatch EM, Anderson DJ, Hetzer MW. Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus. 2012;3:88–100. doi: 10.4161/nucl.18954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raab M, Gentili M, de Belly H, Thiam HR, Vargas P, Jimenez AJ, Lautenschlaeger F, Voituriez R, Lennon-Dumenil AM, Manel N, et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 2016;352:359–362. doi: 10.1126/science.aad7611. [DOI] [PubMed] [Google Scholar]
- 45.Gaillard H, Garcia-Muse T, Aguilera A. Replication stress and cancer. Nat Rev Cancer. 2015;15:276–289. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- 46.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Margolis SR, Wilson SC, Vance RE. Evolutionary Origins of cGAS-STING Signaling. Trends Immunol. 2017 doi: 10.1016/j.it.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 48.Irianto J, Xia Y, Pfeifer CR, Athirasala A, Ji J, Alvey C, Tewari M, Bennett RR, Harding SM, Liu AJ, et al. DNA Damage Follows Repair Factor Depletion and Portends Genome Variation in Cancer Cells after Pore Migration. Curr Biol. 2017;27:210–223. doi: 10.1016/j.cub.2016.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang H, Wang H, Ren J, Chen Q, Chen ZJ. cGAS is essential for cellular senescence. Proc Natl Acad Sci U S A. 2017;114:E4612–E4620. doi: 10.1073/pnas.1705499114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, Olova N, Sutcliffe H, Rainger JK, Leitch A, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 2017 doi: 10.1038/nature23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minocherhomji S, Ying S, Bjerregaard VA, Bursomanno S, Aleliunaite A, Wu W, Mankouri HW, Shen H, Liu Y, Hickson ID. Replication stress activates DNA repair synthesis in mitosis. Nature. 2015;528:286–290. doi: 10.1038/nature16139. [DOI] [PubMed] [Google Scholar]
- 52.Ly P, Cleveland DW. Interrogating cell division errors using random and chromosome-specific missegregation approaches. Cell Cycle. 2017:1–7. doi: 10.1080/15384101.2017.1325047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghezraoui H, Piganeau M, Renouf B, Renaud JB, Sallmyr A, Ruis B, Oh S, Tomkinson AE, Hendrickson EA, Giovannangeli C, et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell. 2014;55:829–842. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Biehs R, Steinlage M, Barton O, Juhasz S, Kunzel J, Spies J, Shibata A, Jeggo PA, Lobrich M. DNA Double-Strand Break Resection Occurs during Non-homologous End Joining in G1 but Is Distinct from Resection during Homologous Recombination. Mol Cell. 2017;65:671–684 e675. doi: 10.1016/j.molcel.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang Y, Jiang L, Yi Q, Lv L, Wang Z, Zhao X, Zhong L, Jiang H, Rasool S, Hao Q, et al. Lagging chromosomes entrapped in micronuclei are not ‘lost’ by cells. Cell Res. 2012;22:932–935. doi: 10.1038/cr.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Royou A, Gagou ME, Karess R, Sullivan W. BubR1- and Polo-coated DNA tethers facilitate poleward segregation of acentric chromatids. Cell. 2010;140:235–245. doi: 10.1016/j.cell.2009.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karg T, Elting MW, Vicars H, Dumont S, Sullivan W. The chromokinesin Klp3a and microtubules facilitate acentric chromosome segregation. J Cell Biol. 2017;216:1597–1608. doi: 10.1083/jcb.201604079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanda T, Otter M, Wahl GM. Coupling of mitotic chromosome tethering and replication competence in epstein-barr virus-based plasmids. Mol Cell Biol. 2001;21:3576–3588. doi: 10.1128/MCB.21.10.3576-3588.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kanda T, Otter M, Wahl GM. Mitotic segregation of viral and cellular acentric extrachromosomal molecules by chromosome tethering. J Cell Sci. 2001;114:49–58. doi: 10.1242/jcs.114.1.49. [DOI] [PubMed] [Google Scholar]
- 60.Chiu YF, Sugden AU, Fox K, Hayes M, Sugden B. Kaposi’s sarcoma-associated herpesvirus stably clusters its genomes across generations to maintain itself extrachromosomally. J Cell Biol. 2017 doi: 10.1083/jcb.201702013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brouwer I, Sitters G, Candelli A, Heerema SJ, Heller I, de Melo AJ, Zhang H, Normanno D, Modesti M, Peterman EJ, et al. Sliding sleeves of XRCC4-XLF bridge DNA and connect fragments of broken DNA. Nature. 2016;535:566–569. doi: 10.1038/nature18643. [DOI] [PubMed] [Google Scholar]
- 62.Orthwein A, Fradet-Turcotte A, Noordermeer SM, Canny MD, Brun CM, Strecker J, Escribano-Diaz C, Durocher D. Mitosis inhibits DNA doublestrand break repair to guard against telomere fusions. Science. 2014;344:189–193. doi: 10.1126/science.1248024. [DOI] [PubMed] [Google Scholar]
- 63.Lee DH, Acharya SS, Kwon M, Drane P, Guan Y, Adelmant G, Kalev P, Shah J, Pellman D, Marto JA, et al. Dephosphorylation enables the recruitment of 53BP1 to double-strand DNA breaks. Mol Cell. 2014;54:512–525. doi: 10.1016/j.molcel.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garsed DW, Marshall OJ, Corbin VD, Hsu A, Di Stefano L, Schroder J, Li J, Feng ZP, Kim BW, Kowarsky M, et al. The architecture and evolution of cancer neochromosomes. Cancer Cell. 2014;26:653–667. doi: 10.1016/j.ccell.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 65.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 66.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 67.Li Y, Schwab C, Ryan S, Papaemmanuil E, Robinson HM, Jacobs P, Moorman AV, Dyer S, Borrow J, Griffiths M, et al. Constitutional and somatic rearrangement of chromosome 21 in acute lymphoblastic leukaemia. Nature. 2014;508:98–102. doi: 10.1038/nature13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 69.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kloosterman WP, Tavakoli-Yaraki M, van Roosmalen MJ, van Binsbergen E, Renkens I, Duran K, Ballarati L, Vergult S, Giardino D, Hansson K, et al. Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep. 2012;1:648–655. doi: 10.1016/j.celrep.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 71.Yang L, Luquette LJ, Gehlenborg N, Xi R, Haseley PS, Hsieh CH, Zhang C, Ren X, Protopopov A, Chin L, et al. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell. 2013;153:919–929. doi: 10.1016/j.cell.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McDermott DH, Gao JL, Liu Q, Siwicki M, Martens C, Jacobs P, Velez D, Yim E, Bryke CR, Hsu N, et al. Chromothriptic cure of WHIM syndrome. Cell. 2015;160:686–699. doi: 10.1016/j.cell.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Voullaire LE, Slater HR, Petrovic V, Choo KH. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am J Hum Genet. 1993;52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- 74.Wandall A, Tranebjaerg L, Tommerup N. A neocentromere on human chromosome 3 without detectable alpha-satellite DNA forms morphologically normal kinetochores. Chromosoma. 1998;107:359–365. doi: 10.1007/s004120050319. [DOI] [PubMed] [Google Scholar]
- 75.Tyler-Smith C, Gimelli G, Giglio S, Floridia G, Pandya A, Terzoli G, Warburton PE, Earnshaw WC, Zuffardi O. Transmission of a fully functional human neocentromere through three generations. Am J Hum Genet. 1999;64:1440–1444. doi: 10.1086/302380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LE, et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol. 2013;15:1056–1066. doi: 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ceccaldi R, Rondinelli B, D’Andrea AD. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. 2017;18:495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ottaviani D, LeCain M, Sheer D. The role of microhomology in genomic structural variation. Trends Genet. 2014;30:85–94. doi: 10.1016/j.tig.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 81.Sfeir A, Symington LS. Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem Sci. 2015;40:701–714. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]