DRAGON: A Member of the Repulsive Guidance Molecule-Related Family of Neuronal- and Muscle-Expressed Membrane Proteins Is Regulated by DRG11 and Has Neuronal Adhesive Properties (original) (raw)
Abstract
DRG11, a transcription factor expressed in embryonic dorsal root ganglion (DRG) and dorsal horn neurons, has a role in the development of sensory circuits. We have used a genomic binding strategy to screen for the promoter region of genes regulated by DRG11. One gene with a promoter region binding to the DNA binding domain of DRG11 encodes a novel membrane-associated [glycosyl-phosphatidylinositol (GPI)-anchored] protein that we call DRAGON. DRAGON expression is transcriptionally regulated by DRG11, and it is coexpressed with DRG11 in embryonic DRG and spinal cord. DRAGON expression in these areas is reduced in DRG11 null mutants. DRAGON is expressed, however, in the neural tube before DRG11, and unlike DRG11 it is expressed in the brain and therefore must be regulated by other transcriptional regulatory elements. DRAGON shares high sequence homology with two other GPI-anchored membrane proteins: the mouse ortholog of chick repulsive guidance molecule (mRGM), which is expressed in the mouse nervous system in areas complementary to DRAGON, and DRAGON-like muscle (DL-M), the expression of which is restricted to skeletal and cardiac muscle. A comparative genomic analysis indicates that the family of RGM-related genes—mRGM, DRAGON, and DL-M—are highly conserved among mammals, zebrafish, chick, and Caenorhabditis elegans but not Drosophila. DRAGON, RGM, and DL-M mRNA expression in the zebrafish embryo is similar to that in the mouse. Neuronal cell adhesion assays indicate that DRAGON promotes and mRGM reduces adhesion of mouse DRG neurons. We show that DRAGON interacts with itself homophilically. The dynamic expression, ordered spatial localization, and adhesive properties of the RGM-related family of membrane-associated proteins are compatible with specific roles in development.
Keywords: development, GPI anchor, neuron, spinal cord, brain, muscle
Introduction
Dorsal root ganglion (DRG) sensory neurons initiate proprioceptive, tactile, thermal, and nociceptive sensations. Each of these neurons has a particular termination pattern in the spinal cord. Proprioceptive afferents terminate in the ventral horn, low-threshold cutaneous mechanoreceptors terminate in the deep dorsal horn, and high-threshold nociceptors and thermoreceptor afferents terminate in the superficial dorsal horn (Willis and Coggeshall, 1991). The specific connectivity between the central terminals of primary afferents and second-order neurons in the different laminas of the spinal cord determines the circuits that mediate transfer of the sensory inflow. How are these circuits established?
During spinal cord development, neuronal progenitors in the neural tube ventricular zone are defined dorsoventrally by midline signals originating from the roof and floor plates. Gradients of the floor plate signal sonic hedgehog determine development of ventral horn neuron precursors, whereas roof plate TGF superfamily signals determine dorsal horn neuron progenitors (Jessell, 2000; Lee et al., 2000; Altmann and Brivanlou, 2001; Goulding et al., 2002). Neurons eventually forming the dorsal horn are born on embryonic day (E) 11.5 from progenitors expressing the basic helix-loop-helix transcription factors Math1, neurogenin (Ngn1), and Mash 1, and establish six identifiable mantle layer cell types (Gowan et al., 2001; Goulding et al., 2002). These cells can be differentiated into two groups on the basis of expression of the transcription factor Lbx1: a dorsal group (dl1-3, TGFβ-dependent; Lbx1-) that migrates ventrally to form neurons in the deep dorsal horn with axons ascending to the brain, and a ventral group (dl4-6, TGFβ-independent; Lbx1+) that migrates dorsolaterally to form the superficial laminas of the dorsal horn, the substantial gelatinaosa (Pierani et al., 2001; Gross et al., 2002; Matise, 2002; Muller et al., 2002). Several homeobox genes downstream of Lbx1—Rnx/Tlx1, Tlx3, and DRG11—determine differentiation of association neurons in the substantia gelatinosa. Null mutations of each of these, as well as LBx1, produce defects in the substantia gelatinosa and in nociceptor termination in the dorsal horn (Chen et al., 2001; Gross et al., 2002; Muller et al., 2002; Qian et al., 2002).
DRG neurons are derived from the neural crest, which is induced from the dorsal neural tube by Wnt and TGFβ (Wilson et al., 2001). Ngn1 and Ngn2 are required for differentiation of sensory neurons from neural crest progenitors (Anderson et al., 1997; Zirlinger et al., 2002). Ia proprioceptors and motor neurons both express the ETS transcription factors ER81 and Pea3, which are necessary for establishment of synaptic contact (Lin et al., 1998; Livet et al., 2002). DRG11, in addition to its expression in dorsal horn association neurons, is also expressed in small-diameter DRG neurons. Consequently, a role has been proposed for DRG11 similar to that of ETS in the monosynaptic reflex: formation of synaptic contact between nociceptor sensory neurons and neurons in the substantia gelatinosa (Saito et al., 1995; Chen et al., 2001).
How may DRG11 contribute to synaptic connectivity? We have addressed this by screening for genes that have promoters regulated by DRG11. One DRG11-regulated gene that we identify is DRAGON, encoding a GPI-anchored protein. DRAGON shares high homology with two other GPI-anchored proteins, mouse ortholog of chick repulsive guidance molecule (mRGM) (Monnier et al., 2002) and DL-M, constituting a family of RGM-related proteins.
Materials and Methods
Plasmid construction. The DNA binding domain (amino acids 31-90) of mouse DRG11 cDNA was amplified by PCR and subcloned into pGEX-KG (Amersham Biosciences, Arlington Heights, IL) downstream of glutathione _S_-transferase (GST). The luciferase plasmid construct was generated by subcloning the 363 bp fragment of DRAGON promoter in the PGL3-promoter vector with the Firefly luciferase reporter (Promega, Madison, WI). The DRG11 expression construct was obtained by subcloning mouse DRG11 cDNA (D. J. Anderson, California Institute of Technology, Pasadena, CA) into pcDNA4/HisB (Promega). DRAGON expression constructs were obtained by subcloning DRAGON cDNA into pcDNA4/HisB (Promega) or pSectagHis2B (Invitrogen, San Diego, CA) expression vectors. DRAGON-alkaline phosphatase (AP) fusion protein was generated by subcloning a DRAGON cDNA fragment excluding the GPI anchor (and the DRAGON antibody epitope; see below) into the mammalian expression vectors pAP-tag4 (GeneHunter) at the sites _Hind_III-_Bgl_II, and transfected into human embryonic kidney (HEK) 293T cells. DRAGON-Fc was generated by subcloning DRAGON cDNA without its GPI anchor into the mammalian expression vector pIgplus (R & D Systems, Minneapolis, MN) in frame with the Fc portion of the human IgG. This allowed us to express a soluble DRAGON-Fc fusion protein. DRAGON-Fc collected in the media of stably transfected HEK293 cells was purified using HiTrap Protein A Affinity Columns (Amersham Biosciences) and eluted with 100 mm glycine-HCl, pH 3.0. The elution fraction was neutralized with 0.3 m Tris-HCl, pH 9. Purified Dragon-Fc was run on SDS-PAGE gel and immunoblotted with anti-Dragon antibody and anti-Fc antibody.
Isolation of DRG11-binding fragments. Ten micrograms of a mouse cytidyl guanosyl (CpG) island plasmid library (Human Genome Mapping Project Resource Center) were incubated with 10 pmol of recombinant mouse DRG11-DNA binding domain, and the screening was conducted as described previously (Watanabe et al., 1998).
Mouse in situ hybridization. In situ hybridization was performed on fresh-frozen mouse embryo sections (12 μm) using 35S-dATP-labeled, 48 bp oligonucleotides (Karchewski et al., 1999). Digoxygenin-labeled in situ hybridization was performed on DRG11-/- and wild-type mouse embryos sections as described previously (Amaya et al., 2000).
Whole-mount in situ hybridization. Zebrafish whole-mount in situ hybridizations were performed as described previously (Harland, 1991; Thisse et al., 1993) using digoxygenin-labeled probes and BM Purple (Roche, Hertforshire, UK) substrate.
Luciferase assay. Two hundred nanograms of the reporter construct, DRG11 expression construct, and renilla luciferase construct were transfected into 60% confluent six-well plates of HEK293T mammalian cells (1 μg total DNA, GFP construct was used to reach 1 μg of DNA). After 48 hr, cells were harvested, and activity was monitored using Dual-Luciferase (Promega); the renilla luciferase was used to normalize the transfection levels among samples.
Pull-down assay. Of the 363 bp DRAGON promoter construct, 0.4 μg was incubated with 1 μg of the GST-DRG11-DNA binding domain (DBD) fusion protein or 1 μg of GST in 250 μl of reaction buffer (10 mm HEPES, pH 7.3, 0.1 mm EDTA, 0.05% NP-40, 1 mg/ml BSA, 2 mm DTT, 10 mm NaCl) for 2 hr on ice. Twenty-five microliters of 50% glutathione-Sepharose beads were then added to the reaction and incubated for 30 min at 4°C. The reaction was spun down briefly and washed three times with 50 mm Tris, pH 8, 0.5% NP-40, and 150 mm NaCl. The pellet was then resuspended in 10 mm Tris, pH 8, 10% glycerol, 0.5% SDS, denatured at 85°C for 15 min, and visualized on 1% agarose gel.
Northern blot. Total RNA (10-12 μg per sample) was separated on 1.5% formaldehyde-agarose gels and blotted onto GeneScreen Plus membrane (NEN, Boston, MA).
Western blot. DRGs or HEK293T cells transfected with DRAGON were homogenized-sonicated in a lysis buffer (200 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5% NP-40, and 10% glycerol) containing a mixture of proteinase inhibitors (Roche). Protein samples were separated on a SDS-PAGE gradient gel (4-15%; Bio-Rad, Hercules, CA) and transferred to a polyvinylidene difluoride filter (Immobilon-P; Millipore, Bedford, MA). The blots were blocked with 6% skim milk (Difco, Detroit, MI) in TBS-T (Tris-buffered saline, 0.15% Tween 20) for 1 hr and incubated with DRAGON antibody (1:6000) overnight at 4°C. The blots were incubated with HRP-conjugated anti-rabbit secondary antibody (1:2000; Amersham Biosciences) for 1 hr at room temperature, developed in ECL solution (PerkinElmer Life Sciences, Emeryville, CA) for 1 min, and exposed onto x-ray films (Amersham Biosciences) for 1-10 min.
Immunohistochemistry. Mouse embryos (E12.5-17.5) were dissected, embedded in OCT compound (Sakura, Tokyo, Japan), and frozen (-80°C). Cryostat sections (16 μm) were cut and stored at -80°C. For immunohistochemistry, sections were fixed (4% paraformaldehyde, 1× PBS) for 30 min at 4°C, washed with PBS, and blocked (1% BSA, 0.3% Triton, 1× PBS) for 1 hr at room temperature. Slides were incubated overnight with DRAGON antibody (1:2000) in the blocking buffer, washed with PBS, and incubated with FITC or Cy3-conjugated secondary antibody (Vector Laboratories, Burlingame, CA; 1:200) at room temperature. After washes with PBS, sections were mounted (Vectashield) and visualized under fluorescence microscopy.
Adhesion assays. Adhesion assays on HEK293T cells were performed as described previously (Campagna et al., 1995). HEK293T cells, nontransfected or transfected with DRAGON or mRGM, were plated on eight-well slides and allowed to grow to confluence (48 hr). Freshly dissociated postnatal day 14 or adult rat DRG neurons (∼4000) were plated on the HEK monolayers and allowed to settle (2-3 hr), washed, fixed (4% paraformaldehyde), and immunostained for DRAGON and neuronal-specific nuclear protein (NeuN) (Chemicon, Temecula, CA) to visualize neuronal cell bodies. Only areas of the HEK monolayer that were at least 90% confluent were scored for neurons.
For adhesion assay on coated plates, laminin (Sigma, St. Louis, MO), poly-d-lysine (Sigma), or DRAGON-Fc was dissolved in distilled water at 10 μg/ml, 100 μg/ml, and 100 ng/ml respectively, and used to coat 96-well tissue culture plates at 4°C overnight. For coating with both poly-d-lysine and laminin, the plates were coated with laminin for 4 hr at room temperature after a previous incubation with poly-d-lysine at 4°C overnight. After coating, the wells were washed with PBS, and 100 μl of neurobasal medium (Invitrogen) containing DRG neurons (3 × 104 cells/ml) was added to each well, incubated for 3 hr at 37°C, and washed three times with PBS. The adhering neurons were fixed with 100 μl of freshly diluted 1% glutaraldehyde in PBS for 10 min at room temperature and incubated with 100 μl of freshly filtered and diluted 0.1% crystal violet in distilled water for 30 min at room temperature. After washing with PBS, the cells were incubated with 50 μl of 0.5% Triton X-100 at 4°C overnight, and optical density was read at 595 nm.
Cotransfection and immunoprecipitation. HEK293T cells were plated in six-well culture plates and cultured at 37°C overnight. The cells were transfected with 2 μg of both DRAGON cDNA and DRAGON-AP or AP cDNA using lipofectamine plus reagent (Invitrogen) and incubated for 48 hr. After washing with PBS, cells were sonicated for 5 sec in lysis buffer (200 mm Tris-HCl, pH 8, 100 mm NaCl, 1 mm EDTA, 0.5% NP-40, and 10% glycerol) at 4°C.
For immunoprecipitation, cell protein samples were precleared with 15 μl of protein G+A agarose beads (Oncogene) at 4°C for 1 hr. The beads were removed by centrifugation and supernatant was collected. Anti-myc antibody (1.5 μg/ml) was added to the supernatant and incubated at 4°C overnight. The immune complex was pulled down by addition of 15 μl of protein G+A agarose beads at 4°C for 2 hr and centrifugation (4000 rpm, 20 min). The pellet was washed with Triton buffer, and samples were subjected to Western blot analysis.
Binding of GST-DRAGON fusion protein. A DRAGON cDNA fragment excluding the GPI anchor (and the epitope for DRAGON antibody) was amplified by PCR and subcloned in the bacterial expression vector pGEX-KG at the sites _Eco_RI-_Hind_III, and a GST-DRAGON fusion protein was prepared. HEK293T cells were plated in eight-well chamber slides, cultured overnight at 37°C, transfected with 2 μg of DRAGON cDNA using lipofectamine plus reagent, and incubated for 48 hr. The cells were then incubated with 0.5 μm GST-DRAGON fusion protein or GST for 3 hr at 37°C and washed with PBS. After fixation with 3.7% paraformaldehyde for 10 min at room temperature, the cells were incubated with primary antibodies (DRAGON 1:2000 or mouse anti-GST 1:700) at 4°C overnight and with secondary antibodies (anti-rabbit FITC or anti-mouse Cy3 1:1000) for 5 hr at 4°C. The staining was visualized using confocal microscopy.
Results
Genomic screening strategy for DRG11 responsive genes
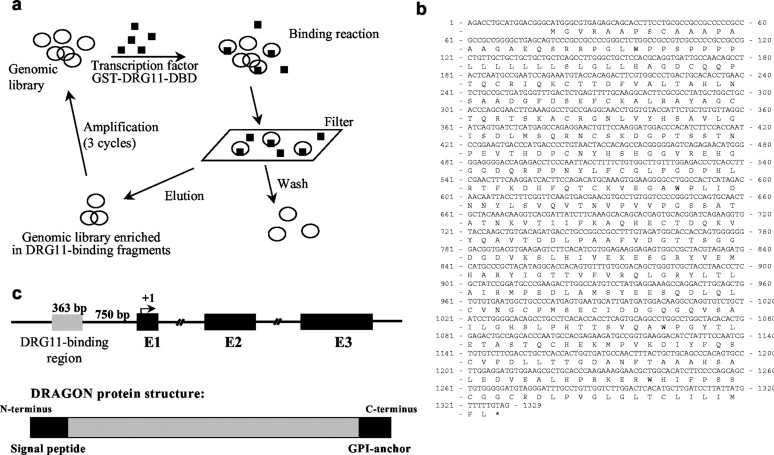
To isolate genes with promoter regions that are responsive to DRG11, a genomic binding site strategy exploiting a mouse CpG island library was used. CpG islands are genomic regions that unlike most vertebrate genomic DNA are CpG dinucleotide rich with unmethylated Cs, and they include promoter regions extending to the first exon (Cross et al., 1999). A GST-DRG11 fusion protein was produced from a DRG11 cDNA fragment that incorporates its DBD. Binding between the DRG11-DBD fusion protein and the genomic library was used to enrich for DRG11-interacting DNA fragments (Fig. 1_a_). The selected genomic fragments were sequenced and analyzed for flanking open reading frames. Among the most abundant clones obtained was a 363 bp DNA fragment located 750 bp upstream of a putative open reading frame for a novel gene. Sequence analysis found the DRG11-binding DNA fragment located in the promoter region of this gene (Fig. 1). Genomic database analysis, combined with RT-PCR and rapid amplification of cDNA ends of mouse DRG and spinal cord cDNA libraries, enabled us to find an open reading frame encoding a 1311 bp cDNA that we call DRAGON, after its regulation by DRG11. The nucleotide and predicted 436 amino acid sequence of DRAGON are shown in Figure 1, b and c.
Figure 1.
a, Genomic screening strategy with a CpG island library. The plasmid DNA was bound with GST-DRG11-DBD and passed through a nitrocellulose filter. DRG11-bound plasmids were eluted and amplified in bacterial cultures. The DRG11-bound plasmids were concentrated over three cycles. b, Full coding cDNA sequence of DRAGON with its open reading frame and putative protein sequence. DRAGON protein has an N-terminal signal peptide and a C-terminal GPI anchor but no other highly conserved domains. c, Top, Schematic representation of the DRAGON gene with its promoter region and three exons. Bottom, DRAGON protein structure with its N-terminal signal peptide andC-terminal GPI anchor.
DRAGON and its homologs: an RGM-related family
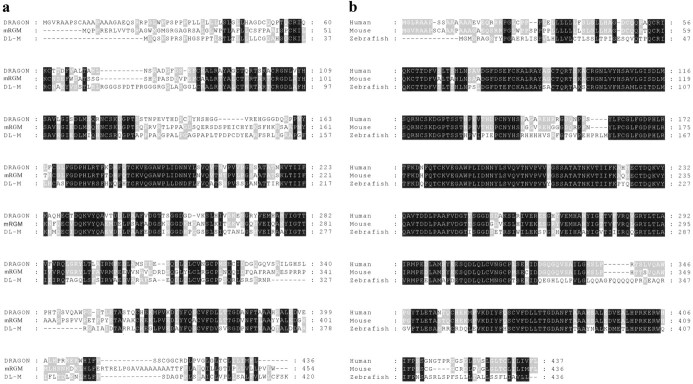
DRAGON is a putative GPI-anchored protein with an N-terminal signal peptide sequence, a C-terminal hydrophobic domain (Fig. 1_c_), and no significant homologies with other conserved protein domains. Two genes highly homologous to DRAGON were found in mouse genomic databases: mRGM (Monnier et al., 2002) and DL-M, named on the basis of its expression pattern in the mouse embryo. mRGM and DL-M possess 50-60% homology to DRAGON, and both have a putative signal peptide and GPI anchor. The predicted amino acid sequences of mRGM and DL-M, as well as sequence alignments indicating areas of homology with DRAGON, are shown in Figure 2_a_. Human homologs of DRAGON, RGM, and DL-M were identified in the human genome database (Fig. 2_b_). Zebrafish orthologs for all three genes were also identified and cloned (Fig. 2_b_) (data not shown). The gene family is conserved in Fugu, but no Drosophila homologs were found. A single Caenorhabditis elegans EST transcript with high homology to portions of DRAGON was identified.
Figure 2.
Amino acid sequence alignments of mRGM and DL-M with DRAGON show a high homology among the members of the newly identified gene family. Black boxes indicate perfect homology among the three members, whereas gray boxes indicate homology among two members.
Regulation of DRAGON expression by DRG11
DRG11 is first expressed at E12-12.5 in the mouse spinal cord by postmitotic neurons adjacent to the ventricular zone in the alar plate, as well as in scattered lateral cells (Saito et al., 1995; Chen et al., 2001). At later developmental stages, expression of DRG11 decreases medially and increases in the substantia gelatinosa. Expression of DRG11 in the DRG begins at E11-12 (Saito et al., 1995; Chen et al., 2001).
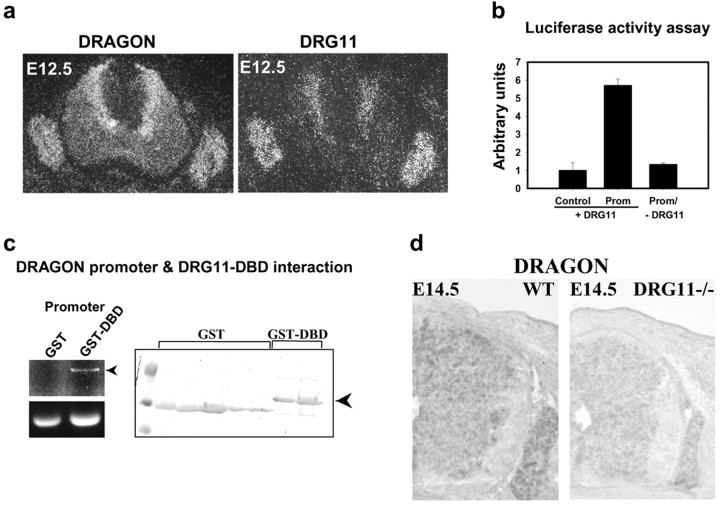
In situ hybridization reveals that at E12.5 DRG11 and DRAGON expression in the DRG and dorsal horn overlaps (Fig. 3_a_). In the DRG most neurons express both DRG11 and DRAGON. In the spinal cord DRG11 and DRAGON are expressed in the same medial region adjacent to the ventricular zone (Fig. 3_a_). To test for DRG11-dependent enhancer activity, we subcloned the fragment into a reporter vector upstream of the luciferase gene. DRG11 triggers a sixfold increase in luciferase activity compared with control (Fig. 3_b_), revealing the presence of one or several DRG11 response elements in the DRAGON 363 bp promoter fragment. No induction of luciferase activity is detected in the absence of DRG11 (Fig. 3_b_). A pull-down assay confirms direct interaction of DRG11 with the 363 bp promoter fragment of DRAGON (Fig. 3_c_). DRAGON mRNA expression is substantially reduced in the spinal cord and DRG of DRG11 -/- mice (Chen et al., 2001) compared with wild-type littermates (Fig. 3_d_).
Figure 3.
a, In situ hybridization showing DRAGON and DRG11 mRNA expression in the mouse DRG and spinal cord at E12.5. b, DRG11-dependent enhancer activity of the DRAGON promoter fragment. Luciferase reporter gene (control or with DRAGON promoter) activity in HEK293 cells is shown in the presence or absence of DRG11. c, Pull-down experiment using either GST or GST-DBD. The purified proteins (right panel) were incubated with the DRAGON promoter fragment and “pulled down” using glutathione-Sepharose. Only GST-DBD fusion protein pulled down the promoter fragment as assessed by agarose gel electrophoresis. d, In situ hybridization showing a decrease in DRAGON mRNA expression in the DRG and the spinal cord of DRG11-/- mouse at E14.5, compared with wild type.
DRAGON, mRGM, and DL-M expression and distribution
DRAGON mRNA is expressed mainly in embryonic and adult mouse DRGs, spinal cord, and brain (Fig. 4). DRAGON expression starts early in development (at least E7) (data not shown), much earlier than DRG11 (E11.5), suggesting that its expression is also regulated by other transcriptional regulatory elements. DRAGON is also expressed in areas of the nervous system where DRG11 is not expressed (outside of the dorsal horn and DRG; see below).
Figure 4.
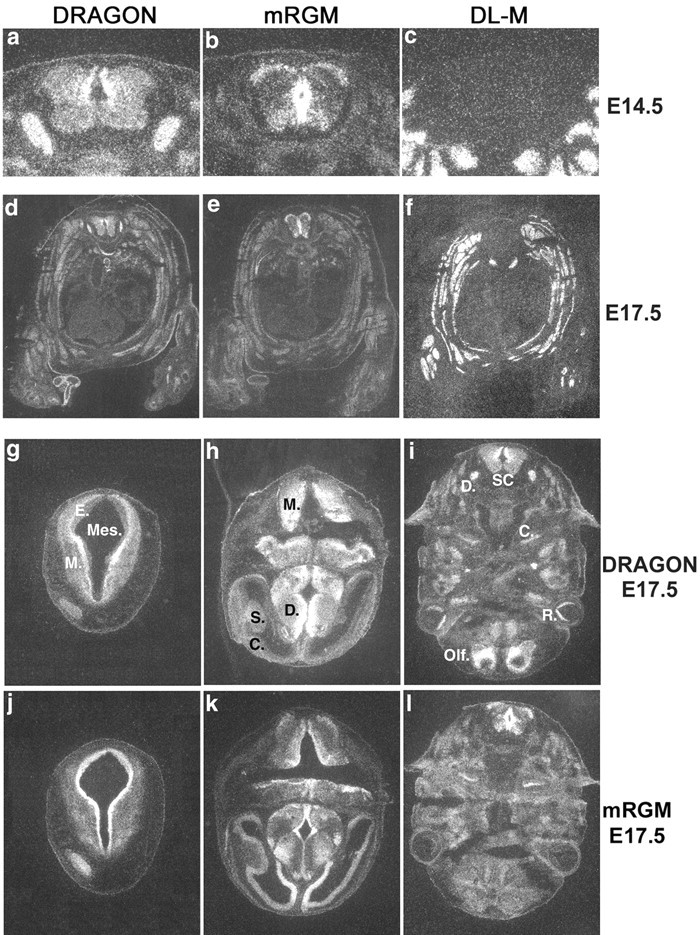
Expression of DRAGON family member mRNA in mouse embryonic tissues. _a_-c, Expression of DRAGON and mRGM but not DL-M mRNA in mouse embryonic E14.5 spinal cord. Only DRAGON is expressed in the DRG. _d_-f, Transverse sections of whole-mouse E17.5 embryo showing DRAGON, DL-M, and mRGM mRNA expression. DL-M is expressed exclusively in skeletal muscle. _g_-i, Expression of DRAGON mRNA in transverse sections of mouse E14.5 embryo head. In g: Mes, mesencephalic vesicle; E, ependymal layer; M, mantle layer. In h: M, myelencephalon; D, diencephalon; S, striatum; C, cortex. In i: D, DRG; S.C., spinal cord; C, cochlea; R, retina; Olf., future olfactory lobe. _j_-l, Expression of mRGM mRNA in transverse section of mouse E14.5 embryo head.
The relative tissue distribution pattern of DRAGON, mRGM, and DL-M mRNA in mouse embryos (E14.5) detected by in situ hybridization indicates that DRAGON and mRGM, but not DL-M, are expressed primarily in the nervous system and that DRAGON and mRGM expression in the nervous system is essentially non-overlapping (Fig. 4). DRAGON is heavily expressed in DRG neurons and in the dorsomedial mantle layer of the spinal cord, with lower expression laterally and ventrally. mRGM shows no expression in the DRG but strong expression in the spinal cord and brain. In the spinal cord, mRGM is expressed in the midline, extending from the roof to the floor plate around the central canal in the ependymal layer, medial and ventral to DRAGON (Fig. 4_a,b_). mRGM-expressing neurons are also present in the marginal layer and ventral horn (Fig. 4_b_). A complementary DRAGON and mRGM expression pattern is also present in embryonic brain (Fig. 4_g_-l). DRAGON is expressed in the alar plate of the myelencephalon, in the marginal layer of the mesencephalon, with lower intensity laterally, in the basal plate of the pons, and in the cerebellar primordia. mRGM is expressed in the ependymal layers of the myelencephalon, mesencephalon, and pons. mRGM but not DRAGON is expressed in the telencephalic cortex, most intensely medially. DRAGON is heavily expressed in the diencephalon, except in the ependymal layer, where mRGM is heavily expressed. DRAGON is homogeneously expressed in the striatum, whereas mRGM is expressed only on its medial surface (Fig. 4). DRAGON, but not mRGM, is expressed in the cortex of the future olfactory lobe, retina, and olfactory epithelium. Both DRAGON and mRGM are expressed in the cochlea (Fig. 4). DL-M is expressed exclusively in skeletal and cardiac muscle (Fig. 4_c,f_).
DRAGON protein expression
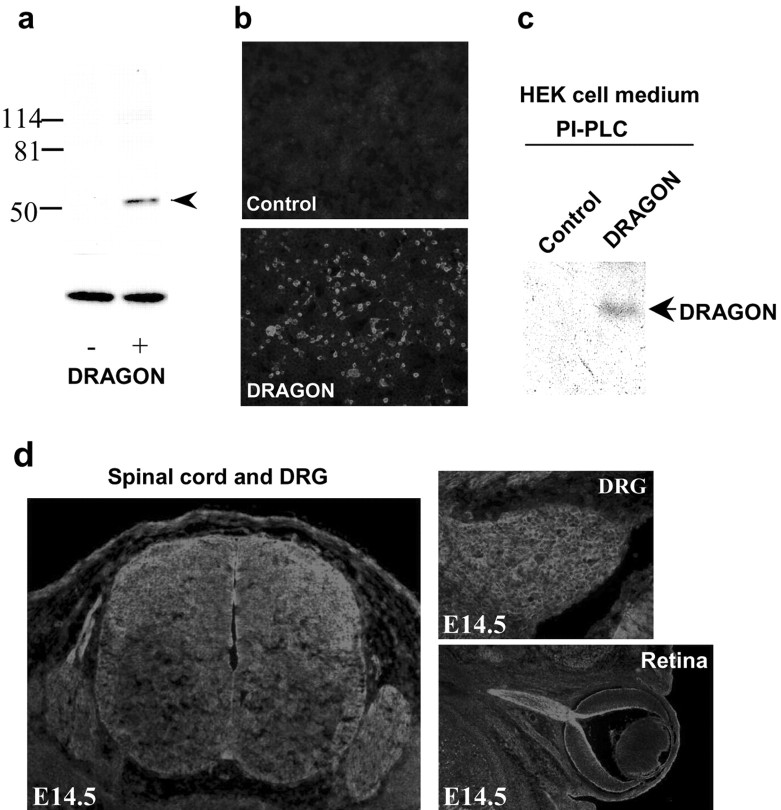
A rabbit polyclonal antibody was raised against the peptide sequence TAAAHSALEDVEALHPRKC (molecular weight, 2019.01), which is present in the C terminus of DRAGON upstream of its hydrophobic tail. The antibody binds with high affinity to recombinant DRAGON expressed in HEK293T transfected cells, recognizing a band of 50-55 kDa in Western blots (Fig. 5_a,b_). Western blots of protein extracts from neonatal and adult DRG and DRG primary cultures show a similar band with an additional lower band of 35-40 kDa, indicating possible proteolytic cleavage of endogenous DRAGON (data not shown), similar to that found for chick RGM (Monnier et al., 2002). Treatment of HEK293T cells expressing DRAGON with phosphatidylinositol-specific phospholipase C (PI-PLC) results in its release into the culture medium, as shown by Western blot analysis (Fig. 5_c_), confirming that DRAGON is GPI anchored. Immunohistochemistry reveals expression of DRAGON in areas where DRAGON mRNA is found: the DRG, spinal cord, and brain (Fig. 5_d_). DRAGON is expressed at high levels in the retinal ganglion cells of the developing mouse (E14.5), extending to the optic nerve (Fig. 5_d_).
Figure 5.
DRAGON protein expression. a, Western blot analysis using a rabbit polyclonal anti-DRAGON antibody of protein extracts from HEK293 cells transfected (+) or not (-) with a DRAGON expression vector. A distinct band of ∼50 kDa is recognized by the antibody in the transfected cells (ERK loading control). b, Immunocytochemical staining of HEK cells expressing DRAGON (bottom) compared with nontransfected cells (Control). c, Western blot analysis of the medium from cultured DRAGON expressing HEK cells pretreated, or not, with PI-PLC. A band corresponding to DRAGON is detected in PI-PLC-treated medium samples. d, DRAGON expression in mouse embryonic E14.5 spinal cord, DRG, and retina.
Zebrafish expression
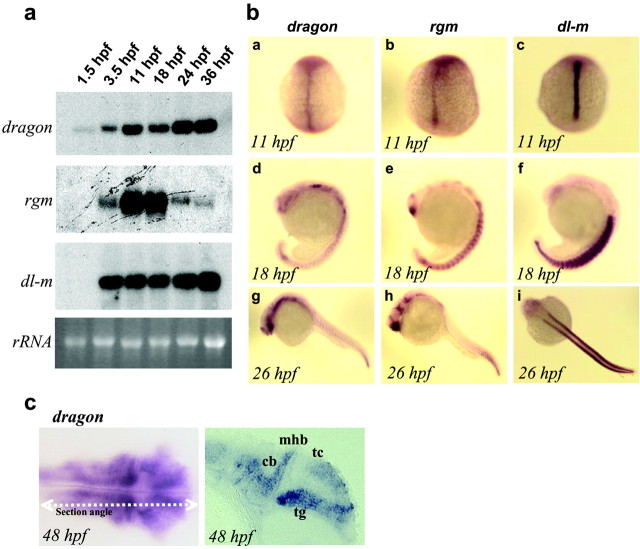
We identified and cloned zebrafish orthologs of DRAGON (dragon), RGM (rgm), and DL-M (dl-m). The sequence and domain structure of all three genes are highly conserved with the mouse genes (65-75% homology), and Northern blot analysis shows a single transcript in each case (Fig. 6_a_). Zebrafish DRAGON (dragon) mRNA is present at the two- to four-cell stage, before initiation of zygotic transcription, suggesting a maternal-early developmental role. After the mid-blastula transition, the levels of dragon mRNA increase and are then maintained at a high level for up to 36 hr, the latest stage examined. rgm mRNA is not detectable at very early developmental stages, first appearing at 10 hr of development (three-somite stage), with a peak at 18 hr and then a decrease (Fig. 6_a_). dl-m mRNA is transcribed zygotically with strong expression starting at the three-somite stage and is then maintained for up to 36 hr (Fig. 6_a_).
Figure 6.
Developmental expression of zebrafish dragon, rgm, and dl-m. a, Northern blot analysis showing zebrafish dragon, rgm, and dl-m mRNAs at different developmental stages as indicated (1.5-36 hpf). The bottom panel shows the corresponding 18S rRNA bands. b, In situ hybridization patterns of zebrafish dragon (a, d, g), rgm (b, e, h), and dl-m (c, f, i) mRNAs at the indicated developmental stages. c, A dorsal view and a longitudinal section of a 48-hr-old zebrafish embryo after in situ hybridization with a dragon probe. Significant staining is observed in the tectum (tc) and the tegmentum (tg) of the midbrain, the cerebellum (cb), and the hindbrain. mhb, Midbrain hindbrain boundary; hpf, hours postfertilization, 28.5°C.
In situ hybridization in the zebrafish embryo reveals differential but overlapping patterns of expression for dragon, dl-m, and rgm (Fig. 6_b_). At the three- to five-somite stages, all three genes are expressed along the midline (Fig. 6_b_). dragon and rgm signals are relatively less intense in the midline at this stage, whereas dl-m is very strong. By the 18-somite stage, dragon expression is resolved to sensory structures such as the olfactory and optic placodes, the eye lens and optic stalk, and the ventral thalamus. It is strong in the trigeminal ganglion, the hindbrain, and the tail bud. Diffuse expression is observed in the somites. rgm also appears in the midline at the three- to five-somite stages; however, the midline expression appears to be stronger toward the posterior end of the axis. By the 18-somite stage, rgm expression is strong in the ventral forebrain, in the ventral thalamus, and in the hypothalamus. In the hindbrain it is expressed in a segmental manner that partially correlates with the rhombomeres. The somites also show signal and the tail-bud region is strongly stained. By 26 hr of development, the trunk of the embryo shows relatively weak staining, whereas the hypothalamus, hindbrain, and tail-bud signals remain strong. During early development dl-m shows high expression in the notochord and the adjacent adaxial cells, the earliest cells to develop into muscle fibers (Thisse et al., 1993), after which it continues to be expressed strongly in somites (Fig. 6). To closely correlate the expression of dragon with neuroanatomical substructures, we sectioned 48-hr-old embryos after whole-mount in situ hybridization. At this stage, dragon expression is restricted to the brain (Fig. 6_c_). A longitudinal section reveals strong dragon expression in the tectum, the tegmentum, the midbrain-hindbrain boundary, the cerebellum, and the hindbrain. In a manner reminiscent of an anteroposterior gradient of rgm expression in the chick, dragon in the zebrafish at this stage appears to show greater expression in the posterior tegmentum.
DRAGON and neuronal adhesion
Several cell surface GPI-anchored proteins, such as NCAM (neuronal cell adhesion molecule), ephrins, and tenascin, act as neuronal and non-neuronal cell adhesion molecules, binding to molecules expressed on neighboring cells or in the extracellular matrix (Klein, 2001). To determine whether DRAGON has adhesive properties, we tested whether it influences adhesion of DRG neurons. Recombinant expression of DRAGON in HEK293 cells significantly enhances (twofold) the adhesion of dissociated cultured DRG neurons to these cells, as indicated by the number of neurons present on a layer of DRAGON-expressing HEK cells versus control HEK cells (Fig. 7). Pretreatment of the transfected HEK cells with PI-PLC before the addition of DRG neurons reduces their adhesive properties to basal levels (Fig. 7_d_). mRGM expression in HEK293 results in a significant reduction (twofold) of DRG neuron adhesion to the cells (Fig. 7_e_), showing that DRAGON and RGM exert opposite adhesive effects on primary sensory neurons.
Figure 7.
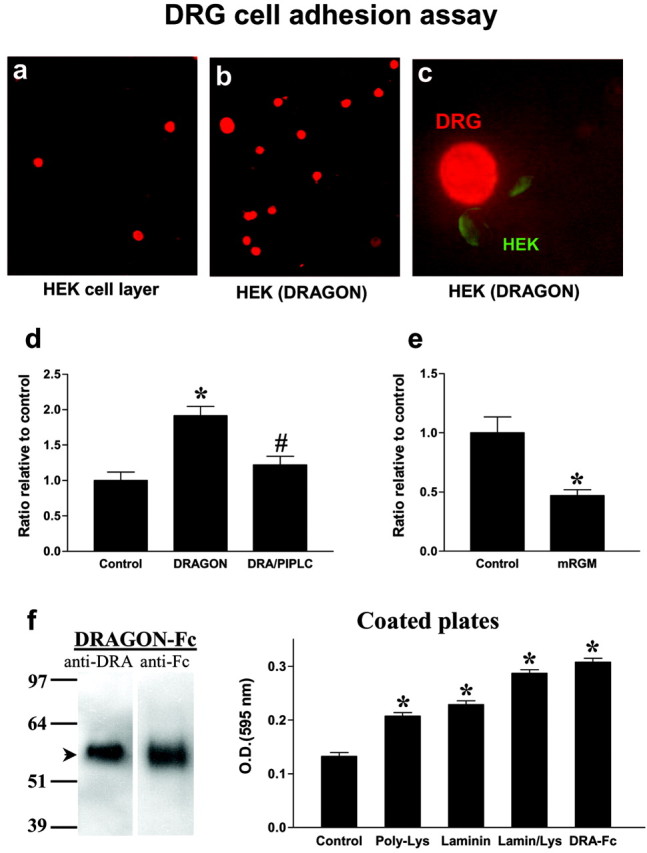
DRAGON expression enhances DRG neuron adhesion to HEK293 cells. a, b, P14 neonatal DRG neurons were plated on a monolayer of confluent HEK cells or DRAGON-transfected HEK cells. The culture slides were washed, fixed, and immunostained for DRG neurons using anti-NeuN. Monolayers at least 90% confluent were scored for neurons, and primary data were determined as neurons adhered per high power microscopy (400×) field. c, Double immunolabeling using anti-NeuN and anti-DRAGON indicates a direct interaction between DRAGON-expressing HEK cells and DRG neurons. d, Ratio of DRG neurons relative to control showing 1.9-fold increase in DRG neuron adhesion to HEK cells when they express DRAGON. Pretreatment of DRAGON-expressing HEK cells with PI-PLC significantly reduces DRG adhesion. *p < 0.05 relative to control; #p < 0.05 relative to DRAGON-transfected cells; Student's t test. e, Ratio of adhesion of DRG neurons relative to control showing twofold reduction in DRG neuron adhesion to HEK cells when they express mRGM. *p < 0.05 relative to control; Kruskal-Wallis test. f, Left, Western blot showing DRAGON-Fc detected by anti-DRAGON and anti-Fc antibodies. Right, Tissue culture plates coated with laminin (10 μg/ml), poly-d-lysine (100 μg/ml), laminin/poly-lysine, or DRAGON-Fc (100 ng/ml) show higher DRG neuron adhesion assessed by optical density measurement (595 nm) of lysate of crystal violet-stained DRG neurons compared with uncoated wells (control). Note that DRAGON-Fc-coated wells show the most pronounced adhesive properties *p < 0.05 relative to control; t test.
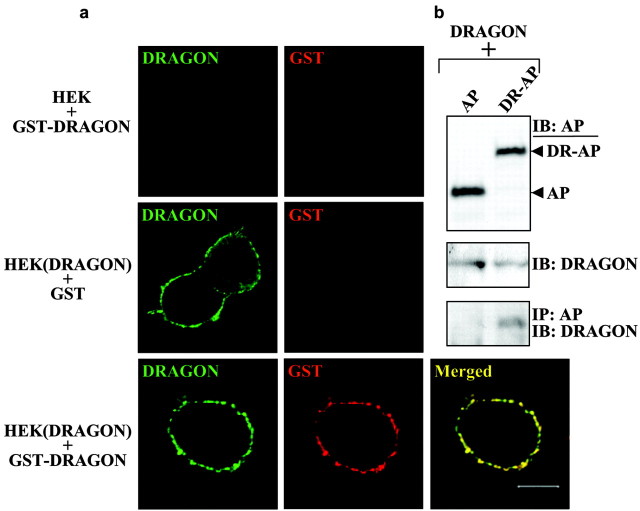
To show that the DRG neuronal adhesion to HEK cells is mediated by DRAGON, we generated a soluble DRAGON-Fc fusion protein similar to those for other molecules involved in adhesion and repulsion such as L1, the ephrins, Robo, and tenascin (Hivert et al., 2002; Rigato et al., 2002; Fuller et al., 2003; Roonprapunt et al., 2003). Purified DRAGON-Fc is detectable by both anti-DRAGON and anti-Fc antibodies (Fig. 7_f_). Tissue culture plates were coated with DRAGON-Fc, poly-d-lysine, or laminin before adding DRG neurons. Quantification of adhering DRG neurons to the coated plates indicates that DRAGON enhances neuronal cell adhesion with an efficiency comparable with that of poly-d-lysine and laminin combined (Fig. 7_f_). Because DRAGON is expressed in DRG neurons, these results may reflect homophilic or heterophilic interactions. To test whether DRAGON can promote adhesion in a homophilic manner, we assessed the interaction between native DRAGON and an AP-tagged version of DRAGON that is not detected by DRAGON antibody. Coexpression of both constructs followed by immunoprecipitation reveals a significant physical interaction between the two proteins, indicating that DRAGON can interact homophilically (Fig. 8). To further confirm a homophilic DRAGON-DRAGON interaction, we used a soluble tagged DRAGON (GST-DRAGON), which is not recognized by DRAGON antibody, to study the binding of soluble DRAGON to HEK cell-expressed DRAGON (native). Binding of GST-DRAGON to HEK cells expressing DRAGON and its colocalization with recombinant native DRAGON were assessed by immunocytochemistry and confocal microscopy using anti-GST and anti-DRAGON antibodies. These experiments reveal a colocalization of the soluble GST-DRAGON with membrane-anchored DRAGON but not with nontransfected HEK cells (Fig. 8).
Figure 8.
DRAGON interacts homophilically. a, GST or soluble GST-DRAGON was added to control (HEK) and DRAGON-transfected (HEK (DRAGON)) HEK293T cells. Recombinant DRAGON expressed by HEK cells was detected using anti-DRAGON antibody (green); GST and GST-DRAGON were detected using anti-GST antibody (red). GST-DRAGON binds to HEK cells if they express DRAGON, and the two molecules colocalize on the cell membrane (bottom). b, Cotransfection of native DRAGON with a tagged version not recognized by the DRAGON antibody (DRAGON-AP fusion protein: DR-AP). The AP tag is detected using anti-myc antibody, and a construct expressing AP alone (AP) was used as negative control. The expression of each protein was detected by immunoblot (IB) using anti-DRAGON or anti-myc antibody. DRAGON and DRAGON-AP interaction was assessed by immunoprecipitation (IP) using anti-myc antibody, followed by IB using anti-DRAGON antibody.
Discussion
We have found a novel family of GPI-anchored membrane-associated RGM-related proteins with distinctive patterns of expression in zebrafish and mouse embryos and with homologs in humans, chick, and C. elegans but not Drosophila. DRAGON, the first member of the family that we identified, was found as a result of a screen for genes regulated by DRG11. DRG11 is a paired homeodomain protein that was isolated from a differential degenerate RT-PCR screen for mammalian sensory neuron-specific transcription factors and is expressed by both DRG primary sensory neurons and neurons in the dorsal horn (Saito et al., 1995). It first appears in the mouse DRG at E11-12 and in dorsal horn neurons a little later at E12-12.5, in that area where small caliber nociceptor sensory neurons terminate (Saito et al., 1995; Chen et al., 2001). Coexpression of DRG11 in these two sets of neurons led to the proposal that DRG11 may determine connectivity between nociceptor primary afferents and their central targets in the superficial dorsal horn (Saito et al., 1995; Chen et al., 2001). The phenotype of the DRG11 knock-out mouse, although complex, supports this hypothesis (Chen et al., 2001).
We used a genomic binding screening strategy to look for genes with expression that is regulated by DRG11 and found DRAGON, on the basis of the binding of a region of its promoter to the DNA binding domain of DRG11 protein. Although we isolated DRAGON by virtue of its regulation by DRG11, it is most unlikely that DRAGON expression depends only on DRG11, because it is expressed before DRG11 in mice and zebrafish embryos and is present in brain areas that do not express DRG11. The expression patterns of mRGM and DL-M are quite different from DRG11, indicating that it is very unlikely that this transcription factor regulates the expression of these two genes. What regulates expression of DRAGON in the early neural tube and brain and whether its expression in the DRG and dorsal horn beyond E12 depends only on DRG11 now need to be explored. DRAGON and RGM do not share the same distribution in either the mouse or the zebrafish (Figs. 4, 6), but both reveal gradient-like patterns in diverse parts of the CNS. Zebrafish RGM (unlike zebrafish DRAGON) is expressed in a gradient in the tectum, in agreement with its chick ortholog, suggesting common functional features of the gene family across species.
Chick RGM is expressed in the tectum and causes collapse of temporal but not nasal retinal ganglion cell growth cones (Monnier et al., 2002). We find, however, that DRAGON does not produce any detectable embryonic DRG neuron growth cone collapse, and it is not repulsive in embryonic and neonatal DRG neuritis. Adhering neurons analyzed in the coculture assays for 24 and 48 hr survive and grow neurites that make direct contact with DRAGON-expressing HEK cells and then continue growing (data not shown). DRAGON does promote attachment of neonatal DRG neurons to HEK293 cells, however, indicating a potential role for the molecule in cell-cell adhesion. Protein interaction experiments indicate that DRAGON could act to promote adhesion in a homophilic manner. The complementary expression pattern of DRAGON and mRGM in the spinal cord and brain suggests a possible functional interaction between the two molecules. Their different effects on DRG neuron adhesion indicate that they may act in an opposite manner, at least on some targets. The binding partners for these membrane proteins themselves may be producing homophilic interactions, other GPI proteins, or indeed quite different ligands.
A cell-cell adhesion function for these GPI-linked membrane-associated proteins may contribute to neuron-neuron (axon guidance and the establishment of synaptic contacts) or neuron-glial interactions within the nervous system (DRAGON and RGM). If DL-M also has a role in cell-cell interactions, it may contribute to motor unit patterning or act in the formation of the neuromuscular junction, as has been shown for the ephrins (Feng et al., 2000). GPI-linked membrane-associated proteins have other functions too; they act, for example, as co-receptors for transmembrane receptors, such as the Nogo receptor, or as ligands for other receptors (Fournier et al., 2001). The presence of a low molecular weight band for DRAGON on Western blot of native DRG tissue indicates its possible cleavage, in which case it may have a role as a diffusible extracellular signal molecule if it is a substrate for a protease.
The identification of DRAGON indicates the utility of the genomic binding strategy for isolating the targets of developmentally expressed transcription factors. There is considerable data on the identity, timing, and spatial location of many developmentally regulated transcription factors as well as the consequences of their deletion. Very little is known, however, about how they effect their actions on neurons. This approach could be applied to find downstream targets for transcription factors such as the ETS-domain transcription factors (e.g., ER81, Pea3), the contribution of which to the connectivity of sensory neurons (Ia muscle spindle afferents) and their central targets (motor neurons) is well documented and supported by genetic models (Lin et al., 1998; Arber et al., 2000).
Footnotes
This work was supported by National Institutes of Health Grants HD38533 and NS38253 (C.J.W.). We thank David Anderson for the DRG11 expression construct, Zhou-Feng Chen for the DRG11 mutant mice, and Andrew Allchorne, Stefan Bloechlinger, and Rhona Seiffers for their contributions to the cloning and characterization of DRAGON.
Correspondence should be addressed to Clifford J. Woolf, Neural Plasticity Research Group, Department of Anesthesia and Critical Care, Massachusetts General Hospital and Harvard Medical School, 149 13th Street, Boston, MA 02129. E-mail: woolf.clifford@mgh.harvard.edu.
Copyright © 2004 Society for Neuroscience 0270-6474/04/242027-10$15.00/0
*
A.S., L.A.K., and S.-J.J. contributed equally to this work.
References
- Altmann CR, Brivanlou AH (2001) Neural patterning in the vertebrate embryo. Int Rev Cytol 203: 447-482. [DOI] [PubMed] [Google Scholar]
- Amaya F, Decosterd I, Samad TA, Plumpton C, Tate S, Mannion RJ, Costigan M, Woolf CJ (2000) Diversity of expression of the sensory neuron-specific TTX-resistant voltage-gated sodium ion channels SNS and SNS2. Mol Cell Neurosci 15: 331-342. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Groves A, Lo L, Ma Q, Rao M, Shah NM, Sommer L (1997) Cell lineage determination and the control of neuronal identity in the neural crest. Cold Spring Harb Symp Quant Biol 62: 493-504. [PubMed] [Google Scholar]
- Arber S, Ladle DR, Lin JH, Frank E, Jessell T (2000) ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell 101: 485-498. [DOI] [PubMed] [Google Scholar]
- Campagna JA, Ruegg MA, Bixby JL (1995) Agrin is a differentiation-inducing “stop signal” for motoneurons in vitro. Neuron 15: 1365-1374. [DOI] [PubMed] [Google Scholar]
- Chen ZF, Rebelo S, White F, Malmberg AB, Baba H, Lima D, Woolf CJ, Basbaum AI, Anderson DJ (2001) The paired homeodomain protein DRG11 is required for the projection of cutaneous sensory afferent fibers to the dorsal spinal cord. Neuron 31: 59-73. [DOI] [PubMed] [Google Scholar]
- Cross SH, Clark VH, Bird AP (1999) Isolation of CpG islands from large genomic clones. Nucleic Acids Res 27: 2099-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Laskowski MB, Feldheim DA, Wang H, Lewis R, Frisen J, Flanagan JG, Sanes JR (2000) Roles for ephrins in positionally selective synaptogenesis between motor neurons and muscle fibers. Neuron 25: 295-306. [DOI] [PubMed] [Google Scholar]
- Fournier AE, GrandPre T, Strittmatter SM (2001) Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature 409: 341-346. [DOI] [PubMed] [Google Scholar]
- Fuller T, Korff T, Kilian A, Dandekar G, Augustin HG (2003) Forward EphB4 signaling in endothelial cells controls cellular repulsion and segregation from ephrinB2 positive cells. J Cell Sci 116: 2461-2470. [DOI] [PubMed] [Google Scholar]
- Goulding M, Lanuza G, Sapir T, Narayan S (2002) The formation of sensorimotor circuits. Curr Opin Neurobiol 12: 508-515. [DOI] [PubMed] [Google Scholar]
- Gowan K, Helms AW, Hunsaker TL, Collisson T, Ebert PJ, Odom R, Johnson JE (2001) Crossinhibitory activities of Ngn1 and Math1 allow specification of distinct dorsal interneurons. Neuron 31: 219-232. [DOI] [PubMed] [Google Scholar]
- Gross MK, Dottori M, Goulding M (2002) Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron 34: 535-549. [DOI] [PubMed] [Google Scholar]
- Harland RM (1991) In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol 36: 685-695. [DOI] [PubMed] [Google Scholar]
- Hivert B, Liu Z, Chuang CY, Doherty P, Sundaresan V (2002) Robo1 and Robo2 are homophilic binding molecules that promote axonal growth. Mol Cell Neurosci 21: 534-545. [DOI] [PubMed] [Google Scholar]
- Jessell TM (2000) Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet 1: 20-29. [DOI] [PubMed] [Google Scholar]
- Karchewski LA, Kim FA, Johnston J, McKnight RM, Verge VM (1999) Anatomical evidence supporting the potential for modulation by multiple neurotrophins in the majority of adult lumbar sensory neurons. J Comp Neurol 413: 327-341. [DOI] [PubMed] [Google Scholar]
- Klein R (2001) Excitatory Eph receptors and adhesive ephrin ligands. Curr Opin Cell Biol 13: 196-203. [DOI] [PubMed] [Google Scholar]
- Lee KJ, Dietrich P, Jessell TM (2000) Genetic ablation reveals that the roof plate is essential for dorsal interneuron specification. Nature 403: 734-740. [DOI] [PubMed] [Google Scholar]
- Lin JH, Saito T, Anderson DJ, Lance-Jones C, Jessell TM, Arber S (1998) Functionally related motor neuron pool and muscle sensory afferent subtypes defined by coordinate ETS gene expression. Cell 95: 393-407. [DOI] [PubMed] [Google Scholar]
- Livet J, Sigrist M, Stroebel S, De Paola V, Price SR, Henderson CE, Jessell TM, Arber S (2002) ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron 35: 877-892. [DOI] [PubMed] [Google Scholar]
- Matise M (2002) A dorsal elaboration in the spinal cord. Neuron 34: 491-493. [DOI] [PubMed] [Google Scholar]
- Monnier PP, Sierra A, Macchi P, Deitinghoff L, Andersen JS, Mann M, Flad M, Hornberger MR, Stahl B, Bonhoeffer F, Mueller BK (2002) RGM is a repulsive guidance molecule for retinal axons. Nature 419: 392-395. [DOI] [PubMed] [Google Scholar]
- Muller T, Brohmann H, Pierani A, Heppenstall PA, Lewin GR, Jessell TM, Birchmeier C (2002) The homeodomain factor lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron 34: 551-562. [DOI] [PubMed] [Google Scholar]
- Pierani A, Moran-Rivard L, Sunshine MJ, Littman DR, Goulding M, Jessell TM (2001) Control of interneuron fate in the developing spinal cord by the progenitor homeodomain protein Dbx1. Neuron 29: 367-384. [DOI] [PubMed] [Google Scholar]
- Qian Y, Shirasawa S, Chen CL, Cheng L, Ma Q (2002) Proper development of relay somatic sensory neurons and D2/D4 interneurons requires homeobox genes Rnx/Tlx-3 and Tlx-1. Genes Dev 16: 1220-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigato F, Garwood J, Calco V, Heck N, Faivre-Sarrailh C, Faissner A (2002) Tenascin-C promotes neurite outgrowth of embryonic hippocampal neurons through the alternatively spliced fibronectin type III BD domains via activation of the cell adhesion molecule F3/contactin. J Neurosci 22: 6596-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roonprapunt C, Huang W, Grill R, Friedlander D, Grumet M, Chen S, Schachner M, Young W (2003) Soluble cell adhesion molecule L1-Fc promotes locomotor recovery in rats after spinal cord injury. J Neurotrauma 20: 871-882. [DOI] [PubMed] [Google Scholar]
- Saito T, Greenwood A, Sun Q, Anderson DJ (1995) Identification by differential RT-PCR of a novel paired homeodomain protein specifically expressed in sensory neurons and a subset of their CNS targets. Mol Cell Neurosci 6: 280-292. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH (1993) Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development 119: 1203-1215. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Inoue S, Hiroi H, Orimo A, Kawashima H, Muramatsu M (1998) Isolation of estrogen-responsive genes with a CpG island library. Mol Cell Biol 18: 442-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis WD, Coggeshall RE (1991) Sensory mechanisms of the spinal cord. New York: Plenum.
- Wilson SI, Rydstrom A, Trimborn T, Willert K, Nusse R, Jessell TM, Edlund T (2001) The status of Wnt signaling regulates neural and epidermal fates in the chick embryo. Nature 411: 325-330. [DOI] [PubMed] [Google Scholar]
- Zirlinger M, Lo L, McMahon J, McMahon AP, Anderson DJ (2002) Transient expression of the bHLH factor neurogenin-2 marks a subpopulation of neural crest cells biased for a sensory but not a neuronal fate. Proc Natl Acad Sci USA 99: 8084-8089. [DOI] [PMC free article] [PubMed] [Google Scholar]