Sequence Analysis of the Protein Kinase Gene Family in Human Testicular Germ-Cell Tumors of Adolescents and Adults (original) (raw)
. Author manuscript; available in PMC: 2020 May 11.
Published in final edited form as: Genes Chromosomes Cancer. 2006 Jan 1;45(1):42–46. doi: 10.1002/gcc.20265
Abstract
The protein kinase gene family is the most frequently mutated in human cancer. Previous work has documented activating mutations in the KIT receptor tyrosine kinase in testicular germ-cell tumors (TGCT). To investigate further the potential role of mutated protein kinases in the development of TGCT and to characterize the prevalence and patterns of point mutations in these tumors, we have sequenced the coding exons and splice junctions of the annotated protein kinase family of 518 genes in a series of seven seminomas and six nonseminomas. Our results show a remarkably low mutation frequency, with only a single somatic point mutation, a K277E mutation in the STK10 gene, being identified in a total of more than 15 megabases of sequence analyzed. Sequencing of STK10 in an additional 40 TGCTs revealed no further mutations. Comparative genomic hybridization and LOH analysis using SNP arrays demonstrated that the 13 TGCTs mutationally screened through the 518 protein kinase genes were uniformly aneuploid with consistent chromosomal gains on 12p, 8q, 7, and X and losses on 13q, 18q, 11q, and 4q. Our results do not provide evidence for a mutated protein kinase implicated in the development of TGCT other than KIT. Moreover, they demonstrate that the general prevalence of point mutations in TGCT is low, in contrast to the high frequency of copy number changes.
Introduction
Testicular germ-cell tumors (TGCT) are the most commonly diagnosed tumors in males aged 20–40 years and account for ∼2% of all cancers in males diagnosed per year (Parkin et al., 2005). The tumors can be predominantly divided into two histological subtypes, nonseminomatous germ-cell tumors (NSGCT) and seminomatous germ-cell tumors (SGCT), based on morphological characteristics. NSGCT can be further subdivided into embryonal carcinoma (EC), teratoma (TE), choriocarcinoma, and yolk sac tumors (YST). TGCTs are remarkable for their responsiveness to chemotherapy, in particular platinum agents, and are the only solid tumors curable with chemotherapy when distant metastases are present (Masters and Koberle, 2003). In addition, the SGCT are highly sensitive to irradiation (Honecker, 2004). However, there remains a small subset of patients with a poor prognosis, the basis of which is not clearly understood. The somatic genetics of testicular cancers at the chromosomal level has been a subject of considerable interest and several general features have been reported. These include marked aneuploidy in nearly all cases, in the hypo to hypertriploid range, as well as specific copy number gains of chromosome arm 12p in virtually all cases (Chaganti and Houldsworth, 2000). Much less is known about point mutations in specific cancer genes and the spectrum of mutations in this tumor type. RAS gene mutations have been reported in <5% of cases (http://www.sanger.ac.uk/cosmic/). TP53 gene mutations are also rare, predominantly related to progression of the disease (Skotheim and Lothe, 2003). This may explain (at least in part) the chemosensitivity of TGCTs through _TP53_-mediated apoptosis in response to DNA damage induced by platinum drugs (Masters and Koberle, 2003). The KIT receptor tyrosine kinase has been reported to be mutated in ∼5% of TGCTs (Tian et al., 1999) (http://www.sanger.ac.uk/genetics/CGP/cosmic/). The finding of mutations in the kinase domain of KIT in a relatively high proportion of bilateral cases suggests that mutations may occur in the gonadal ridge early in embryogenesis in precursor cells that give rise to eventual in situ lesions. Abrogation of strict control over renewal and differentiation within the pluripotent stem cell compartment is likely an important initiating event in the development of eventual invasive disease (Looijenga et al., 2003). Given the role of protein kinases in governing these processes, the previously role of KIT mutations, and with a view to more fully explore the mutational landscape of this highly chemosensitive tumor type, we have undertaken sequencing of the coding exons and splice junctions of the protein kinase gene family in a series of TGCT.
Materials and Methods
Samples
Thirteen TGCTs (7 SGCT and 6 NSGCT) were obtained at surgery under approved patient consents at collaborating hospitals in Rotterdam and surroundings (the Netherlands). All tumors were reviewed by an experienced pathologist (JWO) for confirmation of diagnosis and for assessment of relative tumor content. Samples with more than 80% tumor cells were included in the analyses. Patient and tumor characteristics are given in Table 1. A further series of 40 cancers (20 SGCT and 20 NSGCT) were analyzed through genes in which somatic mutations were found in the primary screen (see later).
Table 1. Testicular Germ-Cell Tumor Case Characteristics.
| CGP numbera | Age | Histology |
|---|---|---|
| Main series | ||
| PD1381a | 29 | SGCT |
| PD1382a | 31 | SGCT |
| PD1383a | 50 | SGCT |
| PD1384a | 72 | SGCT |
| PD1385a | 28 | SGCT |
| PD1386a | 31 | SGCT |
| PD1388a | 22 | SGCT |
| PD1389a | 26 | NSGCT (EC, TE, YST) |
| PD1391a | 37 | NSGCT (EC, TE, YST) |
| PD1392a | 30 | NSGCT (TE, YST) |
| PD1394a | 31 | NSGCT (EC, YST, TE) |
| PD1395a | 32 | NSGCT (YST) |
| PD1396a | 26 | NSGCT (EC, YST, TE) |
| Follow-up | ||
| PD0836a | 34 | SGCT (bilateral) |
| PD0837a | 30 | SGCT (bilateral) |
| PD0838a | 37 | SGCT (bilateral) |
| PD0839a | 19 | SGCT |
| PD0840a | 50 | SGCT |
| PD0841a | 40 | SGCT |
| PD0842a | 34 | SGCT |
| PD0843a | 41 | SGCT |
| PD0844a | 44 | SGCT (bilateral) |
| PD0845a | 28 | SGCT |
| PD0846a | 33 | SGCT |
| PD0847a | 30 | SGCT |
| PD0848a | 28 | SGCT |
| PD0849a | 29 | SGCT |
| PD0850a | 57 | SGCT |
| PD0851a | 32 | SGCT |
| PD0852a | 27 | SGCT |
| PD0853a | 32 | SGCT |
| PD0854a | 40 | SGCT |
| PD0855a | 36 | SGCT |
| PD0856a | 39 | NSGCT (YST) |
| PD0857a | 30 | NSGCT (EC) |
| PD0858a | 28 | NSGCT (EC) |
| PD0859a | 28 | NSGCT (EC) |
| PD0860a | 16 | NSGCT (EC) |
| PD0861a | 32 | NSGCT (TE) |
| PD0862a | 25 | NSGCT (TE) |
| PD0863a | 24 | NSGCT (TE) |
| PD0864a | 20 | NSGCT (TE) |
| PD0865a | 30 | NSGCT (TE) |
| PD0866a | 22 | NSGCT (TE) |
| PD0867a | 31 | NSGCT (YST) |
| PD0868a | 39 | NSGCT (EC, TE, YST) |
| PD0869a | 22 | NSGCT (EC, TE, YST) |
| PD0870a | 26 | NSGCT (EC, YST) |
| PD0871a | 27 | NSGCT (EC, YST) |
| PD0872a | 25 | NSGCT (TE, YST) |
| PD0873a | 26 | NSGCT (TE, YST) |
| PD0874a | 25 | NSGCT (EC, YST) |
| PD0875a | 40 | NSGCT (EC, TE, YST) |
Genes and PCR Primers
Coding exons of the family of protein kinase genes were amplified as previously described (Stephens et al., 2005). Sequences of all amplimers can be obtained from http://www.sanger.ac.uk/genetics/CGP/Kinases/. The total length of coding sequence considered in this work was 1,361 kb/sample, of which 444 kb encoded the protein kinase domains. 423 kb of the kinase domains and 1,252 kb of the complete coding sequence (95 and 92%, respectively) were successfully covered in amplimer design.
PCR and DNA Sequencing
Amplification of sequencing templates and direct sequencing on ABI3730s were done as previously described (Stephens et al., 2005). Briefly, DNA from tumors was amplified by PCR and bidirectionally sequenced. Sequence traces were compared with normal DNA sequences used as reference traces using Mutation Surveyor followed by manual analysis of all suspected variants. Sequence variants were compared with dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) and data from our other mutational screens to identify variants previously reported as polymorphisms. Novel variants were then assessed as rare polymorphisms versus somatic mutations by sequencing the matching normal DNA of the case harboring the variant. If the variant was not observed in the matching normal germline DNA, it was considered to be a somatic mutation. Confirmation of these results was undertaken in every case by further resequencing of both the tumor and matching normal from fresh amplification products. Any kinase gene with a verified somatic mutation was further evaluated by resequencing the entire coding sequence of the gene in a follow-up series of 40 cases as described earlier. More than 90% of designed exons were successfully analyzed in more than 90% of samples in the main series.
Copy Number
Genome-wide analysis of copy number was performed on the paired normal/tumor samples using Affymetrix Human GeneChip mapping 10K array (Xba 131) as previously described (Bignell et al., 2004).
Quantitative PCR of KIT
Quantitative PCR was carried out using the relative quantitation comparative Ct method in separate tubes (ABI User bulletin#2) on the ABI 7900HT sequence detection system. The target sequence was designed to exon 3 of KIT (GAAGCCTCTTCCCAAGGACT, TTTCCGACAGCACTGACTTG, 137 bp) with the reference PCR product being designed to exon 18 of APP (TCAGGTTGA-CGCCGCTGT, TTCGTAGCCGTTCTGCTGC, 67 bp), both PCR products were tested to confirm the presence of a single clean product by gel electrophoresis and melt curve analysis on the ABI 7900HT sequence detection system. PCRs were preformed using the QuantiTect Syber Green PCR Kit (Qiagen, Germany) as per manufacturer’s instruction, using 50 μl PCR volume, 20 ng DNA, and 2.5 ng/μl primer. All PCR’s were carried out in triplicate and the final quantification determined relative to a normal diploid DNA sample.
Results
Thirteen TGCT were sequenced through the coding exons and splice junctions of the protein kinase gene family amounting to a total of ∼1.3 megabases of sequence per sample and more than 15 megabases for the entire study. Only one somatic mutation was identified in the screen, in case PD1386a, a SGCT from a 31 year old. A heterozygous somatic missense mutation (A829G, K277E) was found in the serine/threonine kinase STK10 (http://www.sanger.ac.uk/genetics/CGP/Kinases/). STK10 encodes a polo-like kinase that has been implicated in positive cell cycle regulation through its activation of PLK1. PLK1 is a kinase involved in several key aspects of cell cycle progression, including centrosomal maturation and chromosomal segregation (Myer et al., 2005). The mutation was in a highly conserved residue distal to the activation segment within the kinase domain of STK10 and may therefore be of functional consequence. However, it is unlikely that STK10 mutations play a substantial role in TGCT via mutational activation, as no additional mutations in the gene were found upon sequencing a further 40 TGCTs. No mutations were identified in KRAS, NRAS, or TP53, confirming the low frequency of mutations previously reported (Chaganti and Houldsworth, 2000; Skotheim and Lothe, 2003).
No KIT mutations were detected in the primary series of 13 TGCTs. To further assess the role of this protein kinase, we also analyzed the KIT coding sequence in the follow-up series of 40 cases. Three KIT missense mutations were identified. A heterozygous G2446T mutation was identified in PD0838a, a SGCT from a 37 year old with bilateral disease, resulting in an aspartate to tyrosine substitution (D816Y) at codon 816. A heterozygous T2466G mutation was identified in PD0841a, a SGCT from a 40 year old, resulting in an asparagine to lysine substitution (N822K) at codon 822. A heterozygous G2485C mutation was identified in PD0864a, a NSGCT (a TE) from a 20 year old, resulting in an alanine to proline substitution (A829P) at codon 829. All mutations are in the activation segment of the kinase domain. D816 is the most frequently mutated codon reported in TGCT and N822K has previously been reported as a mutation in a SGCT and acute myeloid leukemia. (http://www.sanger.ac.uk/cosmic/). A829P is also likely to represent an oncogenic mutation given the conservation of the activation segment residues and the nonconservative nature of the substitution.
KIT has also been reported to be involved in cancers through amplification and copy number gains (Sihto et al., 2005). We therefore assessed KIT copy number/amplification status using a competitive PCR strategy. One case in the main series of 13 tumors, a SGCT in a 31 year old (PD1382a), showed ∼5-fold amplification (data not shown). Further evaluation of KIT amplification in the follow-up series yielded no cancers with overt KIT amplification. Taking both mutation and amplification into account, these data show that KIT alterations are involved in ∼7.5% (4/53) of TGCT cases, consistent with previous reports (http://www.sanger.ac.uk/cosmic/).
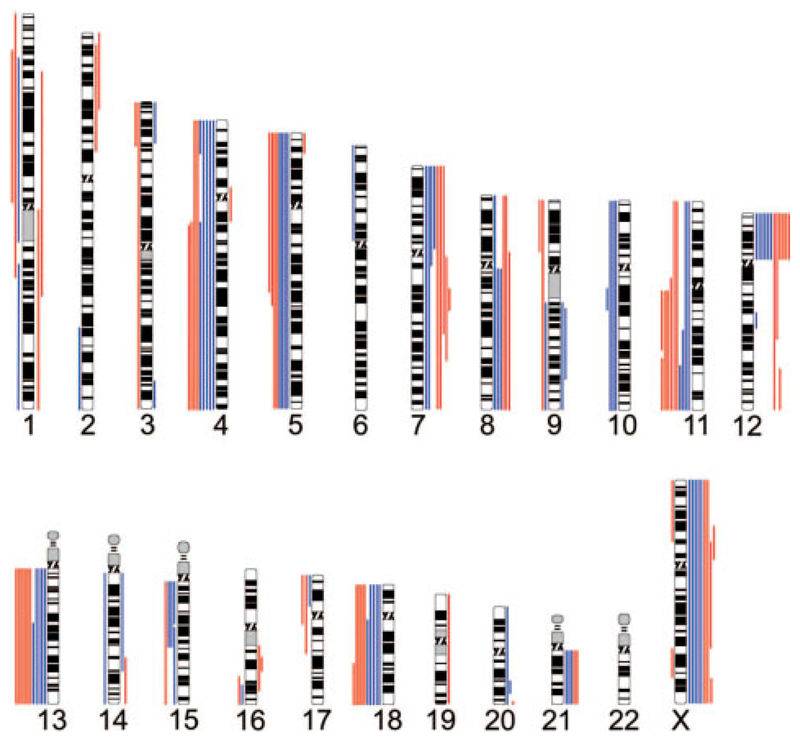
TGCTs have been previously reported to exhibit aneuploidy in all cases. To further characterize the main series for chromosomal-scale changes, we performed genome-wide copy number analysis using Affymetrix Xba 131 SNP arrays. All cancers under investigation exhibited multiple copy number changes, in contrast to the data for point mutations (Fig. 1). Patterns of gains and losses were consistent with other previously published data on TGCTs (Chaganti and Houldsworth, 2000; Skotheim and Lothe, 2003; Oosterhuis and Looijenga, 2005). Gains of 12p were observed in 100% of cases (12/12, 1 failed in analysis). Other common alterations included losses of 13q, 18q, 11q, and 4q. Common gains, in addition to 12p, were found for 8q, 7, and the X chromosome. In general, the copy number data report few distinct differences between NSGCT and SGCTs, although the numbers examined are relatively small.
Figure 1.
Copy number alterations detected using the Affymetrix Genechip Human Mapping 10K Array (Xba 131) in the main series of TGCTs (data from 12/13, PD1384 analysis failed). Copy number decreases are shown to the left of the chromosome ideograms, while gains are shown to the right. Data for the SGCT samples are shown in red with the NSGCT in blue.
Discussion
While this study suggests that activating point mutations of protein kinases are unlikely to be responsible for the development of most TGCTs, the results obtained are nonetheless remarkable. This is by far the largest evaluation of sequence ever presented from this tumor type and the data show that 12/13 cancers under investigation have no somatic mutations at all in 1.3 megabases of DNA examined. In total, only a single somatic mutation was identified in more than 15 megabases of cancer DNA. It remains possible that mutations exist in the small minority of exons that we were unable to analyze or that a small number have been missed. However, the prevalence of somatic point mutations in TGCT appears low.
Although very few comparable datasets are currently available for other tumor types, some general comparisons can be made. Published data from colorectal cancer indicate an average somatic mutation frequency of approximately one mutation per megabase (Wang et al., 2002). Our own data have shown that most non-small-cell lung cancers have at least one and often several somatic mutations per megabase (Davies et al., in press). In contrast, in similar protein kinase gene screens, we found no mutations in lung carcinoids, and breast cancers displayed a mixed pattern, with a few cancers showing very high numbers of mutations and others showing none (Davies et al., in press; Stephens et al., 2005). Taken together, these studies suggest that there are considerable differences in somatic point mutation prevalence between different cancer types.
The somatic mutational burden of a cancer is the sum of the mutations acquired at each round of DNA replication in the cellular lineage from the fertilized egg to the cell that is the progenitor of the final neoplastic clonal expansion. Therefore, the diversity of somatic mutation prevalence is potentially a function of the number of mitoses in this lineage and the mutation rate at each mitosis. The latter may be influenced by endogenous mutagenic processes, exogenous exposures, and defects in DNA repair/apoptotic pathways. The reason for the low frequency of somatic mutations in TGCTs when compared, for example, with that in lung carcinomas is currently unknown. However, it may reflect (at least in part) the fact that normal progenitors of TGCTs are not part of a surface epithelium constantly exposed to exogenous mutagens as are normal lung epithelial cells.
Other facets of normal testicular germ cell and TGCT biology may also affect the mutation frequency. Although previous studies have demonstrated a decreased efficiency of nucleotide excision repair in TGCTs (which might be expected to increase the frequency of somatic mutations), maintenance of the apoptotic response to DNA damage may result in removal of cells with DNA damage (Koberle et al., 1999; Masters and Koberle, 2003). The latter may also be a feature of normal germ-cell precursors in which rates of apoptosis are quite high, presumably to remove defective sperm and hence to reduce the risk of transmitting genetic defects to offspring (Oosterhuis and Looijenga, 2005). The finding of low mutation frequencies in tumors that exhibit high sensitivity to chemotherapy may be consistent with a model of chemosensitivity driven by the interplay of inefficient DNA repair in the context of intact apoptotic responses.
The results presented here provide further insights into the nature of cancer genomes, their heterogeneity, and the forces shaping them. In TGCT, copy number changes are relatively common and point mutations relatively rare when compared with some other cancer types. It may be, therefore, that many of the causative somatic genetic events in TGCT development occur through chromosomescale alterations resulting in changes of gene dosage rather than through point mutations.
Acknowledgements
The authors thank Wendy Haynes for help in manuscript preparation.
Supported by: Wellcome Trust.
References
- Bignell GR, Huang J, Greshock J, Watt S, Butler A, West S, Grigorova M, Jones KW, Wei W, Stratton MR, Futreal PA, et al. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res. 2004;14:287–295. doi: 10.1101/gr.2012304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti RSK, Houldsworth J. Genetics and biology of adult human male germ cell tumors. Cancer Res. 2000;60:1475–1482. [PubMed] [Google Scholar]
- Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, Teague J, Butler A, Edkins S, Stevens C, Parker A, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- Honecker F. Pathobiological implications of the expression of markers of testicular carcinoma in situ by foetal germ cells. J Pathol. 2004;203:849–857. doi: 10.1002/path.1587. [DOI] [PubMed] [Google Scholar]
- Koberle B, Masters JRW, Hartley JA, Wood RD. Reduced repair of cisplatin-induced DNA damage in testicular germ cell tumours due to a specific protein defect. Curr Biol. 1999;9:273–276. doi: 10.1016/s0960-9822(99)80118-3. [DOI] [PubMed] [Google Scholar]
- Looijenga LHJ, de Leeuw H, van Oorschot M, van Gurp RJHLM, Stoop H, Gillis AJM, de Gouveia Brazao CA, Weber RFA, Kirkels WJ, van Dijk T, von Lindern M, et al. Stem cell factor receptor (c-kit) codon 816 mutations predict development of bilateral testicular germ-cell tumors. Cancer Res. 2003;63:7674–7678. [PubMed] [Google Scholar]
- Masters JRW, Koberle B. Curing metastatic cancer: lessons from testicular germ-cell tumours. Nat Rev Cancer. 2003;3:517–525. doi: 10.1038/nrc1120. [DOI] [PubMed] [Google Scholar]
- Myer DL, Bahassi eL M, Stambrook PJ. The Plk3-Cdc25 circuit. Oncogene. 2005;24:299–305. doi: 10.1038/sj.onc.1208278. [DOI] [PubMed] [Google Scholar]
- Oosterhuis JW, Looijenga LHJ. Testicular germ-cell tumours in a broader perspective. Nat Rev Cancer. 2005;5:210–222. doi: 10.1038/nrc1568. [DOI] [PubMed] [Google Scholar]
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- Sihto H, Sarlomo-Rikala M, Tynninen O, Tanner M, Andersson LC, Franssila K, Nupponen NN, Joensuu H. KIT and platelet-derived growth factor receptor α tyrosine kinase gene mutations and kit amplifications in human solid tumors. J Clin Oncol. 2005;23:49–57. doi: 10.1200/JCO.2005.02.093. [DOI] [PubMed] [Google Scholar]
- Skotheim RI, Lothe R. The testicular germ cell tumour genome. APMIS. 2003;111:136–151. doi: 10.1034/j.1600-0463.2003.11101181.x. [DOI] [PubMed] [Google Scholar]
- Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, Bignell G, Teague J, Smith R, Stevens C, O’Meara S, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- Tian Q, Frierson HF, Krystal GW, Moskaluk CA. Activating c-kit gene mutations in human germ cell tumors. Am J Pathol. 1999;154:1643–1647. doi: 10.1016/S0002-9440(10)65419-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T-L, Rago C, Silliman N, Ptak J, Markowitz S, Willson JKV, Parmigiani G, Kinzler KW, Vogelstein B, Velculescu VE. Prevalence of somatic alterations in the colorectal cancer cell genome. Proc Natl Acad Sci USA. 2002;99:3076–3080. doi: 10.1073/pnas.261714699. [DOI] [PMC free article] [PubMed] [Google Scholar]