Anatomical Profiling of Nuclear Receptor Expression Reveals a Hierarchical Transcriptional Network (original) (raw)
. Author manuscript; available in PMC: 2018 Nov 1.
SUMMARY
In multicellular organisms, the ability to regulate reproduction, development, and nutrient utilization coincided with the evolution of nuclear receptors (NRs), transcription factors that utilize lipophilic ligands to mediate their function. Studying the expression profile of NRs offers a simple, powerful way to obtain highly relational information about their physiologic functions as individual proteins and as a superfamily. We surveyed the expression of all 49 mouse NR mRNAs in 39 tissues, representing diverse anatomical systems. The resulting data set uncovers several NR clades whose patterns of expression indicate their ability to coordinate the transcriptional programs necessary to affect distinct physiologic pathways. Remarkably, this regulatory network divides along the following two physiologic paradigms: (1) reproduction, development, and growth and (2) nutrient uptake, metabolism, and excretion. These data reveal a hierarchical transcriptional circuitry that extends beyond individual tissues to form a meganetwork governing physiology on an organismal scale.
INTRODUCTION
Nuclear receptors (NRs) represent the largest family of transcription factors found in metazoans (Mangelsdorf et al., 1995). The success of members of this group stems in part from their ability to function as ligand-dependent sensors for a diverse set of fat-soluble hormones, vita-mins, and dietary lipids (Chawla et al., 2001). Included in this family are receptors for endocrine steroids (i.e., corticosteroids, progesterone, androgens, and estrogens), fat-soluble vitamins A and D, thyroid hormone, fatty acids, oxysterols, bile acids, and numerous xenobiotic lipids derived from the diet. Additional members of this family are called orphan receptors because their ligands remsain unknown. Together, NRs govern expression of genes involved in a broad range of reproductive, developmental, metabolic, and immune response programs. NRs also encompass one of the most successful targets for drugs currently available or being developed to treat a multitude of therapeutic indices, including hypertension, cancer, diabetes, cardiovascular disease, cholesterol gallstone disease, and the metabolic syndrome (Gronemeyer et al., 2004; Shulman and Mangelsdorf, 2005).
Although much has been learned about the physiology of certain receptors in a limited number of well-studied target tissues, surprisingly little is known about NR function in the majority of tissues throughout the body as a whole. Furthermore, relatively little is known about the complexity of interactions that may exist between receptors in various tissues and, consequently, about whether such interactions may direct a higher order process to govern dynamic physiological parameters. Indeed, the ability of NRs to crossreact with each other’s DNA binding sites and to govern similar regulatory cascades implies the existence of such a higher-order transcriptional regulatory network. This raises the question of whether understanding this network can be obtained not only by studying the expression of individual receptors but also by studying expression of the nuclear receptor superfamily as a whole using a systems biology approach. One simple but elegant way to define the blueprint of NR action on a system-wide scale is to look at the expression of the entire NR transcriptome. Given that physiology is a process occurring across both spatial and temporal coordinates, this expression blueprint would be expected to have both an anatomical and circadian rhythm component.
The Nuclear Receptor Signaling Atlas (NURSA; www.nursa.org) is an interdisciplinary consortium focused on creating a web-based, minable, dynamic database of nuclear receptor and coregulator sequence information, expression analysis, and functional data. NURSA aims to provide tools that may catalyze progress in a number of interdisciplinary fields. Therefore, as members of the group, we first set out to establish the anatomical expression profiles for the 49 members of the NR superfamily in 39 tissues from two different strains of the most widely used mouse models. In a companion paper in this issue of Cell, Yang et al. (2006) describe the circadian expression of NRs in four metabolically relevant tissues. The resulting data sets offer an unprecedented view of NR function in space and time. The work presented here suggests that hierarchical, integrated NR networks coordinately relay cues directing tissue-specific and metabolic readouts. This resource should provide new avenues for more advanced investigation into the biological role of each receptor individually and together as a superfamily.
RESULTS AND DISCUSSION
Anatomical Expression Profiling of the NR Superfamily
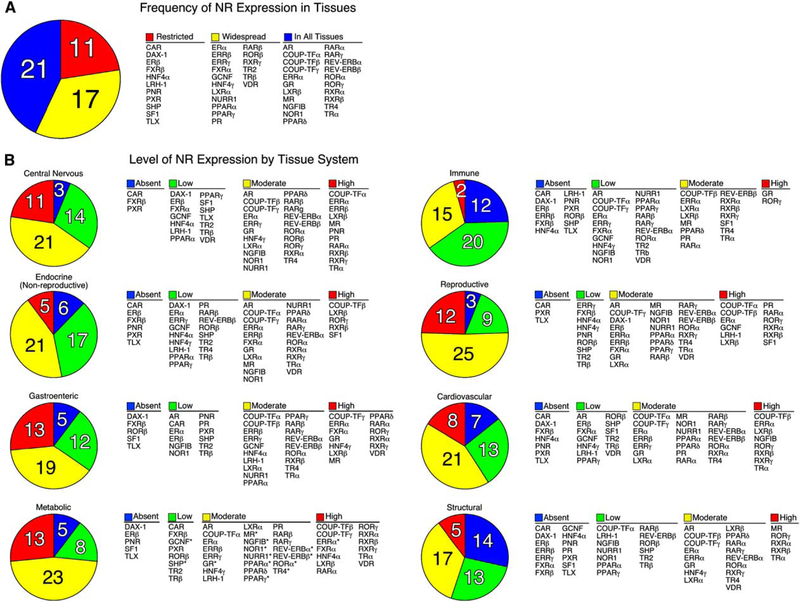
TaqMan-based real-time quantitative RT-PCR (QPCR) was chosen to survey NR expression because of its broad dynamic range of detection, quantitative reliability and reproducibility, and high-throughput capacity. Transcript levels of the major isoforms of the 49 known mouse NRs were profiled in 39 different tissues from two strains of mice, 129×1/SvJ and C57/Bl6. Male mice were used for all tissues except female reproductive tissues, reflecting the common use of male mice in most experimental systems. The two mouse strains were chosen because of their wide use in genetic manipulation and their metabolically diverse phenotypes. For example, 129×1/SvJ mice are relatively resistant to lipid-related disorders in spite of high plasma triglycerides and cholesterol levels, while C57/Bl6 mice are susceptible to diet-induced obesity, atherosclerosis, and type II diabetes (MGD, 2003). The anatomical profile of all 49 NRs in the two strains of mice is depicted on Poster S1, and the entire data set is available on the NURSA website at <www.nursa.org/datasets.cfm?doi=10.1621/datasets.02001>. Analysis of these data revealed nearly complete identity in the expression profile of NRs between the two mouse strains, although 12% of the tissues exhibited differences greater than 2-fold between strains (see accompanying poster). At an organismal level, the tissue-frequency profile showed that 21 NRs were expressed in all tissues, 17 NRs existed in more than half but not all tissues, and 11 NRs were restricted to less than 50% of tissues in the adult animal (Figure 1A). Receptors with relatively widespread expression included the endocrine steroid, thyroid hormone, vita-min A, and vitamin D receptors. Thus, in addition to their well-known roles in classic endocrine target tissues, the broad range of expression of these receptors implies a higher level of complexity and function than might have been expected. Receptors with highly restricted expression profiles included constitutive androstane receptor (CAR); dosage-sensitive sex reversal-adrenal hypoplasia congenita on the X chromosome, gene 1 (DAX-1); estrogen receptor β (ERβ); farnesoid X receptor β (FXRβ); hepatocyte nuclear factor 4a (HNF4α); liver receptor homolog-1 (LRH-1); photoreceptor-cell nuclear receptor (PNR); pregnane X receptor (PXR); short heterodimer partner (SHP); steroidogenic factor 1 (SF1); and tailless homolog orphan receptor (TLX). Within this restricted set the CNS, endocrine, and enterohepatic tissue systems were represented, highlighting the specialized function of these receptors as tissue-specific transcription factors.
Figure 1. Distribution of C57/BL6 Mouse Nuclear Receptor mRNA.
(A) The number of NRs expressed in various tissues is indicated in the pie chart and their names are shown in the table on the right. NRs were grouped into three categories based on their tissue distribution: Restricted (expressed in less than 50% of tissues), Widespread (expressed in more than 50% but not all tissues), and In All Tissues (expressed in 100% of tissues).
(B) The levels of NR expression in different tissue systems are indicated by the pie charts and their names are shown in the tables to the right. Normalized NR mRNA-expression levels were defined as Absent if the Ct value was ≥34, Low if the level was below 0.1 arbitrary units, Moderate if the level was between 0.1 and 1, and High if the level was >1.0 arbitrary units. Tissues systems were categorized as follows: central nervous system (eye, brainstem, cerebellum, cerebrum, corpus striatum, olfactory bulb, spinal cord, hypothalamus, and pituitary); nonreproductive endocrine system (adrenal, thyroid, and pancreas); gastroenteric system (tongue, stomach, duodenum, jejunum, ileum, colon, and gall bladder); metabolic system (liver, kidney, brown and white adipose, and muscle); immune system (spleen and thymus); reproductive system (ovary, uterus, epididymus, preputial gland, prostate, seminal vesicles, testis, and vas deferens); cardiovascular system (aorta, heart, and lungs); and structural system (bone and skin). Note that tissues were pooled from male mice (n = 6) except for ovary and uterus. An asterisk (*) indicates NRs that have a strong circadian expression and were lowest at time of analysis (see accompanying manuscript by Yang et al., 2006 in this issue of Cell).
When grouped according to tissue system (e.g., CNS, endocrine, gastroenteric, metabolic, immune, reproductive, cardiovascular, and structural), the distribution and level of expression of individual NRs becomes apparent (Figure 1B). Receptors that were expressed in most tissues at moderate to high levels (i.e., relative mRNA values ≥ 0.1 arbitrary units) included the chicken ovalbumin upstream promoter-transcription factor γ (COUP-TFγ), estrogen-related receptor α, (ERRα), glucocorticoid receptor (GR), liver X receptor β (LXRβ), and retinoid X receptors α and β (RXRα, RXRβ). Interestingly, LXRβ and RXRβ exhibited the highest level of NR expression found in all mouse tissues (i.e., relative mRNA value of~1.0 or higher). Several other receptors (e.g., PNR, HNF4 α and γ, and SF1) maintained relatively high levels of expression (relative mRNA value ≥ 2.0), but only in a few tissues. It is worth noting that these profiles represent a fixed point in time (ZT 0) and that expression of many receptors is markedly influenced by circadian expression as evidenced in the companion paper by Yang et al. (2006). In addition, because of the heterogeneous population of cells in some tissues, it is more difficult to interpret low levels of expression exhibited by receptors in certain organs or tissue samples. For example, TLX expression, which is limited to the brain, was detected by QPCR in whole brain sections at low levels. However, when analyzed at the level of individual cells, TLX expression is known to be highly expressed, but only in a subset of adult neural stem cells (Shi et al., 2004).
Hierarchical Clustering of NR Expression: The Reproductive and CNS Axes
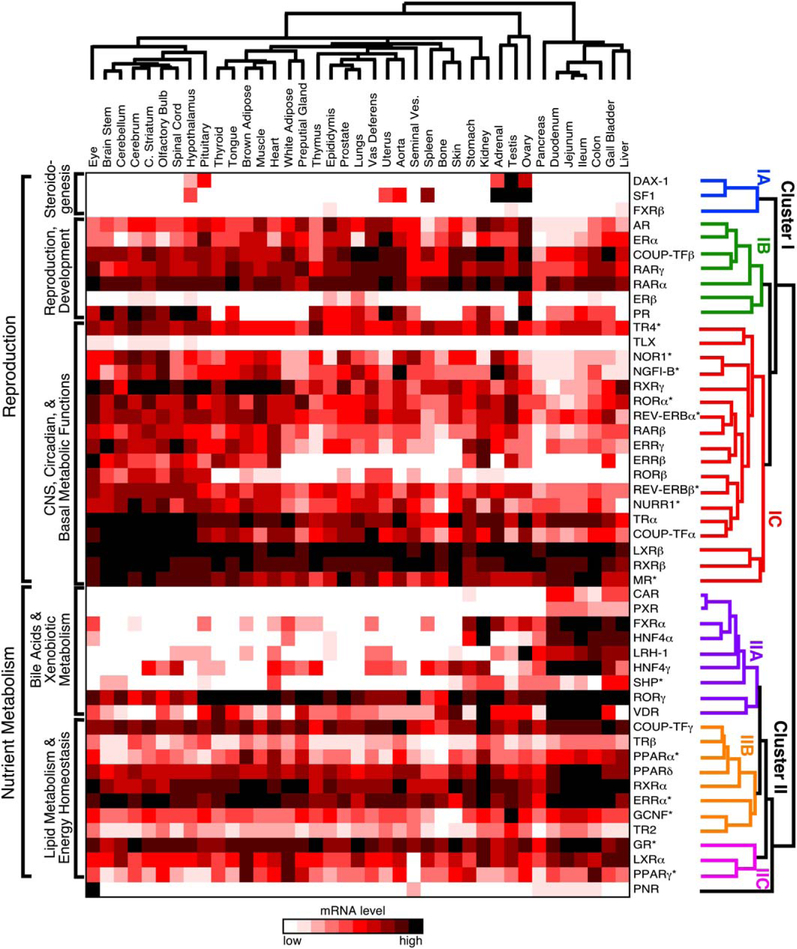
To study the potential relationship between receptor expression, function, and physiology, the QPCR results were analyzed by hierarchical, unsupervised clustering of receptor expression by tissue (Figure 2). The resulting dendrogram on the receptor axis reveals the existence of a higher-order network tying nuclear receptor function to physiology. Unexpectedly at the top of this hierarchy, the dendrogram branches into two major clusters (labeled I and II). Further branching subdivides the two superior clusters into six subordinate clusters, three in cluster I (labeled IA, IB, and IC) and three in cluster II (labeled IIA, IIB, and IIC).
Figure 2. Unsupervised Hierarchical Clustering of Nuclear Receptors Relative to Tissue Expression Pattern.
The mRNA tissue-distribution profile of the NR superfamily in the C57/BL6 mouse was evaluated by unsupervised hierarchical clustering using Matrix 1.28 software as described in Experimental Procedures. Individual NRs segregate into two main clusters (labeled I and II) and six subclusters (labeled IA, IB, IC, IIA, IIB, and IIC) that define higher-order functional relationships. The dendrogram of NR clusters is color-coded for clarity. The asterisk (*) indicates NRs that have a strong circadian expression and were lowest at time of analysis.
Cluster IA forms a distinct group of the following three NRs: SF1, DAX-1, and FXRβ. SF1 and DAX-1 are the key competence factors that function at the top of a transcriptional cascade to direct sexual differentiation and steroido-genesis, consistent with their highly restricted expression in the CNS, reproductive organs, and adrenals (Beuschlein et al., 2002; Parker and Schimmer, 2002). The role of FXRβ, which is not found in humans, remains unknown (Otte et al., 2003). At present, the relevance of its clustering with SF1 and DAX-1 is not clear.
The NRs in cluster IB function at the next level of the sex-steroid transcription cascade. This cluster contains all of the endocrine steroid hormone receptors (androgen receptor [AR], estrogen receptor α [ERα], estrogen receptor β [ERβ], progesterone receptor [PR]) that govern gender-specific traits and sexual reproduction. The cluster also includes two of the retinoic acid receptors (RARα and RARγ) known for their crucial roles in development and COUP-TFβ. COUP-TFβ is believed to modulate retinoic acid signaling by antagonizing its action at various stages of development (Beland and Lohnes, 2005; Qiu et al., 1996). Although retinoids are known to be required for gonadal function (Chung et al., 2004; Cupp et al., 1999; Lufkin et al., 1993), the unambiguous grouping of these receptors suggests a stronger functional link between retinoid and sex-steroid hormone signaling than was anticipated.
Cluster IC encompasses NRs that are expressed either predominantly or most abundantly in the CNS and, in some cases, receptors that are crucial in maintaining global basal metabolism. This group includes TLX, COUP-TFα, and the NR4A orphan receptors (NGFI-B, NOR1, and NURR1), which are all known for their roles in differentiation of neural cells and peripheral tissues. For example, TLX is required for adult neural stem cell maintenance (Shi et al., 2004); COUP-TFα is a primary mediator of neurogenesis, axonal patterning, and organo-genesis (Park et al., 2003); and the NR4A orphan receptors are required for differentiation of dopaminergic neurons (Perlmann and Wallen-Mackenzie, 2004; Ponnio and Con neely, 2004) and lymphocytes (Winoto and Littman, 2002). The NR4A members also are immediate-early genes in a variety of signaling pathways and are believed to be early transcriptional regulators of adipogenesis and macrophage activation (Barish et al., 2005; Fu et al., 2005). Testicular orphan receptor 4 (TR4) is also in this group, and its loss of function has been implicated in abERRαnt cerebellar development (Chen et al., 2005); however, its physiologic functions have not been elucidated definitively. Cluster IC also includes NRs whose expression in the CNS and periphery is coupled to circadian clocks and metabolism, such as REV-ERBα and β, RORa and β, ERRβ and γ, and the NR4A subfamily. This surprising relationship is especially interesting since many aspects of basal metabolism are under circadian control (see accompanying paper by Yang et al., 2006). Other NRs in this cluster, such as thyroid hormone receptor α (TRα), the mineralocorticoid receptor (MR), and LXRβ, also govern maintenance of cardiovascular function. TRα is responsible for modulating heart and basal metabolic rates (Forrest and Vennstrom, 2000; Marrif et al., 2005), MR is the primary rheostat for blood pressure (Gomez-Sanchez et al., 1996; Le Menuet et al., 2004), and LXRβ modulates expression of proteins that stimulate reverse-cholesterol transport and remodeling of serum lipoproteins (Tontonoz and Mangelsdorf, 2003). Thus, in addition to their abundance in brain, these receptors are highly expressed in all tissues. Finally, this cluster also includes the retinoid X receptors, RXRβ and RXRγ, which serve as obligate heterodimer partners to a number of endocrine and lipid sensing receptors (Chawla et al., 2001). Together this subcluster appears to define a clade of NRs involved in development, higher-order CNS functions, and maintenance of peripheral metabolism.
Hierarchical Clustering of NR Expression: The Nutrient Metabolism and Immune System Axes
In contrast to cluster I, the NRs in cluster II are predominantly expressed within the gastro/enterohepatic axis and key metabolic tissues (e.g., adipose and muscle). Cluster IIA includes NRs that function at the top of a transcriptional cascade governing nutrient uptake and the maintenance of the protective barrier along the enterohepatic tract that is exposed to dietary components. Several NRs in this subcluster, including FXRα, liver receptor homolog-1 (LRH-1), and the FXR target gene SHP are required for governing the metabolism of bile acids, which are crucial for facilitating absorption of fat-soluble nutrients (Edwards et al., 2002; Lu et al., 2001). The FXR/SHP/LRH-1 axis has also been implicated in regulation of lipid and glucose metabolism as a consequence of nutrient intake (Duran-Sandoval et al., 2005; Watanabe et al., 2004; Zhang et al., 2004) and in preserving innate immunity in the gut (Inagaki et al., 2006). The xenobiotic receptors, PXR and CAR, are closely related to FXR and protect the body from dietary xenobiotic lipids as well as promote the clearance of steroids and potentially harmful endogenous bioactive lipids (Dixit et al., 2005; Willson and Kliewer, 2002). Another evolutionarily related NR in this group is VDR, which is crucial for dietary calcium uptake and metabolism (Haussler et al., 1997; Kato, 2000). VDR also functions as a sensor of toxic bile acids in the colon (Makishima et al., 2002) and may protect against colorectal cancer (Kallay et al., 2001). The inclusion of the hepatocyte nuclear factor-4 orphan receptors (HNF4α and γ) reflects the role of these NRs in gastro/enterohepatic developmental programs, carbohydrate metabolism, and insulin action (Chen et al., 1994; Hayhurst et al., 2001; Shih and Stoffel, 2002). RORγ is the final NR in this sub-cluster, although its relationship to other members in the IIA cluster is not yet understood. However, RORg is required for lymph node development and thymopoeisis (He, 2000; Winoto and Littman, 2002).
The NRs in cluster IIB function at the second level of the nutrient regulatory network and comprise a select subset of receptors that governs the utilization of diet-derived lipid nutrients as fuel. Not surprisingly, the majority of NRs in this category form a group that is involved in energy expenditure. For example, the peroxisome proliferator-activated receptors, PPARα and PPARδ, bind to various lipid metabolites and thereby activate oxidation of fatty acids (Evans et al., 2004). ERRα is one of the key transcriptional regulators for inducing oxidative gene expression and driving mitochondrial energy utilization (Luo et al., 2003; Schreiber et al., 2004; Willy et al., 2004). Likewise, TRβ mediates many of the enterohepatic and peripheral tissue responses to thyroid hormone that expend energy to increase body temperature and that increase fatty acid and cholesterol metabolism (Gullberg et al., 2000; Weiss et al., 1998). RXRα is the predominant RXR subtype expressed in metabolic tissues, where it serves as the heterodimeric partner for the other receptors in this category (Chambon, 1996; Chawla et al., 2001). Three other orphan receptors, COUP-TFγ, TR2, and GCNF, are also included in this category, although their roles in adult peripheral tissues remain unclear. Of these three receptors, GCNF has been the most studied and shown to govern early developmental programs in stem cells, neurogenesis, and gonadal cells (Cooney et al., 2001). Likewise, COUP-TFγ (also called EAR2) and TR2 have been characterized for their roles in CNS development (Lee et al., 2002; Warnecke et al., 2005). It is of interest to note that COUPTFg has recently been implicated in the regulation of circa-dian clock genes in the forebrain that govern food-driven entrainment (Warnecke et al., 2005), implying another NR link to nutrient uptake and metabolism.
The third category (cluster IIC) contains three NRs (PPARγ, LXRα, and GR) that govern specialized aspects of fuel utilization, including fat storage, cholesterol metabolism, and glucose metabolism. In general, these receptors reflect the fed and fasted states of the organism. LXRα and PPARγ sense the fed state by responding to dietary cholesterol and fat, promoting the effects of insulin, and facilitating the storage of fat as a fuel source (Chawla et al., 2001; Evans et al., 2004; Kalaany et al., 2005). On the other hand, glucocorticoid activation of GR antagonizes the effects of insulin by promoting lipolysis and gluconeogenesis in the fasted state (Opherk et al., 2004; Wilson and Foster, 1992; Wintermantel et al., 2004). Another intriguing aspect that links these three receptors is their profound anti-inflammatory actions, implicating the existence of an important evolutionary link between the regulation of innate immunity and nutrient metabolism (Ogawa et al., 2005).
PNR is the last NR included in cluster II and, because of its uniquely restricted expression, it falls into its own subcluster. Although it is predominantly expressed in the eye, PNR is also detected at low levels in seminal vesicles and all along the enteric tract. PNR is required for photo-cell development and function (Akhmedov et al., 2000; Haider et al., 2000; Kobayashi et al., 1999), but whether its low level of expression in other tissues implies other functions is unknown.
The above findings indicate that NRs in cluster I are predominantly involved in reproduction and CNS function, while cluster II receptors are predominantly involved in nutrient metabolism and immunity. The division of NRs along these two physiologic axes reveals a striking supra-regulatory relationship that groups a number of receptor clades into higher-order transcriptional networks. These networks phenocopy known physiologic pathways and hint at the molecular underpinning of metabolic homeostasis.
NR Expression Profiles Imply Common Mechanisms for Their Transcriptional Regulation
The observation that several NR subgroups share virtually identical profiles of expression along with related physiology suggests integral patterns may be dictated by regulatory elements in the promoters of these genes. Expression patterns of three of the most related subgroups provide telling examples (Figure 3). One distinctive group includes the enterohepatic receptors, CAR, FXRα, HNF4α, _HNF4_γ, LRH-1, PXR, SHP, and VDR (Figure 3A); a second group includes the CNS receptors, _COUP-TF_α, NURR1, _ROR_β, TLX, and _TR_α (Figure 3B); and a third group includes the steroidogenic-tissue receptors, DAX-1 and SF1 (Figure 3C). In addition to the obvious functional relationship shared by these receptors, the remarkable similarity of their expression profiles clearly implies that common regulatory factors may govern expression of each receptor within these groups. By mining the regulatory regions of the genes for each of these receptors, it may be possible in the future to identify common _cis_-acting motifs and thereby reveal the primary transcriptional components that link a particular physiologic pathway to its anatomical and perhaps even developmental network.
Figure 3. Tissue-Selective Expression of Nuclear Receptors Defines a Common Set of Regulatory Queues.
Expression of NRs that are restricted to specific tissue systems implies the existence of common transcriptional mechanisms to regulate their expression. Examples are shown for NRs expressed in the C57/Bl6 mouse gastroenteric system (A), central nervous system (B), and steroidogenic tissues (C). An asterisk (*) indicates NRs that have a strong circadian expression and were lowest at time of analysis. Representatives of duplicate experiments are shown. Values are plotted as the mean of triplicate measurements ± standard deviation error bars.
Perspectives
Survival of all organisms depends on efficient energy maintenance through acquisition, storage, and utilization and on self-propagation, i.e., reproduction. Both physiologic processes are controlled by deliberate and compulsory actions instigated by the central nervous system signaling to effecter organs in the periphery, which then return information like nutritional status. Within the animal kingdom, this information is relayed through factors (both of endocrine and dietary origin) that are diffused or actively transported from cells, that traverse the body through the bloodstream and that eventually elicit their actions on other tissues. Nuclear receptors permit the integration and communication of such signals between central and peripheral organs because of their established roles as molecular sensors and governors of endocrine-hormone signaling. Understanding how these signaling pathways provide the complexity that is needed to integrate all this information is a major goal of current research. Given that many factors may be required across spatial and temporal distances to govern similar regulatory networks raises a number of key questions. For example, how and why are certain actions elicited in particular tissues and at particular times and not others? And is this activity specification controlled by common sets of cis and trans regulatory components? In the study presented here and in the companion paper by Yang et al. (2006), we utilized QPCR to interrogate the anatomical and circadian expression profiles of the NR superfamily as a means to begin probing these questions in depth. The quantitative power, reproducibility, and integrity of the QPCR data set provide distinct advantages of this technique over other high-throughput profiling methods (e.g., gene chip microarrays), which are often compromised by poor quantitation, incomplete representation of candidate genes, and by the inherent bias of the software that is used to interrogate the data. Indeed, direct comparison of the data presented here with other databases (e.g., SymAtlas and Mouse Gene Prediction Database) revealed numerous quantitative and qualitative discrepancies, including a number of genes that were not represented at all. Thus, the present data set provides an unparalleled molecular fingerprint of NRs that should provide an important resource tool to the scientific community.
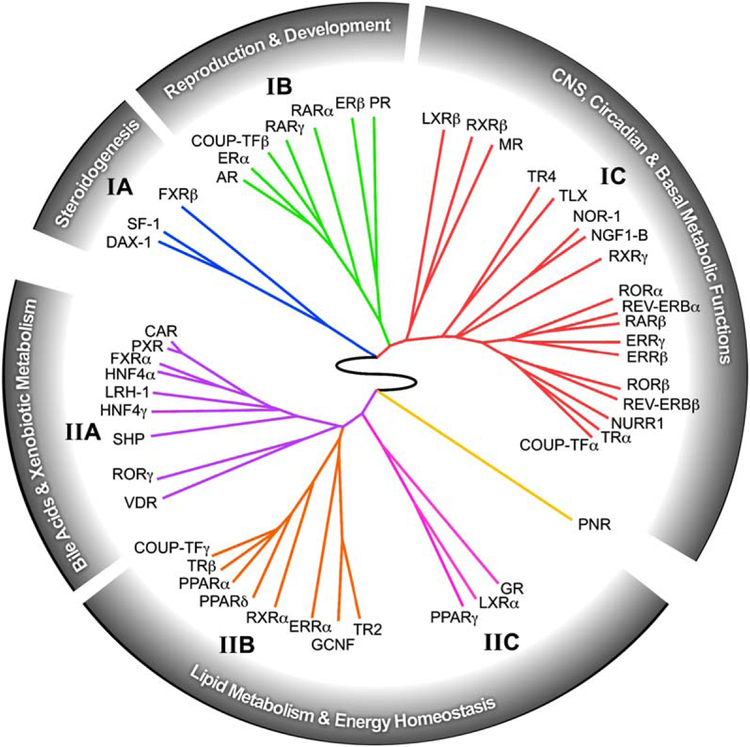
The predictive power of the QPCR data set is shown in the unbiased clustering analysis of the NR expression profile, which could only be accomplished by a comprehensive evaluation of the entire NR superfamily in virtually every tissue system in the animal (Figure 4). Within this ring of NR physiology, a number of interesting receptor associations becomes evident. Perhaps the most remarkable discovery is the inextricable link between NRs and three signature characteristics of animal physiology—reproduction, development, and metabolism. In addition, another prevailing theme revealed by this analysis is the strong nexus that exists between the modulation of nutrient metabolism and immune function, as exemplified by the dual roles of at least 15 different receptors (FXR, GR, LXRs, PPARs, RORs, RXRs, and the NR4A subfamily) in these two physiologic processes. As just one example of the predictive power of this analysis, we note that the identification of GR, LXRα, and PPARγ as a distinct group (cluster IIC) has recently been validated by showing that these receptors function in a combinatorial manner to regulate the evolution of host immune responses (Ogawa et al., 2005). Taken together these data suggest that while individual receptor function is important, the receptors also function together in specific clades as part of higher-order regulatory networks. Thus, investigation of the relationships revealed by this work may lead to the discovery of other combinatorial regulatory networks.
Figure 4. The Nuclear Receptor Ring of Physiology.
The relationship between receptor expression, function, and physiology is depicted as a circular dendrogram using the hierarchical, unsupervised clustering of NR tissue expression distribution profiles obtained from Figure 2. The analysis reveals the existence of a higher-order network tying nuclear receptor function to reproduction, development, central, and basal metabolic functions, dietary-lipid metabolism, and energy homeostasis.
Finally, this work emphasizes the advantage of using QPCR-expression profiling of selected gene arrays (rather than open-ended expression arrays of all genes) to study complex physiologic as well as pathophysiologic processes. Recent studies by us and others have illustrated the utility of this approach for studying adipogenesis, macrophage activation, and cancer (Barish et al., 2005; Fu et al., 2005; Szakacs et al., 2004). One way the QPCR profiling of NRs might be used in the future is to mine information about the regulatory elements required to govern specific clades of receptors that have common expression profiles. The expectation would be that these regulatory elements would reveal the discovery of common sets of transcriptional regulators that not only govern expression of specific NR circuits but also the physiologic pathways encoded within those circuits. The incorporation of further expression-profiling studies on the NR co-activators and corepressors should lead ultimately to the characterization of the master network that governs the interrelationship of each tissue to the organism as a whole.
EXPERIMENTAL PROCEDURES
Animal Care and Use
Six-week-old male and female pure strain C57/Bl6J and 129×1/SvJ mice were purchased from Jackson Laboratory (Bar Harbor, ME). Mice were allowed to reach 8–9 weeks of age on a 12 hr light/dark cycle and fed standard diets (Harlan Teklad #7001; Madison, WI). Females were housed together to ensure a synchronous estrus cycle, although that point was not determined prior to the harvest. All protocols were approved by the University of Texas Southwestern Medical Center Institutional Animal Care and Use Committee.
Tissue Harvest
Male or female mice (n = 6) at 8–9 weeks of age from each strain were sacrificed at lights on (ZT0) by halothane inhalation (Halocarbon Laboratories; River Edge, NJ). Except for female-reproductive tissues, all tissues were from male mice. Mice were exsanguinated via the vena cava, and whole tissues were collected and snap frozen in liquid nitrogen. Tissues were isolated from appropriate anatomical locations according to established laboratory methods or Iwaki et al. (2001). Brown adipose was collected from the dorsal interscapular depression and any surrounding white fat or connective tissue was removed. White adipose was collected from the epididymal area. Skeletal muscle (quadriceps) was isolated from both femurs, and the remaining bone was flushed with PBS to remove bone marrow. The enteric tract, including duodenum, jejunum, ileum, colon, and rectum was sectioned and flushed with PBS, and the intestinal mucosa scraped and collected. The brain was sectioned into pituitary, hypothalamus, brain stem, cerebrum, ol-factory bulb, cerebellum, and corpus striatum, which was the remaining brain after the other sections had been removed (Paxinos and Franklin, 2001). The upper spinal cord was pushed out of the spinal column using an 18G needle. Both uterine horns were collected. Dorsal skin was shaved prior to collection. Pancreas was collected directly into the RNA-isolation reagent and prepared immediately due to its high RNase content. The rest of the snap-frozen tissues were stored at −80°C until RNA extraction.
RNA Isolation and cDNA Preparation
RNA was extracted using RNAStat60 (TelTest; Friendswood, TX) according to the manufacturer’s directions. Modifications were made to the standard protocol to aid in the mechanical disruption of some organs prior to RNA isolation. Skeletal muscle, bone, and skin were crushed into a powder using a Bessmann pulverizer (Fisher Scientific). To minimize differences in lobe-to-lobe mRNA expression, whole livers were also crushed, and the resulting powder homogenized. Total RNA was pooled in equal quantities for each tissue (n = 6). RNA pools from male mice were used for all tissue analyses except those of the ovary and uterus. Genomic DNA contamination was eliminated by DNase treatment using Ambion’s Turbo DNA-free kit (Austin, TX), except for pancreas and seminal vesicle RNA, which were sensitive to degradation. Preparation of cDNA for QPCR assays was performed as previously described (Bookout and Mangelsdorf, 2003) with the following changes: 2.4 μg of RNA was first treated with 2U DNase I and 4.2 mM MgCl2 in a final volume of 40 μl. The reverse-transcription reaction was carried out in 100 ml final volume. Following cDNA synthesis, DEPC-H2O was added to increase the sample volume to 300 μl.
QPCR
Tissue mRNA levels were measured with an ABI 7900HT Sequence Detection System as described previously (Bookout et al., 2005). Prior to pooling RNA samples for nuclear receptor expression analysis, individual tissue samples were assayed for certain difficult-to-dissect tissues to ensure their fidelity. Tissue-specific marker expression was confirmed using the SYBR Green ΔΔCt method as described (Bookout et al., 2005) with the primers summarized in Table S1. Individual RNAs were compared to known, commercially available samples (Ambion, BD Clontech, Harlan Bioproducts, Stratagene).
Analysis of nuclear receptor expression was performed using the TaqMan-based efficiency-corrected ΔCt assay with 10 ng cDNA per reaction for 50 cycles (Bookout et al., 2005). Nuclear receptor mRNAs with cycle times ≥ 34 were determined to be below detection. Primer concentrations were 75 nM for 18S rRNA and 300 nM for nuclear receptor primers, except for DAX-1 and _TR_α, which used predeveloped assays purchased from Applied Biosystems. The primer/probe sets for the 49 mouse nuclear receptors were validated as described (Fu et al., 2005) and are available on our website at www.nursa.org. Universal cDNA standards generated from mouse RNA (BD Clontech) were used for analysis of all receptors except CAR, FXRβ, PXR, SHP, DAX-1, ERβ, LRH-1, PNR, SF1, and TLX, which were too limited in expression to use the universal RNA set. For these receptors tissue-specific total RNA standards were used from liver, ovary, eye, adrenal, and whole brain, as appropriate.
QPCR data were analyzed using ABI instrument software SDS2.1. Baseline values of amplification plots were set automatically and threshold values were kept constant to obtain normalized cycle times and linear regression data. Because PCR efficiencies for each receptor primer set vary, individual receptor PCR efficiencies were determined to permit receptor-to-receptor comparisons. PCR efficiencies were calculated from the slope of the resulting standard curves as reported previously (Fu et al., 2005). Normalized mRNA levels are expressed as arbitrary units and were obtained by dividing the averaged, efficiency-corrected values for nuclear receptor mRNA expression by that for 18S RNA expression for each sample. The resulting values were multiplied by 105 for graphical representation and plotted ± standard deviation (S.D.) from triplicate sample wells.
Hierarchical Clustering
Unsupervised clustering was performed on the normalized RNA levels by calculating Pearson’s centered correlation coefficient (Stekel, 2003) followed by average linkage analysis using Matrix 1.28 software (gift from Dr. Luc Girard). In brief, normalized RNA expression values (in arbitrary units) were input into the Matrix 1.28 software, which then analyzes the data as follows: (1) Pair-wise correlation values are calculated for each receptor-to-receptor pair based on the tissue distribution profile, given the formula (∑(ai−a)(bi−b)/∑(ai−a)2∑(bi−b)2), where a i and b i are data points being compared and a and b are their respective averages. This calculation “centers” the data, meaning that the scale of the y axis is, in essence, ignored. (2) The resulting Pearson coefficients are used to calculate the “distance metric” that is illustrated by the lines connecting each member of a cluster on the heat map. The NR pairs with the highest correlation coefficient segregate together to form a node. The lengths of the lines between the nodes are relative to the strength of their correlations. Tissue-to-tissue associations were clustered first followed by NR-to-NR relationships. The resulting cluster analysis in Figure 2 was then displayed as a wheel dendrogram in Figure 4 using the application Phylodendron (http://iubio.bio.indiana.edu/soft/molbio/java/apps/trees/).
Supplementary Material
Supplemental file 1
Supplemental file 2
ACKNOWLEDGMENTS
We thank Drs. Neil McKenna, Rainer Lanz, Zeljko Jericevic, and David Steffen of the NURSA bioinformatics group (www.nursa.org) for integrating the data into an online resource. We thank the Mango Lab for tissue preparation and critiquing the manuscript, Dr. Carolyn Cummins and Bryan Ong for QPCR data analyses, Dr. Luc Girard for pair-wise correlation and hierarchical clustering code, and Dr. Gerard Manning for rendering the wheel dendrogram. D.J.M. and R.M.E. are investigators of the Howard Hughes Medical Institute (HHMI). This work was funded by HHMI, the Robert A. Welch Foundation (# I-1275), and the National Institutes of Health (Altas Grant #U19DK62434).
Footnotes
Supplemental Data
Supplemental Data include one table and one poster and can be found with this article online at http://www.cell.com/cgi/content/full/126/4/789/DC1/.
The supplemental poster shows a graphical representation of the anatomical and circadian expression profiles of the 49 mouse NRs from this work and the companion paper by Yang et al. (2006). The poster depicts the common and IUPHAR nomenclature for each receptor, along with the chemical structure of known regulatory ligands. The center panel illustrates the phylogenetic tree of the NR superfamily.
REFERENCES
- Akhmedov NB, Piriev NI, Chang B, Rapoport AL, Hawes NL, Nishina PM, Nusinowitz S, Heckenlively JR, Roderick TH, Kozak CA, et al. (2000). A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc. Natl. Acad. Sci. USA 97, 5551–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish GD, Downes M, Alaynick WA, Yu RT, Ocampo CB, Bookout AL, Mangelsdorf DJ, and Evans RM (2005). A Nuclear Receptor Atlas: macrophage activation. Mol. Endocrinol 19, 2466–2477. [DOI] [PubMed] [Google Scholar]
- Beland M, and Lohnes D (2005). Chicken ovalbumin upstream promoter-transcription factor members repress retinoic acid-induced Cdx1 expression. J. Biol. Chem 280, 13858–13862. [DOI] [PubMed] [Google Scholar]
- Beuschlein F, Keegan CE, Bavers DL, Mutch C, Hutz JE, Shah S, Ulrich-Lai YM, Engeland WC, Jeffs B, Jameson JL, and Hammer GD (2002). SF-1, DAX-1, and acd: molecular determinants of adrenocortical growth and steroidogenesis. Endocr. Res 28, 597–607. [DOI] [PubMed] [Google Scholar]
- Bookout AL, and Mangelsdorf DJ (2003). Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl. Recept. Signal 1, e012 10.1621/nrs.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookout AL, Cummins CL, and Mangelsdorf DJ (2005). High-throughput real-time quantitative reverse transcription PCR In Current Protocols in Molecular Biology, Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, and Struhl K, eds. (Hoboken, NJ: John Wiley & Sons, Inc.) [DOI] [PubMed] [Google Scholar]
- Chambon P (1996). A decade of molecular biology of retinoic acid receptors. FASEB J. 10, 940–954. [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, and Mangelsdorf DJ (2001). Nuclear receptors and lipid physiology: opening the X-files. Science 294, 1866–1870. [DOI] [PubMed] [Google Scholar]
- Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, and Darnell JE Jr. (1994). Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 8, 2466–2477. [DOI] [PubMed] [Google Scholar]
- Chen YT, Collins LL, Uno H, and Chang C (2005). Deficits in motor coordination with abERRαnt cerebellar development in mice lacking testicular orphan nuclear receptor 4. Mol. Cell. Biol 25, 2722–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SS, Sung W, Wang X, and Wolgemuth DJ (2004). Retinoic acid receptor alpha is required for synchronization of spermatogenic cycles and its absence results in progressive breakdown of the spermatogenic process. Dev. Dyn 230, 754–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney AJ, Lee CT, Lin SC, Tsai SY, and Tsai MJ (2001). Physiological function of the orphans GCNF and COUP-TF. Trends Endocrinol. Metab 12, 247–251. [DOI] [PubMed] [Google Scholar]
- Cupp AS, Dufour JM, Kim G, Skinner MK, and Kim KH (1999). Action of retinoids on embryonic and early postnatal testis development. Endocrinology 140, 2343–2352. [DOI] [PubMed] [Google Scholar]
- Dixit SG, Tirona RG, and Kim RB (2005). Beyond CAR and PXR. Curr. Drug Metab 6, 385–397. [DOI] [PubMed] [Google Scholar]
- Duran-Sandoval D, Cariou B, Percevault F, Hennuyer N, Grefhorst A, van Dijk TH, Gonzalez FJ, Fruchart JC, Kuipers F, and Staels B (2005). The farnesoid X receptor modulates hepatic carbohydrate metabolism during the fasting-refeeding transition. J. Biol. Chem 280, 29971–29979. [DOI] [PubMed] [Google Scholar]
- Edwards PA, Kast HR, and Anisfeld AM (2002). BAREing it all: the adoption of LXR and FXR and their roles in lipid homeostasis. J. Lipid Res 43, 2–12. [PubMed] [Google Scholar]
- Evans RM, Barish GD, and Wang YX (2004). PPARs and the complex journey to obesity. Nat. Med 10, 355–361. [DOI] [PubMed] [Google Scholar]
- Forrest D, and Vennstrom B (2000). Functions of thyroid hormone receptors in mice. Thyroid 10, 41–52. [DOI] [PubMed] [Google Scholar]
- Fu M, Sun T, Bookout AL, Downes M, Yu RT, Evans RM, and Mangelsdorf DJ (2005). A nuclear teceptor atlas: 3T3-L1 adipogenesis. Mol. Endocrinol 19, 2437–2450. [DOI] [PubMed] [Google Scholar]
- Gomez-Sanchez EP, Zhou M, and Gomez-Sanchez CE (1996). Mineralocorticoids, salt and high blood pressure. Steroids 61, 184–188. [DOI] [PubMed] [Google Scholar]
- Gronemeyer H, Gustafsson JA, and Laudet V (2004). Principles for modulation of the nuclear receptor superfamily. Nat. Rev. Drug Discov 3, 950–964. [DOI] [PubMed] [Google Scholar]
- Gullberg H, Rudling M, Forrest D, Angelin B, and Vennstrom B (2000). Thyroid hormone receptor beta-deficient mice show complete loss of the normal cholesterol 7alpha-hydroxylase (CYP7A) response to thyroid hormone but display enhanced resistance to dietary cholesterol. Mol. Endocrinol 14, 1739–1749. [DOI] [PubMed] [Google Scholar]
- Haider NB, Jacobson SG, Cideciyan AV, Swiderski R, Streb LM, Searby C, Beck G, Hockey R, Hanna DB, Gorman S, et al. (2000). Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat. Genet 24, 127–131. [DOI] [PubMed] [Google Scholar]
- Haussler MR, Haussler CA, Jurutka PW, Thompson PD, Hsieh JC, Remus LS, Selznick SH, and Whitfield GK (1997). The vita-min D hormone and its nuclear receptor: molecular actions and disease states. J. Endocrinol 154 (Suppl), S57–S73. [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, and Gonzalez FJ (2001). Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol 21, 1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YW (2000). The role of orphan nuclear receptor in thymocyte differentiation and lymphoid organ development. Immunol. Res 22, 71–82. [DOI] [PubMed] [Google Scholar]
- Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, et al. (2006). Regulation of mucosal defense in intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci. USA 103, 3920–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki T, Yamashita H, and Hayawaka T (2001). A Color Atlas of Sectional Anatomy of the Mouse (Tokyo: Adthree; ). [Google Scholar]
- Kalaany NY, Gauthier KC, Zavacki AM, Mammen PP, Kitazume T, Peterson JA, Horton JD, Garry DJ, Bianco AC, and Mangelsdorf DJ (2005). LXRs regulate the balance between fat storage and oxidation. Cell Metab. 1, 231–244. [DOI] [PubMed] [Google Scholar]
- Kallay E, Pietschmann P, Toyokuni S, Bajna E, Hahn P, Mazzucco K, Bieglmayer C, Kato S, and Cross HS (2001). Characterization of a vitamin D receptor knockout mouse as a model of colorectal hyperproliferation and DNA damage. Carcinogenesis 22, 1429–1435. [DOI] [PubMed] [Google Scholar]
- Kato S (2000). The function of vitamin D receptor in vitamin D action. J. Biochem. (Tokyo) 127, 717–722. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Takezawa S, Hara K, Yu RT, Umesono Y, Agata K, Taniwaki M, Yasuda K, and Umesono K (1999). Identification of a photoreceptor cell-specific nuclear receptor. Proc. Natl. Acad. Sci. USA 96, 4814–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YF, Lee HJ, and Chang C (2002). Recent advances in the TR2 and TR4 orphan receptors of the nuclear receptor superfamily. J. Steroid Biochem. Mol. Biol 81, 291–308. [DOI] [PubMed] [Google Scholar]
- Le Menuet D, Viengchareun S, Muffat-Joly M, Zennaro MC, and Lombes M (2004). Expression and function of the human mineralocorticoid receptor: lessons from transgenic mouse models. Mol. Cell. Endocrinol 217, 127–136. [DOI] [PubMed] [Google Scholar]
- Lu TT, Repa JJ, and Mangelsdorf DJ (2001). Orphan nuclear receptors as eLiXiRs and FiXeRs of sterol metabolism. J. Biol. Chem 276, 37735–37738. [DOI] [PubMed] [Google Scholar]
- Lufkin T, Lohnes D, Mark M, Dierich A, Gorry P, Gaub MP, Le-Meur M, and Chambon P (1993). High postnatal lethality and testis degeneration in retinoic acid receptor alpha mutant mice. Proc. Natl. Acad. Sci. USA 90, 7225–7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Sladek R, Carrier J, Bader JA, Richard D, and Giguere V (2003). Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha. Mol. Cell. Biol 23, 7947–7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, and Mangelsdorf DJ (2002). Vitamin D receptor as an intestinal bile acid sensor. Science 296, 1313–1316. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, and Evans RM (1995). The nuclear receptor superfamily: the second decade. Cell 83, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrif H, Schifman A, Stepanyan Z, Gillis MA, Calderone A, Weiss RE, Samarut J, and Silva JE (2005). Temperature homeostasis in transgenic mice lacking thyroid hormone receptor-alpha gene products. Endocrinology 146, 2872–2884. [DOI] [PubMed] [Google Scholar]
- MGD (Mouse Genome Database). (2003). Mouse Genome Informatics Website (Bar Harbor, ME: The Jackson Laboratory; ). http://www.informatics.jax.org/. [Google Scholar]
- Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG, and Glass CK (2005). Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell 122, 707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opherk C, Tronche F, Kellendonk C, Kohlmuller D, Schulze A, Schmid W, and Schutz G (2004). Inactivation of the glucocorticoid receptor in hepatocytes leads to fasting hypoglycemia and ameliorates hyperglycemia in streptozotocin-induced diabetes mellitus. Mol. Endocrinol 18, 1346–1353. [DOI] [PubMed] [Google Scholar]
- Otte K, Kranz H, Kober I, Thompson P, Hoefer M, Haubold B, Remmel B, Voss H, Kaiser C, Albers M, et al. (2003). Identification of farnesoid X receptor beta as a novel mammalian nuclear receptor sensing lanosterol. Mol. Cell. Biol 23, 864–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JI, Tsai SY, and Tsai MJ (2003). Molecular mechanism of chicken ovalbumin upstream promoter-transcription factor (COUPTF) actions. Keio J. Med 52, 174–181. [DOI] [PubMed] [Google Scholar]
- Parker KL, and Schimmer BP (2002). Genes essential for early events in gonadal development. Ann. Med 34, 171–178. [PubMed] [Google Scholar]
- Paxinos G, and Franklin KBJ (2001). The Mouse Brain in Stereo-taxic Coordinates, Second Edition (San Diego, CA: Academic Press; ). [Google Scholar]
- Perlmann T, and Wallen-Mackenzie A (2004). Nurr1, an orphan nuclear receptor with essential functions in developing dopamine cells. Cell Tissue Res. 318, 45–52. [DOI] [PubMed] [Google Scholar]
- Ponnio T, and Conneely OM (2004). nor-1 regulates hippocampal axon guidance, pyramidal cell survival, and seizure susceptibility. Mol. Cell. Biol 24, 9070–9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Krishnan V, Pereira FA, Tsai SY, and Tsai MJ (1996). Chicken ovalbumin upstream promoter-transcription factors and their regulation. J. Steroid Biochem. Mol. Biol 56, 81–85. [DOI] [PubMed] [Google Scholar]
- Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, and Kralli A (2004). The estrogen-related receptor alpha (ERRαlpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. USA 101, 6472–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Chichung Lie D, Taupin P, Nakashima K, Ray J, Yu RT, Gage FH, and Evans RM (2004). Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature 427, 78–83. [DOI] [PubMed] [Google Scholar]
- Shih DQ, and Stoffel M (2002). Molecular etiologies of MODY and other early-onset forms of diabetes. Curr. Diab. Rep 2, 125–134. [DOI] [PubMed] [Google Scholar]
- Shulman AI, and Mangelsdorf DJ (2005). Retinoid × receptor heterodimers in the metabolic syndrome. N. Engl. J. Med 353, 604–615. [DOI] [PubMed] [Google Scholar]
- Stekel D (2003). Microarray Bioinformatics (New York: Cambridge University Press; ). [Google Scholar]
- Szakacs G, Annereau JP, Lababidi S, Shankavaram U, Arciello A, Bussey KJ, Reinhold W, Guo Y, Kruh GD, Reimers M, et al. (2004). Predicting drug sensitivity and resistance: profiling ABC transporter genes in cancer cells. Cancer Cell 6, 129–137. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, and Mangelsdorf DJ (2003). Liver X receptor signaling pathways in cardiovascular disease. Mol. Endocrinol 17, 985–993. [DOI] [PubMed] [Google Scholar]
- Warnecke M, Oster H, Revelli JP, Alvarez-Bolado G, and Eichele G (2005). Abnormal development of the locus coeruleus in Ear2(Nr2f6)-deficient mice impairs the functionality of the forebrain clock and affects nociception. Genes Dev. 19, 614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, Moore DD, and Auwerx J (2004). Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J. Clin. Invest 113, 1408–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RE, Murata Y, Cua K, Hayashi Y, Seo H, and Refetoff S (1998). Thyroid hormone action on liver, heart, and energy expenditure in thyroid hormone receptor beta-deficient mice. Endocrinology 139, 4945–4952. [DOI] [PubMed] [Google Scholar]
- Willson TM, and Kliewer SA (2002). PXR, CAR and drug metabolism. Nat. Rev. Drug Discov 1, 259–266. [DOI] [PubMed] [Google Scholar]
- Willy PJ, Murray IR, Qian J, Busch BB, Stevens WC Jr., Martin R, Mohan R, Zhou S, Ordentlich P, Wei P, et al. (2004). Regulation of PPARgamma coactivator 1alpha (PGC-1alpha) signaling by an estrogen-related receptor alpha (ERRαlpha) ligand. Proc. Natl. Acad. Sci. USA 101, 8912–8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JD, and Foster DW (1992). William’s Textbook of Endocrinology, Eighth Edition (Philadelphia: W.B. Saunders Co.). [Google Scholar]
- Winoto A, and Littman DR (2002). Nuclear hormone receptors in T lymphocytes. Cell 109 (Suppl), S57–S66. [DOI] [PubMed] [Google Scholar]
- Wintermantel TM, Berger S, Greiner EF, and Schutz G (2004). Genetic dissection of corticosteroid receptor function in mice. Horm. Metab. Res 36, 387–391. [DOI] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, and Evans RM (2006). Nuclear receptor expression links the circadian clock to metabolism. Cell 126, this issue, 801–810. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Castellani LW, Sinal CJ, Gonzalez FJ, and Edwards PA (2004). Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes Dev. 18, 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental file 1
Supplemental file 2