Drosophila Hyperkinetic Mutants Have Reduced Sleep and Impaired Memory (original) (raw)
Abstract
In mammals, sleep is thought to be important for health, cognition, and memory. Fruit flies share most features of mammalian sleep, and a recent study found that Drosophila lines carrying loss-of-function mutations in Shaker (Sh) are short sleeping, suggesting that the Sh current plays a major role in regulating daily sleep amount. The Sh current is potentiated by a β modulatory subunit coded by Hyperkinetic (Hk). Here, we demonstrate that severe loss-of-function mutations of Hk reduce sleep and do so primarily by affecting the Sh current. Moreover, we prove, using a transgenic approach, that a wild-type copy of Hk is sufficient to restore normal sleep. Furthermore, we show that short-sleeping Hk mutant lines have a memory deficit, whereas flies carrying a weaker hypomorphic Hk allele have normal sleep and normal memory. By comparing six short-sleeping Sh lines with two normal sleeping ones, we also found that only alleles that reduce sleep also impair memory. These data identify a gene, Hk, which is necessary to maintain normal sleep, and provide genetic evidence that short sleep and poor memory are linked.
Keywords: Drosophila, sleep, learning, memory, Shaker, hyperkinetic
Introduction
Sleep is thought to be important for health, cognition, and memory (Horne, 1988; Bonnet and Arand, 1997; Durmer and Dinges, 2005). Many features of sleep are shared between mammals and fruit flies. As in mammals, sleep in Drosophila consists of long periods of behavioral immobility with increased arousal threshold (Hendricks et al., 2000; Shaw et al., 2000), is associated with changes in brain electrical activity (Nitz and Tononi, 2002) and gene expression (Cirelli et al., 2005a; Zimmerman et al., 2006), is reduced by caffeine and stimulants (Shaw et al., 2000; Hendricks et al., 2003a; Andretic et al., 2005), and becomes fragmented with aging (Koh et al., 2006). In both mammals and flies, sleep is homeostatically regulated, because its duration and intensity increase with the duration of previous waking (Huber et al., 2004), and sleep deprivation (SD) results in reduced performance (Huber et al., 2004).
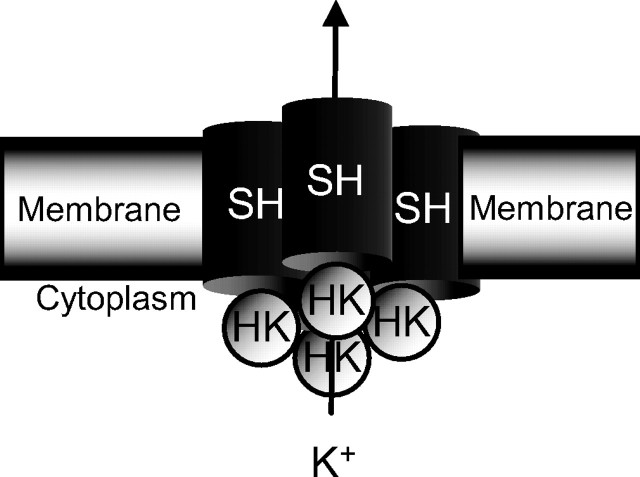
In a recent study, we found that Shmns flies, which carry a point mutation in a conserved Shaker (Sh) domain, sleep only 3–4 h/d, whereas their wild-type controls sleep 8–14 h/d (Cirelli et al., 2005b). After crossing out genetic modifiers accumulated over many generations, we found that other Sh alleles become short sleepers and fail to complement the short sleeping Shmns phenotype, suggesting that the Sh current is responsible for the short sleeping phenotype. The Sh locus encodes the α subunit of a tetrameric potassium channel that passes a voltage-activated fast-inactivating (_I_A) current (Schwarz et al., 1988). Sh orthologs (Kv) occur in vertebrates (Littleton and Ganetzky, 2000) and, in both mammals and flies, play a major role in the control of membrane repolarization and transmitter release (Schwarz et al., 1988). Hyperkinetic (Hk) encodes a β subunit that binds to each α subunit in the Sh tetramer (Fig. 1), and its presence increases current amplitude and alters both voltage dependence and kinetics of inactivation and activation (Chouinard et al., 1995). Hk loss-of-function mutations, however, enhance neuronal excitability (Yao and Wu, 1999). Hk orthologs are also present in mammals, where they also bind to members of the Kv1 gene family (Rettig et al., 1994). These channels are members of the aldo-keto reductase family, and it has been suggested that their redox-sensor is responsible for modulating their activity over long periods of time (Gulbis et al., 1999).
Figure 1.
Schematic showing the Hk β modulatory unit attached on the cytoplasmic side to the Sh pore. A fast-inactivating voltage-dependent potassium (K+) current passes through the pore.
In this study, we tested whether flies carrying severe Hk loss-of-function mutations are short sleeping, and found that this was the case. Moreover, we found that mutations of Hk reduce sleep primarily by affecting the Sh current and that short-sleeping Hk mutant flies are restored to normal sleep after insertion of a wild-type copy of Hk. Thus, we demonstrate, using a transgenic approach, that Hk is required to maintain normal sleep in Drosophila. Finally, we also found that all characterized severe Sh or Hk loss-of-function mutations not only reduce sleep but also impair memory, suggesting a link between the two phenotypes.
Materials and Methods
Animals.
Flies (1–2 weeks old) were cultured and tested at 20°C and 68% humidity on yeast, dark corn syrup, and agar food. Hk1, HkY, and Hk2 were induced by ethyl methanesulfonate and are probably point mutations (Schlimgen, 1991). Flies carrying Hk mutant alleles, Hk+ transgenes, and balancers were crossed at least four generations into the respective genetic background. Df(Sh)B55-W32 refers to the synthetic deletion produced using T(1;Y)B55 and T(1;Y)W32 (Papazian et al., 1987).
Locomotor activity, sleep, and measures of sleep intensity.
Experiments included 1 d of adaptation, 2 baseline days, 1 SD day and 2 recovery days after SD. At the beginning of the experiment, individual flies were placed in the Drosophila Activity Monitor System (DAMS; Trikinetics, Waltham, MA) inside glass tubes with enough food for 1 week of recording. Monitors were housed inside environmental chambers (ThermoForma, Marietta, OH) where temperature and humidity were kept constant. Data analysis was performed by a custom-designed software developed in our laboratory (Huber et al., 2004) and based on Statistica (StatSoft, Tulsa, OK). The data were further analyzed using Matlab (MathWorks, Natick, MA). Sleep and wakefulness were determined for consecutive 1 min epochs. Wakefulness was defined as any period of ≥1 min characterized by activity (≥1 count/min). Based on previous work (Huber et al., 2004), sleep was defined as any period of uninterrupted behavioral immobility (0 counts/min) lasting >5 min. The duration of sleep episodes was calculated by counting the number of consecutive 1 min epochs of sleep.
Escape response to a complex stimulus.
Flies were exposed to a complex stimulus consisting of a combination of noise and vibration. Flies remained inside a DAMS, which was inserted into a custom-made frame specifically designed for the test (Huber et al., 2004). The stimulus was produced by flaps vigorously pushed for a few seconds against the glass tubes housing the flies. Such stimulus was delivered once every hour at either side of the tubes via a computer-controlled motor, and flies were tested for a total of 48 h, including 1 baseline day and the first recovery day after SD. Previous studies have shown that most flies move away from the stimulus (and by doing so cross the infrared beam) if, before its delivery, they had been actively moving around. In contrast, most flies do not show an escape response if they had been immobile for 5 min before the stimulus was delivered (Huber et al., 2004). Thus, the percentage of nonresponsive flies is used as a measure of the arousal threshold to distinguish flies that are awake from sleeping flies.
Escape response to heat.
Single flies were placed inside a heat-box where position and movements of the fly were recorded and displayed on-line (Huber et al., 2004). Flies were first adapted to the chamber for 10 min. Temperature on either side of the chamber was then alternately increased every 1 min (23, 25, 27, 30, 33, 36, 39, 42°C), whereas the other side of the chamber was kept at 21°C. The latency (i.e., the time a fly needed to move to the cooler side of the chamber) was measured for each temperature step. Latencies for all temperature steps were averaged for each fly. Most flies took <8 s to move to the cold side of the chamber. Pilot studies showed that the response to heat does not habituate in flies tested during 2 consecutive baseline days.
SD.
During SD, flies remained in the DAMS, which was placed vertically inside a framed box able to rotate along its major axis under the control of a motor (Huber et al., 2004). The box could rotate 180° clock-wise or counter clock-wise (2–3 rpm). At the nadir of each rotation, the monitor was dropped 1 cm. This caused the flies to fall from their current position to the bottom of the tube. Previous studies (Huber et al., 2004) had shown that this method is effective in reducing total sleep time by >90%. Because locomotor activity during SD was continuously recorded, the extent of sleep loss could be calculated for each individual fly.
Heat box experimental setup.
The conditioning apparatus is a modified version of the one used in Wustmann et al. (1996). It consists of an array of 16 chambers (26 × 4 × 2 mm) operated in parallel, each with Peltier elements on the top and bottom allowing for fast heating and cooling. Experiments were performed in complete darkness. Chambers were cleaned with a pipe cleaner every day before experiments. The standard experiment consisted of three phases: pretraining (10 min), training (9 min), and post-training (10 min), which are described in the legend of Figure 8. Data were derived from flies that met the following behavioral criteria: (1) crossed the midline >10 times (average 1 crossing/min) during pretraining; (2) traveled at least half the chamber during pretraining; (3) crossed over to the heated side at least once during training. Both mutant and wild-type siblings were tested in parallel. After finishing with one group (mutant and wild type), the side that was heated was switched. Flies did not show a preference for either side of the chamber.
Figure 8.
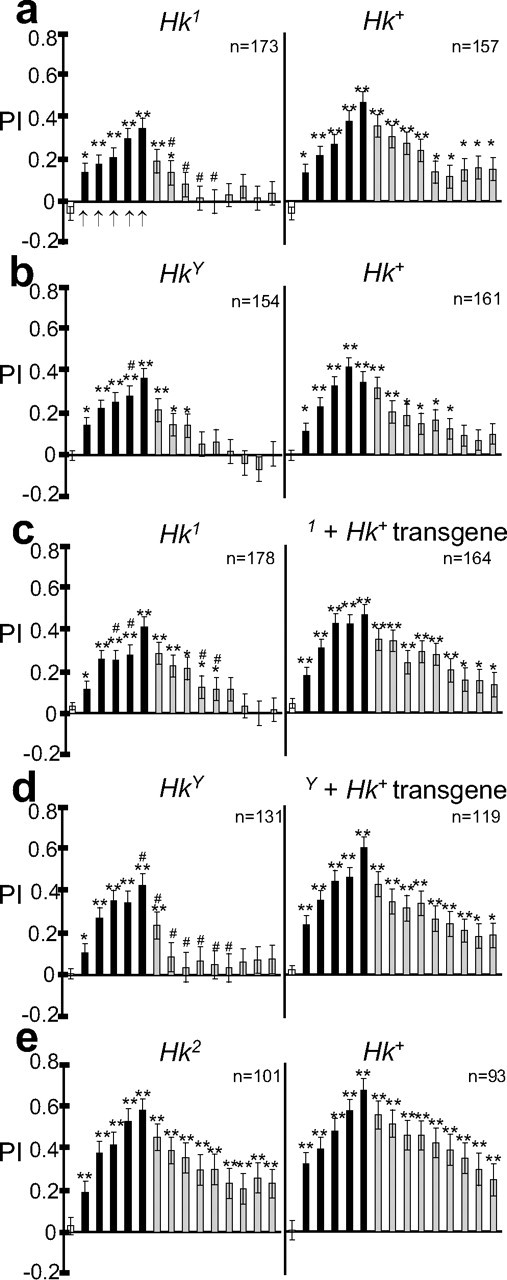
Hk mutations that reduce sleep also impair performance in the heat-box learning paradigm. Performance in the heat box for Hk mutants and their wild-type siblings (a, b, e), and siblings that inherited the _Hk_17K-X94 transgenic construct (c, d), is shown (all data are from males). The performance index (PI; mean ± SEM) is calculated as the difference between the time the fly spent in the nonheated versus heated half of the chamber divided by the total time. PI can range from −1 (all time is spent on the heated side) to +1 (all time is spent on the nonheated side), whereas 0 indicates no preference for either side. In each panel, the white bar on the far left represents the 10 min pretraining period when both sides of the chamber were kept at 22°C, and indicates that flies did not show any side preference during pretraining. From left to right, Each arrow indicates consecutive 1 min training periods, when the punished side was heated to 39°C. Each black bar represents consecutive 1 min periods of testing (both sides at 22°C) immediately after each training period. Gray bars represent consecutive 1 min periods after training has ended. Temperature sensitivity did not differ between mutant and wild-type siblings (data not shown; temperature sensitivity was also tested by measuring the escape response, as described in Materials and Methods, and did not differ between lines). PI measures learning (when testing periods alternate with training periods) or memory (after the training periods have ended). All flies learned to prefer the nonheated side of the chamber, with no significant differences between mutant and wild-type siblings (asterisks indicate periods with a significant preference for the nonheated side; * p < 0.05, **p < 0.001, signrank test). After training, however, short sleeping (Hk1, HkY), but not normal sleeping (Hk2) flies lost their preference for the nonheated side more quickly. Periods significantly different between mutant and wild-type siblings are marked with hash marks (p < 0.05, unpaired Student's t test). Data were collected over ≥3 different days, and in each experiment mutants and wild-type flies were tested in parallel.
Statistical analysis.
Two-way ANOVAs with factors “day” (e.g., baseline vs recovery) and “line” were used to analyze the data. Contrasts were tested by post hoc t test if the main factor or interaction reached significance.
Results
Mutations in Hk reduce sleep
To test whether mutations in Hk result in a short-sleeping phenotype, we first backcrossed the Hk1 and HkY mutations into three different genetic backgrounds (wCS10, CS, w1118) (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). As shown in Figure 2, we found that in all tested genetic backgrounds, _Hk_− males spent 30–54% less time asleep compared with their wild-type siblings (Fig. 2a,b). We then tested _Hk_− females in the CS and _w_1118 background. Again _Hk_− homozygous females had reduced sleep (from 10 to 53% less) compared with their wild-type siblings (Fig. 2c,d). Daily sleep amount did not differ between heterozygous and wild-type females (Fig. 2c), indicating that mutations in Hk act recessively to reduce sleep. During waking, both male and female mutant flies were more active than their wild-type siblings (Fig. 2a,c). However, the decrease in daily sleep negatively correlated with the increase in waking locomotor activity in females (_r_2 = 0.55; F = 0.034) but not in males (_r_2 = 0.018; F = 0.80), suggesting that the two phenotypes could be dissociated (supplemental Fig. 2, available at www.jneurosci.org as supplemental material).
Figure 2.
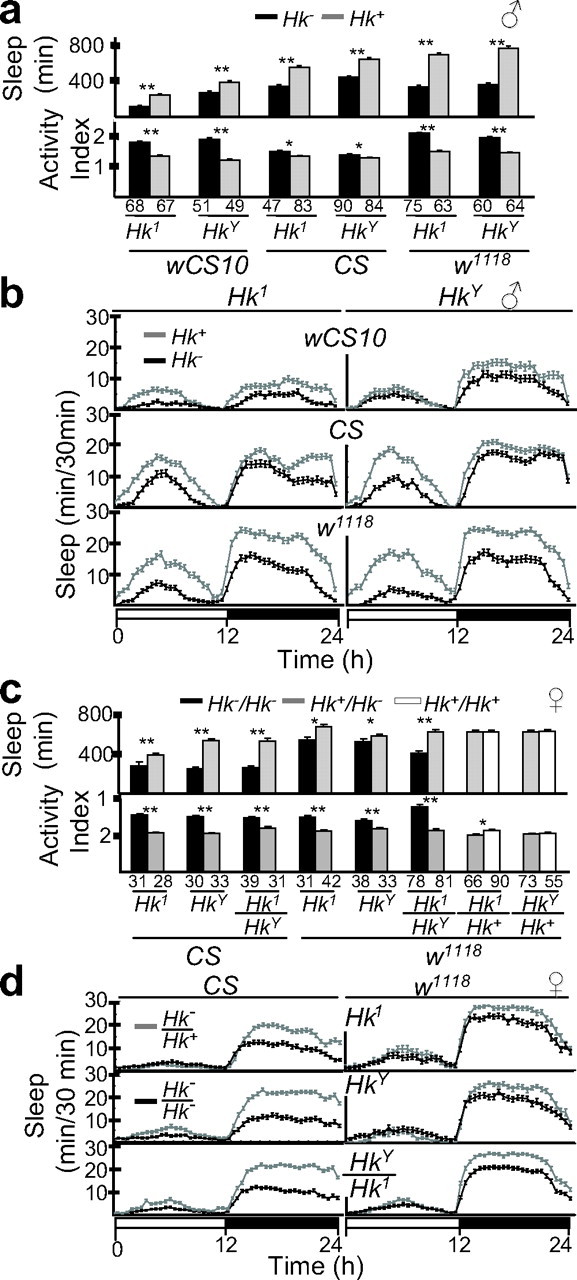
Severe mutations in Hk reduce sleep in male and female flies. a, Males: minutes of sleep over a 24 h period (mean ± SEM) and activity index (number of beam crossings per minute during waking; mean ± SEM). The numbers of tested flies is indicated below each column. b, Males: daily time course (in 30 min intervals; mean ± SEM) of the amount of sleep in Hk1 (left) and HkY (right) flies. White and black bars under the _x_-axis indicate the light and dark period, respectively. The same data set used to calculate total sleep amount and activity index were used to produce the daily sleep time course. c, d, Females: sleep amount, activity index, and daily sleep time course are displayed using the same units used for males. Strains wCS10, CS, and _w_1118 were chosen because they differ significantly in daily sleep amount. Black, gray, and white bars or lines represent mutant, heterozygotes, and homozygous wild-type flies, respectively. Comparisons are made between siblings, shown next to each other. *p < 0.05; **p < 0.001; unpaired Student's t test.
The decrease in daily sleep amount was most consistently associated, in both males and females, with a decrease in the duration of sleep episodes (Fig. 3). In contrast, the effects of Hk mutations on the number of sleep episodes, and on the number and duration of waking bouts, varied significantly depending on sex and genetic background (Fig. 3). As shown in supplemental Figure 2 (available at www.jneurosci.org as supplemental material), the changes in the 24 h sleep amount were positively correlated with the changes in the average duration of sleep bouts (males: _r_2 = 0.97, F <0.001; females: _r_2 = 0.79, F = 0.0034), but not with the changes in the number of sleep bouts (males: _r_2 = 0.63, F = 0.058; females: _r_2 = 0.24, F = 0.22) or in the number or duration of waking episodes.
Figure 3.
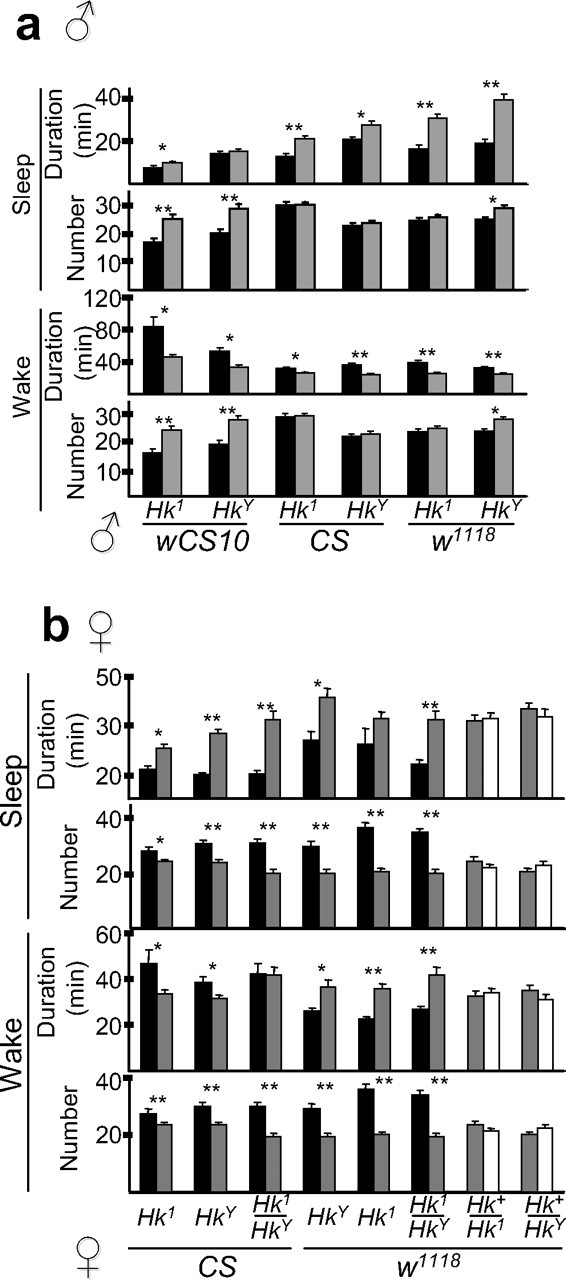
The effects of severe Hk mutations on sleep and waking parameters depend on strain and gender. Duration (in minutes) and number of sleep and waking episodes over a 24 h period (mean ± SEM) in male (a) and female (b) flies (same flies shown in Fig. 2). *p < 0.05; **p < 0.001; Student's t test.
Mutations in Hk are responsible for the short-sleeping phenotype
To determine whether mutations in Hk are responsible for the short-sleeping phenotype, four sets of experiments were performed. First, genetic mapping indicated that both the shaking phenotype and the short-sleeping phenotype were mapped to a locus between cv and v that includes the Hk locus (supplemental Figs. 3, 4, available at www.jneurosci.org as supplemental material). A second experiment tested whether Hk1 and HkY failed to complement each other. Indeed, like homozygous Hk1/Hk1 and HkY/HkY flies, Hk1/HkY flies also slept significantly less than their heterozygous siblings that inherited a wild-type Hk+ allele (Fig. 2c,d). Hk1 and HkY are two independent mutations, and it is unlikely that both of them carry a second mutation in the same gene that are different from Hk but close to the Hk locus. Moreover, in a third experiment, both Hk1 and HkY failed to complement three deficiencies that span the Hk locus for the recessive short-sleeping phenotype (Fig. 4). Thus, genetic mapping, complementation analysis, and cytological mapping confirmed that mutations within the Hk locus reduce sleep and increase waking activity.
Figure 4.
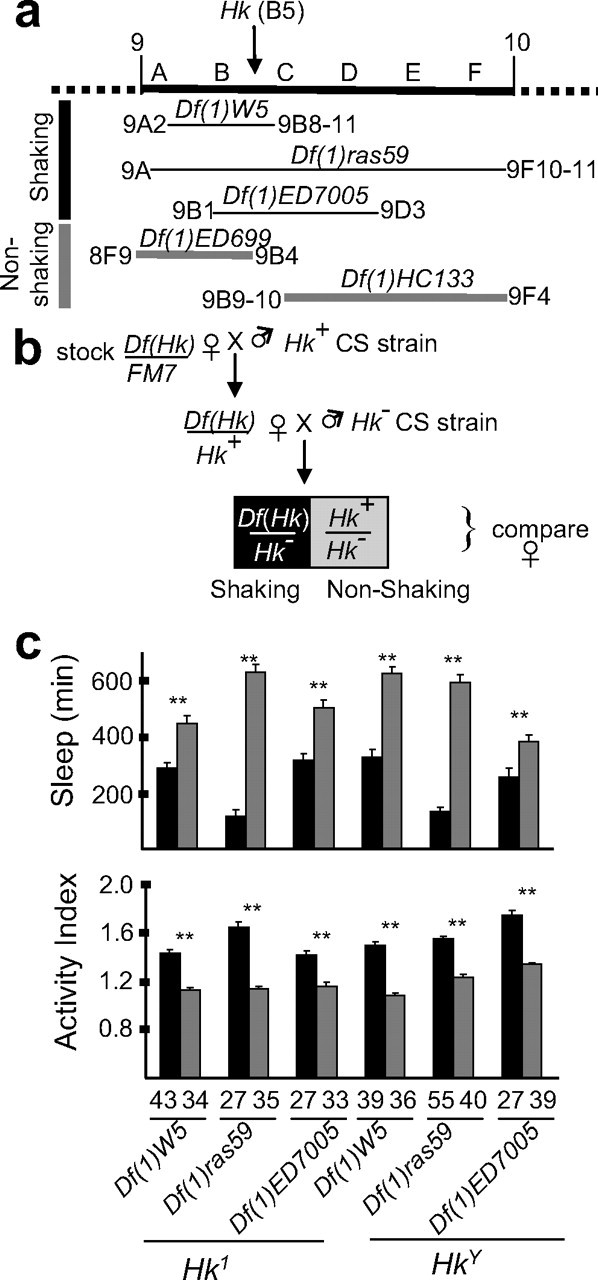
Deficiencies that span the Hk locus fail to complement the short-sleeping phenotype. a, Schematic representation of the region on the X chromosome containing the Hk locus (9B5). As expected, deficiency lines Df(1)W5, Df(1)ras59, and Df(1)ED7005, which include the Hk locus, produced a shaking phenotype when crossed to either Hk1 or HkY, whereas deficiencies that did not include 9B5 (Df(1)ED699, Df(1)HC13) did not. b, Crossing scheme used to perform complementation tests with Hk1 and HkY. c, Daily sleep amounts (mean ± SEM; minutes of sleep per 24 h) and activity index (number of beam crossings per minute during waking). As in b, black bars represent the _Df(Hk)/Hk_− combination, whereas gray bars represent the _Hk_−/Hk+ combination. The deficiency and _Hk_− allele tested are indicated below the siblings pairs. The number of tested flies is indicated below each column. **p < 0.001; unpaired Student's t test.
In the fourth experiment, we tested whether a wild-type Hk+ transgene could rescue the wild-type sleeping phenotype. The wild-type _Hk_17K-X94 transgene is known to rescue the shaking phenotype produced by either the Hk1 or the HkY mutation (Chouinard et al., 1995). We compared sleep in flies carrying a copy of _Hk_17K-X94 in the presence of either a mutant or a wild-type copy of Hk (Fig. 5a). Male _Hk_− mutants that inherited the transgene slept significantly more than their _Hk_− siblings (Fig. 5b,c), and their daily sleep amount did not differ consistently from that of wild-type siblings that inherited the balancer chromosome FM7a (Fig. 5c) or the chromosome with a wild-type Hk+ allele (compare Figs. 2a, 5c). Importantly, the transgene did not consistently increase the daily sleep amount in flies that inherited a wild-type Hk+ allele (Fig. 5c). Thus, a wild-type Hk+ transgene specifically rescues the short sleeping phenotype caused by a mutation in Hk. Whereas the rescue of the short-sleeping phenotype was usually associated with a decrease in waking locomotor activity, this was not the case for Hk1 mutants in the wCS10 background (Fig. 5c), again suggesting that Hk effects on sleep and activity can be dissociated.
Figure 5.
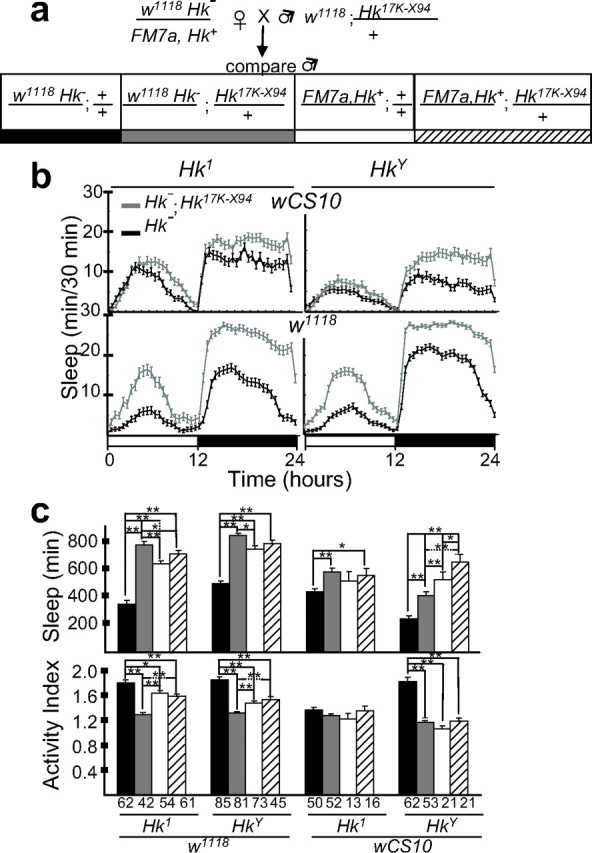
A wild-type Hk transgene rescues the short-sleeping phenotype. a, Crossing scheme. In addition to a 17 kb genomic fragment containing the wild-type Hk+ gene, the _Hk_17K-X94 transgenic construct includes the wild-type w+ transgene, which allowed identification based on eye color in a _w_− mutant (w1118 or wCS10) background. b, Daily time course (in 30 min intervals; mean ± SEM) of the amount of sleep in _Hk_− males (black line), and in _Hk_− males carrying the transgene (gray line). The color code is according to the scheme in panel a. c, Daily sleep amounts (mean ± SEM; minutes of sleep per 24 h) and activity index (number of beam crossings per minute during waking). The numbers of tested flies is indicated below each column. The color code is according to the scheme in panel a. *p < 0.05; **p < 0.001; unpaired Student's t test. The analysis included an FM7a balancer chromosome dominantly marked with Bar1 (B1) in addition to recessive markers (_y_31d _s.c._8 wa vOf). This FM7a balancer has an Hk+ allele, because in female flies it does not produce a shaking phenotype in combination with either Hk1 or HkY. FM7a was selected among all of the X chromosome balancers tested because it does not significantly affect daily sleep amount in an _w_1118 background.
The homeostatic and circadian regulation of sleep are normal in Hk mutants
When subjected to 24 h of SD, _Hk_− and Hk+ flies lost >90% of their baseline sleep, and showed an increase in sleep duration during the 24 h after SD (Fig. 6). Interestingly, the amount of sleep recovered varied depending on genetic background. In all cases, however, the percentage of increase in sleep duration did not differ between the _Hk_− mutants and their wild-type siblings (Fig. 6a). After SD, sleep in _Hk_− flies became more intense, as indicated by an increase in arousal thresholds, but again _Hk_− flies and their wild-type siblings showed similar changes (Fig. 6b). Finally, both sleep-deprived Hk_− and Hk+ siblings were impaired in their ability to move away from a complex stimulus (Fig. 6c). The short-sleeping phenotype persisted under constant darkness, when sleep amounts were even lower than under light/dark conditions (supplemental Fig. 5_a,b, available at www.jneurosci.org as supplemental material). Moreover, under constant darkness, Hk_− flies maintained a rhythmic modulation of locomotor activity with a period of ∼24 h (supplemental Fig. 5_c,d, available at www.jneurosci.org as supplemental material). Thus, Hk mutations affect the daily sleep amount but not the homeostatic or the circadian regulation of sleep, which remain normal.
Figure 6.
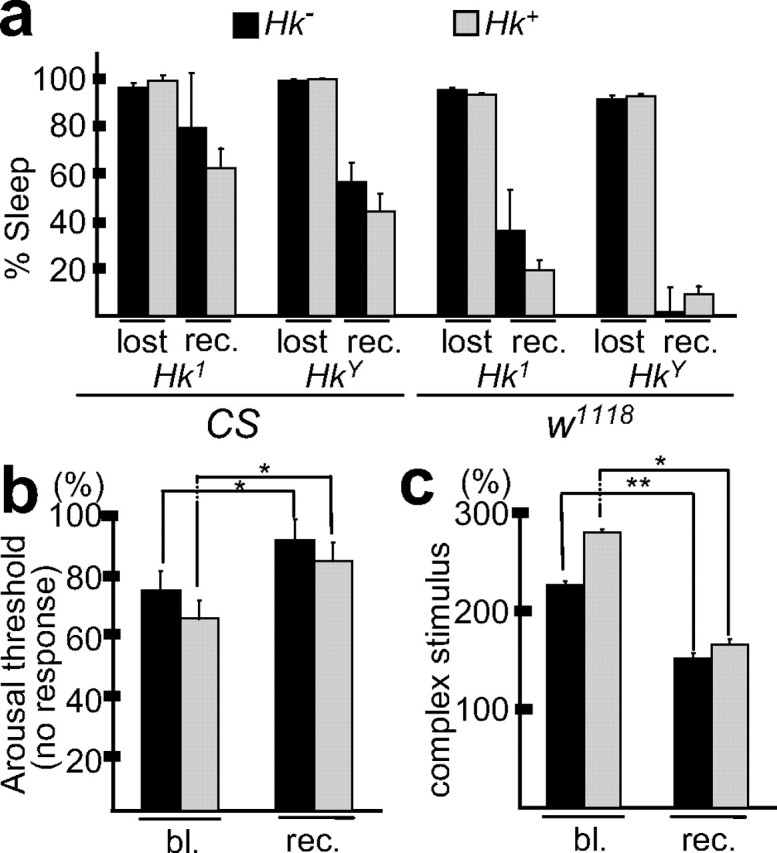
The response to SD is similar in _Hk_− mutants and their wild-type siblings. a, Hk1 and HkY males and their Hk+ siblings were tested in the _w_1118 and CS background. The two left bars in each group represent the percentage of sleep lost during 24 h of SD (compared with baseline sleep equals 100%; mean ± SEM), whereas the two right bars indicate the percentage of sleep recovered during the 24 h period after SD (recovered equals the number of minutes flies overslept relative to baseline during the first 24 h after SD, expressed as the percentage of sleep lost during SD). Only sleep-deprived flies that lost ≥80% of baseline sleep were included in this analysis. With the exception of HkY flies in the _w_1118 background, all flies slept longer the day after SD than during baseline (p < 0.05, paired Student's t test). The amount of sleep lost and regained after SD was not significantly different between _Hk_− mutant and wild-type flies. b, The arousal threshold was measured as the percentage of sleeping flies (immobile for ≥5 min) that did not show an escape response after the delivery of a complex stimulus of low intensity (Huber et al., 2004). A similar percentage (60–70%) of male Hk1 flies and wild-type siblings are nonresponsive during baseline, and their number increases significantly, and to a similar extent, after SD. Values (mean ± SEM) for baseline (bl.) and recovery after SD (rec.) refer to the first 6 h of the dark period. c, To assess performance after SD, awake flies were tested for their ability to respond to a complex stimulus. Performance was measured as the decrease in the latency to the first beam crossing after the delivery of the stimulus relative to the minute before the stimulation. All flies were active (awake) the minute before the delivery of the stimulus. Values (mean ± SEM) refer to the first 6 h of the light period before (bl.) and after (rec.) SD. *p < 0.05; **p < 0.001 (unpaired Student's t test). The analysis in a–c included ≥31 male flies for each line.
Changes in sleep and waking activity primarily occur through Sh
The assumption thus far has been that mutations in Hk reduce sleep by primarily reducing the potassium current through the Sh pore. However, Hk also binds to ether-a-go-go (eag), another voltage-dependent potassium channel (Wilson et al., 1998). If Hk loss-of-function mutations reduce sleep by affecting the Sh pore, then Hk and Sh double mutants would not be expected to produce a stronger phenotype than either mutant alone. However, if Hk loss affects two different pathways, one acting through Hk and the other possibly through eag, then a double mutant should show a stronger sleep phenotype, because both pathways are being affected. We therefore generated double Sh Hk mutants (supplemental Fig. 6, available at www.jneurosci.org as supplemental material) and measured their daily sleep amount. We found that they had reduced sleep, but that their short-sleeping phenotype was not significantly stronger compared with siblings that inherited either an Hk or a Sh mutation alone (Fig. 7). Thus, these results are consistent with the hypothesis that mutations in Hk reduce sleep primarily by affecting the Sh complex. In agreement with this conclusion, mutations in eag alone result in only small changes in the sleep phenotype (D. Bushey, G. Tononi, C. Cirelli, unpublished observations).
Figure 7.
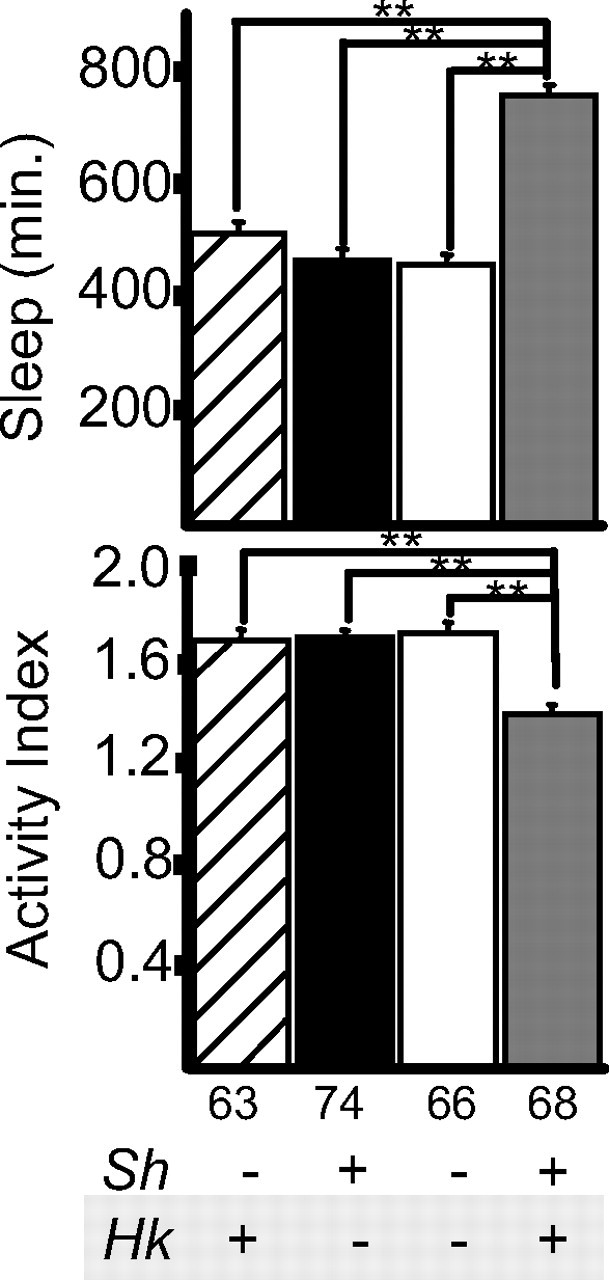
Hk primarily reduces sleep through the Sh complex. Bar graphs represent daily sleep amounts (mean ± SEM; minutes of sleep per 24 h) and activity index (number of beam crossings per minute during waking). The number of flies tested, as well as the Sh and Hk alleles present, are indicated below each bar graph. **p < 0.001; unpaired Student's t test. The analysis in a–c included ≥31 male flies for each line.
Mutations in Hk and Sh reduce performance in a learning and memory paradigm
Behavioral studies in humans suggest a link between sleep and memory (Stickgold, 2005). To investigate this link in flies, we assessed learning and memory in _Hk_− mutants by using a heat-box paradigm (Wustmann et al., 1996; Wustmann and Heisenberg, 1997; Putz and Heisenberg, 2002). In this paradigm, single flies were tested in a rectangular chamber divided in half such that the temperature on either side can be controlled independently. During five training sessions, half of the chamber was heated to 39°C, whereas the other side was maintained at 22°C, a temperature that flies prefer. These training sessions alternated with testing periods when the whole box remained at 22°C (spaced training). During the testing sessions, wild-type flies showed a progressively greater preference for the previously nonheated side of the box, an indication of learning (Fig. 8a,b, right panels, black bars). Moreover, for some time afterward, wild-type flies continued to avoid the previously heated side even after training stopped, an indication of memory (Fig. 8a,b, right panels, gray bars). Hk1 and HkY mutants also learned to prefer the nonheated side of the chamber, like their wild-type siblings (Fig. 8a,b, left panels, black bars). After training, however, Hk1 and HkY mutants quickly lost their preference for the nonheated side (Fig. 8a,b, left, gray bars), suggesting a deficit in short-term memory. Importantly, the wild-type Hk+ transgene that rescued the short-sleeping phenotype was also able to rescue the deficit in short-term memory in Hk mutants (Fig. 8c,d). In contrast, the same transgene did not improve memory when inherited with a functional Hk+ allele (supplemental Fig. 7, available at www.jneurosci.org as supplemental material).
Thus far, we have studied strong hypomorphic Hk mutants. To determine whether the memory impairment was related to the short-sleeping phenotype, we then tested Hk2, a weak hypomorph (Trout and Kaplan, 1970; Kaplan and Trout, 1974; Wang et al., 2000). Consistent with Hk2 being a weak allele, and with the data shown previously indicating that a profound deficit in Hk function is needed to produce a short-sleeping phenotype (Figs. 2c, 4c), Hk2 flies did not sleep less than their wild-type siblings (Fig. 9a,b) and became short sleepers only in combination with a more severe Hk allele (Fig. 9c,d). Moreover, when tested in the heat box, normal-sleeping Hk2 flies also showed normal learning and memory (Fig. 8e).
Figure 9.
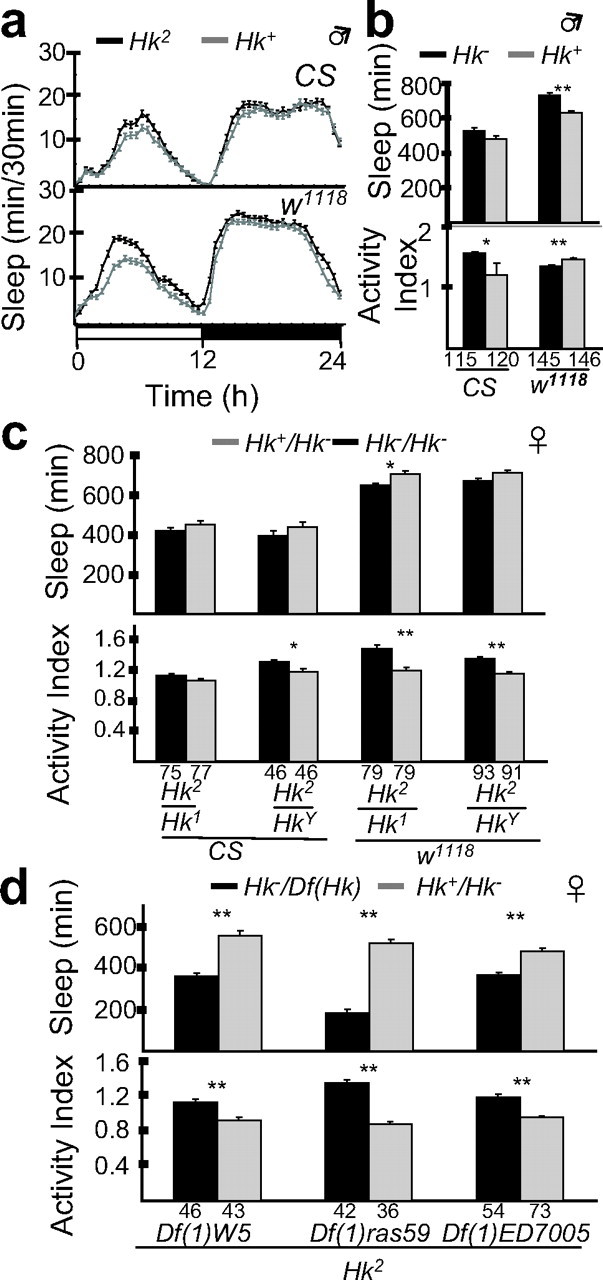
_Hk_2 mutants have a normal daily sleep amount. a, Daily time course (in 30 min intervals; mean ± SEM) of the amount of sleep in Hk2 and Hk+ males in two different genetic backgrounds: CS and _w_1118. The number of tested flies is the same as in b. b, Daily sleep amounts (mean ± SEM; minutes of sleep per 24 h) and activity index (number of beam crossings per minute during waking; mean ± SEM). The number of tested flies is indicated below each column. c, Complementation analysis between Hk2 and either Hk1 or HkY (daily sleep amounts and activity index in heterozygous females). Hk2 heterozygous with either Hk1 or HkY tended to sleep less than their siblings that inherited a wild-type Hk+ allele (Hk+/Hk2), but the difference was significant (p < 0.005) only for Hk2/Hk1 flies in the w1118 background. d, Complementation analysis with deficiencies including the Hk locus (daily sleep amounts and activity index in heterozygous females). The data confirm that Hk2 acts as a weak hypomorph with respect to the other Hk alleles, because it produces a weak sleep phenotype in combination with Hk1 and HkY (c) and a strong sleep phenotype with deficiencies that include Hk (d). *p < 0.05; **p < 0.001; unpaired Student's t test.
To further examine the link between the short-sleeping phenotype and memory decay, we tested previously isolated Sh alleles for their effect on sleep and memory. They included two alleles (ShHD and ShX) whose effects on the _I_A current are unknown, one hypomorphic allele (Sh120) that reduces but does not eliminate _I_A (Zhao et al., 1995), and five alleles (Shmns, Sh133, ShM, Df(Sh), Sh102) that have been shown by molecular and/or electrophysiological methods to completely eliminate _I_A (Wu and Haugland, 1985; Papazian et al., 1987; Haugland and Wu, 1990; Lichtinghagen et al., 1990) (Bushey, Tononi, Cirelli, unpublished observations for Shmns). As shown in Figure 10, we found that all Sh mutants that have completely lost _I_A slept less, whereas Sh120 flies slept normally. Moreover, when tested in the heat-box paradigm, the memory of all short-sleeping Sh mutants (Shmns, Sh133, ShM, Sh102, Df(Sh), ShHD) decayed faster than in their wild-type siblings (Fig. 10). In contrast, the memory decay in normal-sleeping Sh mutants (Sh120, ShX) was not different from their wild-type siblings. Overall, when all tested Hk and Sh mutants were considered, we found a negative correlation between daily sleep amount and memory decay (_r_2 = 0.34; F = 0.037). Two short-sleeping Sh mutations (Shmns, Sh133) also performed poorly during training (Fig. 10, maximum performance index), suggesting that at least some Sh mutations can also affect learning in this paradigm.
Figure 10.
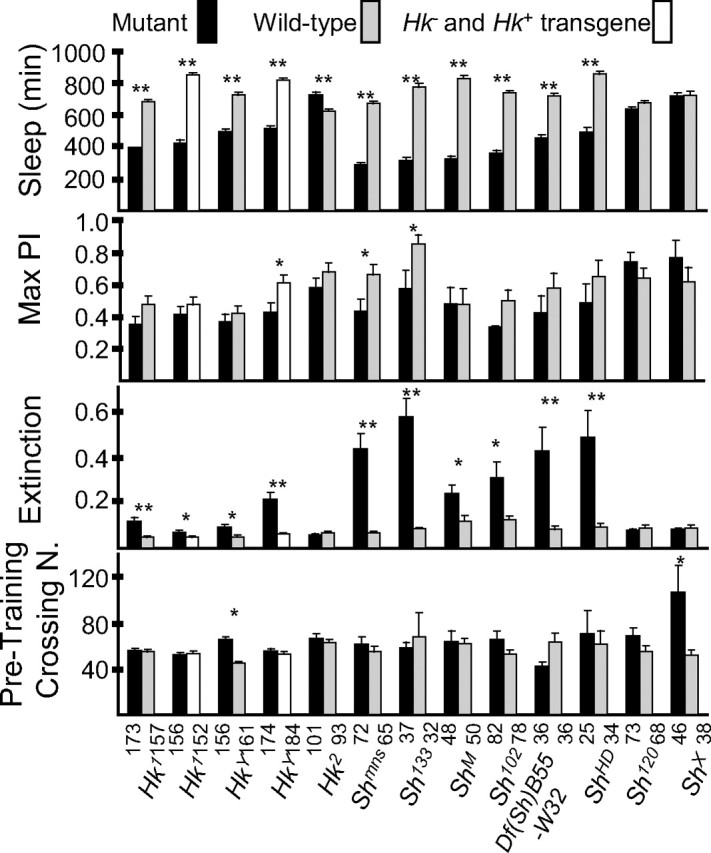
Only Sh mutations that reduce sleep also affect memory in the heat-box learning paradigm. Comparing daily sleep amounts (mean ± SEM; minutes of sleep per 24 h) and performance in the heat box between Sh and Hk mutants to their wild-type siblings. In addition, _Hk_− mutants are also compared with siblings that inherited the wild-type Hk+ transgene _Hk_17K-X94. Only males were tested. Extinction is a measure of memory and reflects how quickly flies lost their preference for the nonheated side after the end of training. To avoid potential confounding factors because of differences in learning, extinction was calculated by dividing the maximum performance index (PI) by the amount of time the flies had a significant preference for the nonheated side: the larger the extinction number, the quicker the flies forgot to stay on the nonheated side. The maximum PI (mean ± SEM), the highest value of PI obtained during the test, is a measure of learning, and in all cases was reached either 1 min after or 1 min before the last training session (see Fig. 8). Most _Sh_− and _Hk_− mutants had maximum PI smaller than their wild-type siblings, but the difference was significant only for Shmns and Sh133. The pretraining crossing number (number of midline crossings during the 10 min pretraining period; mean ± SEM) is a measure of spontaneous locomotor activity in the heat box. Sleep was measured before flies were tested in the heat box. The number of tested flies is indicated below each column. *p < 0.05; **p < 0.001; unpaired Student's t test. N., Number.
Locomotor activity was measured in the heat box as the number of pretraining crossings. Among all Hk and Sh mutants tested, only two were hyperactive relative to their siblings, including the ShX mutant, which had normal sleep and normal memory (Fig. 10). Similar results were found for walking speed (data not shown). Thus, an Sh mutation associated with hyperactivity does not necessarily result in short sleep or poor performance in the heat-box paradigm.
Discussion
In this study, we found that loss-of-function mutations in the β modulatory (Hk) subunit of a highly conserved voltage-dependent potassium channel reduce sleep in Drosophila. As in short-sleeping Sh mutants, the duration of sleep episodes is decreased in Hk mutants, and the homeostatic and circadian regulation of sleep is normal. Moreover, a normal copy of Hk is sufficient to restore normal sleep. All experiments compared mutant and wild-type siblings, which share both the same genetic background and a common environment during development. This is crucial, because we found in previous (Cirelli et al., 2005b) and in this study that stocks can accumulate genetic modifiers able to alter sleep and performance in the heat box.
Hk and Sh mutations affect daily sleep amount without impairing the response to SD, suggesting that the underlying mechanisms can be dissociated. Other genetic manipulations in flies and rodents support this hypothesis. In Drosophila, mutations in the genes coding for the dopamine transporter or the serotonin receptor d5-HT1A cause both a short-sleep phenotype and a reduced response to SD (Kume et al., 2005; Yuan et al., 2006), whereas CREB-deficient flies, in which daily sleep amount is increased, show an increased response to SD (Hendricks et al., 2001). Clock mutations, however, decrease daily sleep amount without affecting the response to SD (Hendricks et al., 2003b). Moreover, the genetic ablation of the mushroom bodies, which decreases daily sleep amount, is not associated with consistent changes in the response to SD (Pitman et al., 2006). Finally, in mice, a decrease in CREB expression is associated with an increase in daily sleep amount, but the response to SD is normal (Graves et al., 2003).
Short-sleeping Hk mutants also show an increase in waking locomotor activity, prompting the question of whether their reduced sleep is a direct consequence of hyperactivity. Several reasons suggest that this is unlikely. If the increased waking activity was the cause of the reduced sleep, the two variables should be strongly negatively correlated. Although waking activity and total sleep amount were negatively correlated in females, there was no correlation in males. Moreover, the wild-type transgene was always able to restore a normal-sleeping phenotype, whether or not waking activity decreased. Furthermore, if the short-sleeping phenotype was a direct consequence of hyperactivity, one would expect sleep in Hk mutants to be highly fragmented. Instead, the duration of sleep episodes decreased without a consistent increase in their number (the number of sleep bouts actually decreased in several lines). Moreover, arousal thresholds did not differ between Hk mutants and wild-type siblings (Fig. 5b), and the same was true for the number of brief awakenings, another measure of sleep fragmentation (data not shown). Finally, it is worth mentioning that Sh mutations, such as Sh120, ShX, and Sh102 show a similar increase in motor activity relative to their wild-type siblings (data not shown), yet only Sh102 is a short sleeper, again suggesting that mutations that affect _I_A and increase locomotor activity do not necessarily also decrease sleep.
Because potassium channels are core regulators of excitable cell function, it could be that mutations in any other potassium channel or related protein, by causing general hyperexcitability, would result in a short-sleeping phenotype. We have recently found that slowpoke, shal, and quiver mutants sleep normally, whereas mutations in eag slightly reduce daily sleep amount, mainly in the second half of the night (Bushey, Tononi, Cirelli, unpublished results). Therefore, mutations affecting potassium currents do not necessarily produce a sleep phenotype, and if they do, they can produce a unique phenotype compared with mutations in Sh and Hk.
Why the Sh current is necessary for normal sleep is not known. Ion channel mutations are pleiotropic. Hk and Sh mutants, for example, shake under ether anesthesia (Kaplan and Trout, 1969), and show an abnormal visually induced jumping response (Kaplan and Trout, 1974). It is likely that these phenotypes, like short sleeping, are because of an increase in neuronal excitability, although possibly in different circuits. Indeed, the shaking phenotype depends on motoneurons of the ventral thoracic ganglion (Ikeda and Kaplan, 1970b), whereas the locus of the jumping response is in the CNS (Ikeda and Kaplan, 1970a). The Drosophila brain areas mediating the short-sleeping phenotype remain unknown, but recent evidence suggests that the mushroom bodies could play an important role (Joiner et al., 2006; Pitman et al., 2006). Our data suggest that the jumping and shaking phenotypes dissociate from the sleeping and memory phenotypes. This is because the shaking phenotype is dominant in females, whereas the learning (Cowan and Siegel, 1986) and sleep phenotypes (Cirelli et al., 2005b) are recessive. Moreover, Hk2 flies show an abnormal jump reflex (Kaplan and Trout, 1974) but sleep and remember normally (present study).
This study also shows that reduced sleep is linked to reduced performance in the heat-box paradigm. Previous studies using courtship and olfactory conditioning have found learning deficits in a single Sh mutant, Sh5 (Cowan and Siegel, 1984,, 1986), which we had previously shown to have a weak short-sleeping phenotype (Cirelli et al., 2005b). _Sh_5 is better classified as a neomorph, rather than a simple hypomorph, because the mutation changes the kinetics of activation and inactivation of the Sh current (Wu and Haugland, 1985; Haugland and Wu, 1990; Lichtinghagen et al., 1990). Our study expands on the previous Sh5 findings by using a different learning paradigm and by including most available loss-of-function Sh alleles, ranging from amorphs to hypomorphs (Wu and Haugland, 1985; Lichtinghagen et al., 1990; Zhao et al., 1995). We are unaware of any previous reports detailing the role of Hk in learning and memory in Drosophila. Interestingly, however, mice deficient for kvbeta1.1, an orthologue of Hk, have impairments in certain types of learning and memory (Giese et al., 1998).
Of all _Hk_− and _Sh_− flies tested, only short-sleeping lines showed a consistently faster decay in memory in the heat-box paradigm. Specifically, all alleles that produced a short-sleeping phenotype (Hk1, HkY, Shmns, Sh133, ShM, Sh102, Df(Sh)B55-W32, ShHD) had faster memory extinction than their siblings, whereas alleles that did not alter the sleep phenotype (Hk2, ShX, Sh120) did not. Furthermore, extinction was quicker with Sh than with Hk mutations. Because the former can completely abolish channel function rather than modulate its activity, like the latter, these data suggest that mutations that completely abolish _I_A have a greater effect on both sleep and memory. Indeed, daily sleep amount and extinction were negatively correlated. This correlation, however, is weak, which may suggest a threshold effect whereby memory is affected only when sleep is substantially reduced.
The memory decay (extinction) was calculated by dividing the peak performance by the amount of time the flies had a significant preference for the nonheated side, to control for differences in learning. However, peak performance and extinction are negatively correlated (_r_2 = 0.60; F = 0.0020), and in two mutant lines, learning performance was significantly lower than in wild-type siblings. Thus, although we found an association between short sleep and faster extinction, an effect on learning cannot be excluded. Interestingly, almost all studies so far have focused on the detrimental effects of sleep loss on memory consolidation (Stickgold, 2005). Consistent with our findings in flies, however, new studies in humans suggest that sleep loss can also impair memory acquisition (Yoo et al., 2007).
These data do not prove a causal relationship between sleep amount and memory. In fact, the link between sleep and memory could be the consequence of parallel, independent effects of the mutations on multiple brain functions. As discussed above, whereas it may be impossible to completely rule out this possibility with recessive phenotypes, the fact that normal sleeping Hk2 flies show the shaking and jumping phenotypes but sleep and remember normally suggests a closer link between sleep and memory. One possibility is that the memory impairment is because of reduced sleep, as suggested by the negative effects of acute SD or chronic sleep restriction on performance, learning, and memory (Van Dongen et al., 2003). Alternatively, the memory deficit may cause the sleep deficit. Thus, several Drosophila memory mutants are also short sleepers (Hendricks et al., 2001) and lesions of the mushroom bodies, which are important for some forms of memory, lead to a reduction of sleep time (Joiner et al., 2006; Pitman et al., 2006). Moreover, exposure to an enriched environment increases daily sleep amount in flies (Ganguly-Fitzgerald et al., 2006), as well as the intensity of sleep in rats (Huber et al., 2007). Thus, the extent of plastic phenomena during waking may be a major determinant of sleep need.
Footnotes
This study was supported by the Canadian Institutes of Health Research, Defense Advanced Research Projects Agency, and National Institute of General Medical Sciences Grant R01 GM075315. We thank Barry Ganetzky for providing fly stocks, Martin Heisenberg and his laboratory for help with the heat box, and Michael Rosbash for the circadian software.
References
- Andretic R, van Swinderen B, Greenspan RJ. Dopaminergic modulation of arousal in Drosophila. Curr Biol. 2005;15:1165–1175. doi: 10.1016/j.cub.2005.05.025. [DOI] [PubMed] [Google Scholar]
- Bonnet MH, Arand DL. Hyperarousal and insomnia. Sleep Med Rev. 1997;1:97–108. doi: 10.1016/s1087-0792(97)90012-5. [DOI] [PubMed] [Google Scholar]
- Chouinard SW, Wilson GF, Schlimgen AK, Ganetzky B. A potassium channel beta subunit related to the aldo-keto reductase superfamily is encoded by the Drosophila hyperkinetic locus. Proc Natl Acad Sci USA. 1995;92:6763–6767. doi: 10.1073/pnas.92.15.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, LaVaute TM, Tononi G. Sleep and wakefulness modulate gene expression in Drosophila. J Neurochem. 2005a;94:1411–1419. doi: 10.1111/j.1471-4159.2005.03291.x. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, Tononi G. Reduced sleep in Drosophila Shaker mutants. Nature. 2005b;434:1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- Cowan TM, Siegel RW. Mutational and pharmacological alterations of neuronal membrane function disrupt conditioning in Drosophila. J Neurogenet. 1984;1:333–344. doi: 10.3109/01677068409107095. [DOI] [PubMed] [Google Scholar]
- Cowan TM, Siegel RW. Drosophila mutations that alter ionic conduction disrupt acquisition and retention of a conditioned odor avoidance response. J Neurogenet. 1986;3:187–201. doi: 10.3109/01677068609106849. [DOI] [PubMed] [Google Scholar]
- Durmer JS, Dinges DF. Neurocognitive consequences of sleep deprivation. Semin Neurol. 2005;25:117–129. doi: 10.1055/s-2005-867080. [DOI] [PubMed] [Google Scholar]
- Ganguly-Fitzgerald I, Donlea J, Shaw PJ. Waking experience affects sleep need in Drosophila. Science. 2006;313:1775–1781. doi: 10.1126/science.1130408. [DOI] [PubMed] [Google Scholar]
- Giese KP, Storm JF, Reuter D, Fedorov NB, Shao LR, Leicher T, Pongs O, Silva AJ. Reduced K+ channel inactivation, spike broadening, and after-hyperpolarization in Kvbeta1.1-deficient mice with impaired learning. Learn Mem. 1998;5:257–273. [PMC free article] [PubMed] [Google Scholar]
- Graves LA, Hellman K, Veasey S, Blendy JA, Pack AI, Abel T. Genetic evidence for a role of CREB in sustained cortical arousal. J Neurophysiol. 2003;23:23. doi: 10.1152/jn.00882.2002. [DOI] [PubMed] [Google Scholar]
- Gulbis JM, Mann S, MacKinnon R. Structure of a voltage-dependent K+ channel beta subunit. Cell. 1999;97:943–952. doi: 10.1016/s0092-8674(00)80805-3. [DOI] [PubMed] [Google Scholar]
- Haugland FN, Wu CF. A voltage-clamp analysis of gene-dosage effects of the Shaker locus on larval muscle potassium currents in Drosophila. J Neurosci. 1990;10:1357–1371. doi: 10.1523/JNEUROSCI.10-04-01357.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, Yin JC, Sehgal A. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–1115. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Kirk D, Panckeri K, Miller MS, Pack AI. Modafinil maintains waking in the fruit fly Drosophila melanogaster. Sleep. 2003a;26:139–146. doi: 10.1093/sleep/26.2.139. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Lu S, Kume K, Yin JC, Yang Z, Sehgal A. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J Biol Rhythms. 2003b;18:12–25. doi: 10.1177/0748730402239673. [DOI] [PubMed] [Google Scholar]
- Horne J. New York: Oxford UP; 1988. Why we sleep: the functions of sleep in humans and other mammals. [Google Scholar]
- Huber R, Hill SL, Holladay C, Biesiadecki M, Tononi G, Cirelli C. Sleep homeostasis in Drosophila melanogaster. Sleep. 2004;27:628–639. doi: 10.1093/sleep/27.4.628. [DOI] [PubMed] [Google Scholar]
- Huber R, Tononi G, Cirelli C. Exploratory behavior, cortical BDNF expression, and sleep homeostasis. Sleep. 2007;30:129–139. doi: 10.1093/sleep/30.2.129. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Kaplan WD. Unilaterally patterned neural activity of gynandromorphs, mosaic for a neurological mutant of Drosophila melanogaster. Proc Natl Acad Sci USA. 1970a;67:1480–1487. doi: 10.1073/pnas.67.3.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Kaplan WD. Patterned neural activity of a mutant Drosophila melanogaster. Proc Natl Acad Sci USA. 1970b;66:765–772. doi: 10.1073/pnas.66.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- Kaplan WD, Trout WE., III The behavior of four neurological mutants of Drosophila. Genetics. 1969;61:399–409. doi: 10.1093/genetics/61.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan WD, Trout WE. Genetic manipulation of an abnormal jump response in Drosophila. Genetics. 1974;77:721–739. doi: 10.1093/genetics/77.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Evans JM, Hendricks JC, Sehgal A. From the cover: a Drosophila model for age-associated changes in sleep:wake cycles. Proc Natl Acad Sci USA. 2006;103:13843–13847. doi: 10.1073/pnas.0605903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25:7377–7384. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JD, Funes P, Dowse HB, Hall JC. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002;3:1. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtinghagen R, Stocker M, Wittka R, Boheim G, Stuhmer W, Ferrus A, Pongs O. Molecular basis of altered excitability in Shaker mutants of Drosophila melanogaster. EMBO J. 1990;9:4399–4407. doi: 10.1002/j.1460-2075.1990.tb07890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littleton JT, Ganetzky B. Ion channels and synaptic organization: analysis of the Drosophila genome. Neuron. 2000;26:35–43. doi: 10.1016/s0896-6273(00)81135-6. [DOI] [PubMed] [Google Scholar]
- Nitz D, Tononi G. Tonic rhythmic activity of rat cerebellar neurons. Exp Brain Res. 2002;146:265–270. doi: 10.1007/s00221-002-1205-7. [DOI] [PubMed] [Google Scholar]
- Papazian DM, Schwarz TL, Tempel BL, Jan YN, Jan LY. Cloning of genomic and complementary DNA from Shaker, a putative potassium channel gene from Drosophila. Science. 1987;237:749–753. doi: 10.1126/science.2441470. [DOI] [PubMed] [Google Scholar]
- Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- Putz G, Heisenberg M. Memories in Drosophila heat-box learning. Learn Mem. 2002;9:349–359. doi: 10.1101/lm.50402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettig J, Heinemann SH, Wunder F, Lorra C, Parcej DN, Dolly JO, Pongs O. Inactivation properties of voltage-gated K+ channels altered by presence of beta-subunit. Nature. 1994;369:289–294. doi: 10.1038/369289a0. [DOI] [PubMed] [Google Scholar]
- Schlimgen AK. Genetics. Madison, WI: Wisconsin-Madison; 1991. The genetic and molecular characterization of Hk, a gene affecting neuronal excitability in Drosophila melanogaster. [Google Scholar]
- Schwarz TL, Tempel BL, Papazian DM, Jan YN, Jan LY. Multiple potassium-channel components are produced by alternative splicing at the Shaker locus in Drosophila. Nature. 1988;331:137–142. doi: 10.1038/331137a0. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- Stickgold R. Sleep-dependent memory consolidation. Nature. 2005;437:1272–1278. doi: 10.1038/nature04286. [DOI] [PubMed] [Google Scholar]
- Trout WE, Kaplan WD. A relation between longevity, metabolic rate, and activity in shaker mutants of Drosophila melanogaster. Exp Gerontol. 1970;5:83–92. doi: 10.1016/0531-5565(70)90033-1. [DOI] [PubMed] [Google Scholar]
- Van Dongen HP, Maislin G, Mullington JM, Dinges DF. The cumulative cost of additional wakefulness: dose-response effects on neurobehavioral functions and sleep physiology from chronic sleep restriction and total sleep deprivation. Sleep. 2003;26:117–126. doi: 10.1093/sleep/26.2.117. [DOI] [PubMed] [Google Scholar]
- Wang JW, Humphreys JM, Phillips JP, Hilliker AJ, Wu CF. A novel leg-shaking Drosophila mutant defective in a voltage-gated K+ current and hypersensitive to reactive oxygen species. J Neurosci. 2000;20:5958–5964. doi: 10.1523/JNEUROSCI.20-16-05958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GF, Wang Z, Chouinard SW, Griffith LC, Ganetzky B. Interaction of the K channel beta subunit, Hyperkinetic, with eag family members. J Biol Chem. 1998;273:6389–6394. doi: 10.1074/jbc.273.11.6389. [DOI] [PubMed] [Google Scholar]
- Wu CF, Haugland FN. Voltage clamp analysis of membrane currents in larval muscle fibers of Drosophila: alteration of potassium currents in Shaker mutants. J Neurosci. 1985;5:2626–2640. doi: 10.1523/JNEUROSCI.05-10-02626.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wustmann G, Heisenberg M. Behavioral manipulation of retrieval in a spatial memory task for Drosophila melanogaster. Learn Mem. 1997;4:328–336. doi: 10.1101/lm.4.4.328. [DOI] [PubMed] [Google Scholar]
- Wustmann G, Rein K, Wolf R, Heisenberg M. A new paradigm for operant conditioning of Drosophila melanogaster. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1996;179:429–436. doi: 10.1007/BF00194996. [DOI] [PubMed] [Google Scholar]
- Yao WD, Wu CF. Auxiliary hyperkinetic beta subunit of K+ channels: regulation of firing properties and K+ currents in Drosophila neurons. J Neurophysiol. 1999;81:2472–2484. doi: 10.1152/jn.1999.81.5.2472. [DOI] [PubMed] [Google Scholar]
- Yoo SS, Hu PT, Gujar N, Jolesz FA, Walker MP. A deficit in the ability to form new human memories without sleep. Nat Neurosci. 2007;10:385–392. doi: 10.1038/nn1851. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Joiner WJ, Sehgal A. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr Biol. 2006;16:1051–1062. doi: 10.1016/j.cub.2006.04.032. [DOI] [PubMed] [Google Scholar]
- Zhao ML, Sable EO, Iverson LE, Wu CF. Functional expression of Shaker K+ channels in cultured Drosophila“giant” neurons derived from Sh cDNA transformants: distinct properties, distribution, and turnover. J Neurosci. 1995;15:1406–1418. doi: 10.1523/JNEUROSCI.15-02-01406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JE, Rizzo W, Shockley KR, Raizen DM, Naidoo N, Mackiewicz M, Churchill GA, Pack AI. Multiple mechanisms limit the duration of wakefulness in Drosophila brain. Physiol Genomics. 2006;27:337–350. doi: 10.1152/physiolgenomics.00030.2006. [DOI] [PubMed] [Google Scholar]