Eukaryotic Translation Initiation Factor 4AIII (eIF4AIII) Is Functionally Distinct from eIF4AI and eIF4AII (original) (raw)
Abstract
Eukaryotic initiation factor 4A (eIF4A) is an RNA-dependent ATPase and ATP-dependent RNA helicase that is thought to melt the 5′ proximal secondary structure of eukaryotic mRNAs to facilitate attachment of the 40S ribosomal subunit. eIF4A functions in a complex termed eIF4F with two other initiation factors (eIF4E and eIF4G). Two isoforms of eIF4A, eIF4AI and eIF4AII, which are encoded by two different genes, are functionally indistinguishable. A third member of the eIF4A family, eIF4AIII, whose human homolog exhibits 65% amino acid identity to human eIF4AI, has also been cloned from Xenopus and tobacco, but its function in translation has not been characterized. In this study, human eIF4AIII was characterized biochemically. While eIF4AIII, like eIF4AI, exhibits RNA-dependent ATPase activity and ATP-dependent RNA helicase activity, it fails to substitute for eIF4AI in an in vitro-reconstituted 40S ribosome binding assay. Instead, eIF4AIII inhibits translation in a reticulocyte lysate system. In addition, whereas eIF4AI binds independently to the middle and carboxy-terminal fragments of eIF4G, eIF4AIII binds to the middle fragment only. These functional differences between eIF4AI and eIF4AIII suggest that eIF4AIII might play an inhibitory role in translation under physiological conditions.
All cellular (except organellar) eukaryotic mRNAs possess a cap structure, m7GpppN (where N is any nucleotide), at the 5′ terminus. The interaction of the cap structure with the eukaryotic initiation factor 4F (eIF4F) is important for efficient translation of the mRNA. eIF4F, in association with the RNA binding protein eIF4B, is thought to unwind the secondary structure in the 5′ untranslated region of an mRNA to facilitate the binding of the 40S ribosome preinitiation complex (for reviews, see references 29, 35, and 52).
eIF4F is a protein complex consisting of three subunits: eIF4E, eIF4G, and eIF4A. eIF4E specifically interacts with the cap structure to position eIF4F at the 5′ terminus of the mRNA. eIF4G is an adapter protein that bridges the mRNA and the 40S ribosome and serves as a scaffold for binding of several proteins, including eIF4E, eIF4A, eIF3, poly(A) binding protein and a serine/threonine kinase, Mnk-1 (18, 20, 22, 27, 44). eIF4A is the prototypic DEAD box protein, which belongs to a large family of proteins, whose members share nine conserved motifs (24). eIF4A is an RNA-dependent ATPase and an ATP-dependent RNA helicase (47). The RNA helicase activity of eIF4A is strongly stimulated by eIF4B (47, 48; for reviews, see references 10 and 29). Furthermore, eIF4A is likely to gain access to the mRNA only as a subunit of eIF4F and recycle through eIF4F (38, 59), because eIF4A in the eIF4F complex exhibits ∼20-fold more efficient RNA helicase activity than the free form of eIF4A (45, 48). Several viral and cellular mRNAs initiate translation in a cap-independent manner, which is accomplished by internal binding of the ribosome at or upstream of the initiation codon (for reviews, see references 3 and 21). eIF4A is required for both cap-dependent and cap-independent translation (38, 41). The essential function of eIF4A in translation has been firmly established, since translation of all mRNAs is abolished in extracts prepared from yeast in which the eIF4A gene is disrupted (5) and since mutants of mammalian eIF4A act as dominant negative inhibitors of translation (38).
Several eukaryotic translation initiation factors are encoded by more than one gene. Two isoforms of eIF4G are expressed in yeast (11), plants (1, 6), and mammals (12, 58). In mammals, eIF4GII is a functional homolog of eIF4GI that exhibits similar biochemical activities (12) but is less sensitive to cleavage by viral proteases (13, 54). Two isoforms of eIF4A exist in yeast (TIF1 and TIF2) and mammals (eIF4AI and eIF4AII). They are identical in yeast (25), and the murine proteins have 91% identity at the amino acid level (33). Furthermore, both isoforms function in translation initiation and are functionally interchangeable. Both can be incorporated into the eIF4F complex with similar kinetics (59). The different functions, if any, of the two eIF4A forms are not known.
A third member of the eIF4A family, eIF4AIII, was isolated from a Nicotiana plumbaginifolia root cDNA library and from a Xenopus embryo library (34, 57). Overexpression of Xenopus eIF4AIII induced epidermis formation in Xenopus embryo cells that would otherwise assume a neural fate, without causing an increase in the rate of overall protein synthesis (57). In this report, we characterize the biochemical activity of human eIF4AIII. We describe that while eIF4AI can bind to two sites on eIF4GI, eIF4AIII can bind only one site. This could explain why eIF4AIII is unable to form a productive preinitiation complex in a toeprinting assay and why it is inhibitory for translation in a reticulocyte lysate.
MATERIALS AND METHODS
Cloning of cDNAs.
Nuk34 cDNA was obtained from a human keratinocyte cDNA library (accession no. P38919). A phylogenetic tree was generated by using the CLUSTAL method with the PAM250 residue weight table in the Megalign program. Sequence alignment of eIF4AIII was performed with the CLUSTAL W multiple-sequence alignment program.
Plasmid construction and protein expression.
The oligonucleotides 5′-TATGGCTAGCCTCGAGG-3′ and 3′-ACCGATCGGAGCTCCCTAG-5′ were inserted into the pET-3b vector which had been digested with _Nde_I-_Bam_HI. This created an _Xho_I site. A PCR-synthesized DNA encoding amino acids (aa) 1 to 50 of eIF4AIII was inserted into the modified pET-3b vector together with an _Eco_RI-_Xho_I fragment of the eIF4AIII cDNA encoding aa 51 to 411 to express the full-length eIF4AIII. The PCR fragment was sequenced in its entirety. The forward primer for PCR was 5′-CGCGGACTCTGACATATGGCGACCACGGCCACGATG-3′, and the reverse primer was 5′-TCCCGCAGGCCCATGGTGTCG-3′. pcDNA3-HA-eIF4AIII was generated by cloning the coding sequence of eIF4AIII into pcDNA3-HA (19) by using _Eco_RI-_Xho_I sites.
Escherichia coli BL-21 (DE3) was transformed with pET3b-eIF4AI or pET3b-eIF4AIII, and protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside. Recombinant eIF4AI was purified by the method of Pause and Sonenberg (39). Recombinant eIF4AIII was purified by HiTrap Blue (Pharmacia) chromatography as specified by the manufacturer. Both eIF4AI and eIF4AIII were passed through HiTrap heparin (Pharmacia) to remove contaminating RNases. Protein preparations were tested for the absence of RNase activity by incubation with a 32P-labeled RNA-12 (used in the helicase assay as described below), which was then resolved by denaturing (7 M urea) polyacrylamide gel electrophoresis (PAGE) with 20% polyacrylamide.
Western blotting and antibodies.
The peptide ATSGSARKRLLKEED, derived from eIF4AIII, was conjugated to keyhole limpet hemocyanin and used to raise antibodies in rabbits. An anti-eIF4AI monoclonal antibody was a kind gift from H. Trachsel.
HeLa, 293, U937, or SW480 cells were lysed in buffer A (100 mM KCl, 0.5 mM EDTA, 20 mM HEPES-KOH [pH 7.6], 10% glycerol, and a cocktail of protease inhibitors [Boehringer Mannheim]) by three cycles of freezing and thawing. Cell extract or eIF4AI or eIF4AIII protein was resuspended in an equal volume of 2× Laemmli sample buffer, boiled for 5 min, and then subjected to sodium dodecyl sulfate (SDS)-PAGE and blotted onto nitrocellulose membranes (overnight transfer). The membranes were blocked for 1 h at room temperature in 5% skim milk and probed with antibodies against eIF4AI (1:10 dilution of hybridoma conditioned medium) or eIF4AIII (1:1000 dilution) for 2 h at room temperature in 5% skim milk. The blots were washed and subsequently incubated at a 1:5,000 dilution in the same buffer for 1 h at room temperature with either sheep anti-mouse immunoglobin conjugated with horseradish peroxidase (Amersham) or donkey anti-rabbit immunoglobin conjugated with horseradish peroxidase (Amersham). After extensive washing, the blots were developed with the Renaissance enhanced chemiluminescence system (Amersham).
Quantitation of eIF4AI and eIF4AIII proteins.
HeLa cells were lysed in buffer A by three cycles of freezing and thawing. The cytoplasmic fraction was obtained by centrifugation (10,000 × g for 10 min). HeLa extract and recombinant protein were mixed and subjected to SDS-PAGE (12.5% polyacrylamide). Western blotting was performed as above except that either 0.03 μCi of 125I-conjugated anti-mouse immunoglobulin G per ml for 1 h (eIF4AI) or 0.1 μCi of 125I-conjugated protein A per ml for 4 h (eIF4AIII) was used. The membrane was then washed and exposed to an X-ray film. Signals obtained for eIF4AI or eIF4AIII protein were quantified with a phosphorimager (Fuji).
ATPase assay.
The ATPase assay was performed by the method of Richter-Cook et al. (46). Briefly, 1 μg of eIF4AI or eIF4AIII was incubated at 37°C for 15 min with 0.3 absorbance at 260 nm units of poly(A) and 100 μM [γ-32P]ATP (3,000 cpm/pmol) in 15 mM HEPES-KOH (pH 7.5)–80 mM KCl–2.5 mM magnesium acetate–1.0 mM dithiothreitol (DTT). Entire reaction mixtures were extracted with the following reagents at 4°C, which were added in the indicated order: (i) 0.5 ml of 20 mM silicotungstate–20 mM sulfuric acid; (ii) 1.2 ml of 1 mM potassium phosphate (pH 7.0); (iii) 0.5 ml of 5% ammonium molybdate–4 M sulfuric acid; (iv) 0.3 ml of 2.5% trichloroacetic acid–50% acetone, and (v) 2 ml of 50% isobutyl alcohol–50% benzene. Inorganic phosphate was sequestered in the upper phase. Aliquots (0.5 ml) were collected, mixed with 5 ml of Ecolite (ICN) scintillation solution, and counted. Each value of released inorganic phosphate represents the average of two independent determinations. ATP hydrolysis in the absence of protein was subtracted as background.
RNA substrate for the helicase assay.
RNA-1 and RNA-12 oligonucleotide (47) were mixed to generate the duplex RNA substrate for the helicase assay: 5′-GGGGAGA(A4C)5UAGCACCGUAAAGCACGC-3′ (RNA-1) and 3′-CGUGGCAUUUCG-5′ (RNA-12). The bottom strand (RNA-12) was synthesized by an RNA synthesizer and purified by PAGE. The top strand (RNA-1), a 50-nucleotide RNA, was synthesized by in vitro transcription with T7 RNA polymerase. The template for transcription was generated by annealing two synthetic DNA oligonucleotides (synthesized by the Case Western Reserve University Molecular Biology Core Facility and purified with an oligonucleotide purification cartridge), 5′-GAATTTAATACGACTCACTATAG-3′ and 3′-CTTAAATTATGCTGAGTGATATCCCCTCT(TTTG)5ATCGTGGCATTTCGTGCG-5′. The RNA was passed over a Bio-Rad P6 spin column to remove salts and unincorporated nucleotides. The eluate was then extracted once with phenol. The aqueous phase was subsequently extracted three times with ether to remove all traces of phenol.
RNA helicase assay.
The helicase assay was performed essentially as described previously (47). To prepare 32P-labeled RNA-12, RNA-12 (40 pmol) was labeled with 10 pmol of [γ-32P]ATP (specific activity, 3,000 Ci/mmol; 10 mCi/ml) and 10 U of T4 polynucleotide kinase (NEB) in a 10-μl reaction mixture. Duplex RNA was generated by annealing 12.5 pmol of RNA-1 and 10 pmol of 32P-labeled RNA-12 in a 20-μl volume containing 10 mM Tris–1 mM EDTA buffer and 100 mM KCl. The samples were heated to 95°C and then slowly cooled to 4°C over 90 min.
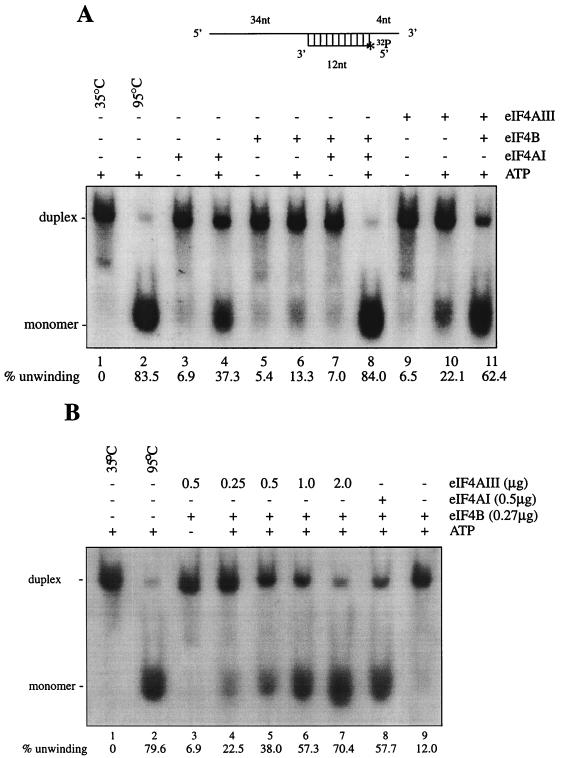
eIF4AI and eIF4B were purified from rabbit reticulocyte lysate by previously described procedures (14). The eIF4AIII used was the nontagged recombinant eIF4AIII. For the helicase assay, eIF4AI (0.38 μg) or eIF4AIII (0.5 to 2 μg) and eIF4B (0.23 μg) were incubated with 1.8 nM 32P-labeled RNA duplex at 35°C for the times indicated in the legend to Fig. 6 in a buffer containing 20 mM HEPES (pH 7.5), 2 mM DTT, 70 mM KCl, 1 mg of bovine serum albumin per ml, 1 mM magnesium acetate, and 1 mM ATP in a final volume of 20 μl. The reactions were stopped by addition of 5 μl of 50% glycerol–2% SDS–20 mM EDTA and 0.01% bromophenol blue and xylene cyanol dyes. The products of the reaction (duplex and released strands) were analyzed on a 12% native polyacrylamide gel and scanned directly with an Ambis radioanalytical scanner. Unwinding efficiency was defined as a percentage based on the ratio of unwound monomer RNA relative to the input duplex RNA (47).
FIG. 6.
RNA helicase activity of eIF4AIII. (A) An RNA duplex was used for the helicase assay, as described in Materials and Methods. eIF4AI (0.38 μg) or eIF4AIII (0.38 μg) was incubated in the presence or absence of eIF4B (0.23 μg) with 20,000 cpm (0.04 pmol) of 32P-labeled duplex RNA for 15 min at 35°C. (B) Dose-dependent helicase activity of eIF4AIII in the presence of eIF4B. nt, nucleotides.
Transient transfection and immunoprecipitation.
HeLa cells (in a 6-cm-diameter tissue culture dish) were infected with recombinant vaccinia virus vTF7-3 and transfected with plasmid DNA (5 μg) by using Lipofectin (GIBCO-BRL) as recommended by the manufacturer. Following transfection, the cells were cultured for 16 h. They were lysed in 1 ml of buffer B (100 mM KCl, 0.5 mM EDTA, 20 mM HEPES-KOH [pH 7.6], 0.4% Nonidet P-40, 10% glycerol, 1 mM phenylemthylsulfonyl fluoride). Following centrifugation, the supernatant was mixed for 5 h at 4°C with 2 μg of anti-HA antibody immobilized on protein G-Sepharose resin. After the mixture was washed with buffer B (0.4 ml three times), immunoprecipitates were collected by centrifugation and proteins were eluted with Laemmli SDS sample buffer and subjected to SDS-PAGE.
In vitro translation.
Translation reactions were performed with rabbit reticulocyte lysate (Promega) in a final volume of 15 μl as follows: lysate (9 μl) was preincubated for 5 min at 30°C with 20 U of RNasin (0.5 μl; Promega), a complete amino acid mixture (1 mM each amino acid; 0.5 μl), and eIF4AIII or eIF4AI (0 to 8 μg) in buffer containing 20 mM HEPES (pH 7.5), 100 mM KCl, 0.1 mM EDTA, 1 mM DTT, and 10% glycerol (4 μl). Uncapped luciferase mRNA (100 ng; 1 μl [Promega]) was added, and incubation was continued for an additional 30 min at 30°C. Luciferase activity was measured with a bioluminometer (BIOORBIT). The value obtained in the absence of added eIF4A was set at 100%.
Ribosomal binding assay.
The ribosomal binding experiments were carried out by the method of Pestova et al. (40), except that eIF4F was replaced with a combination of recombinant eIF4E, eIF4A, and eIF4GI (31). Reaction mixtures (40 μl) containing β-globin mRNA (0.3 μg), His-eIF1 (0.5 μg), His-eIF1A (0.5 μg), eIF2 (3 μg), eIF3 (7 μg), FLAG-eIF4B (1 μg), Met-tRNAiMet (6 pmol), 40S ribosomal subunits (6 pmol), His-eIF4GI (2 μg), His-eIF4E (0.3 μg), and either His-eIF4AI (3 μg) or His-eIF4AIII (3 μg) were incubated for 5 min at 30°C. The reaction mixtures were then subjected to a reverse transcriptase reaction in the presence of [32P]dATP by using a primer preannealed to β-globin mRNA (40). The same primer was used for sequencing a plasmid harboring β-globin cDNA. The primer extension and sequence products were resolved side by side on a sequencing gel.
RESULTS
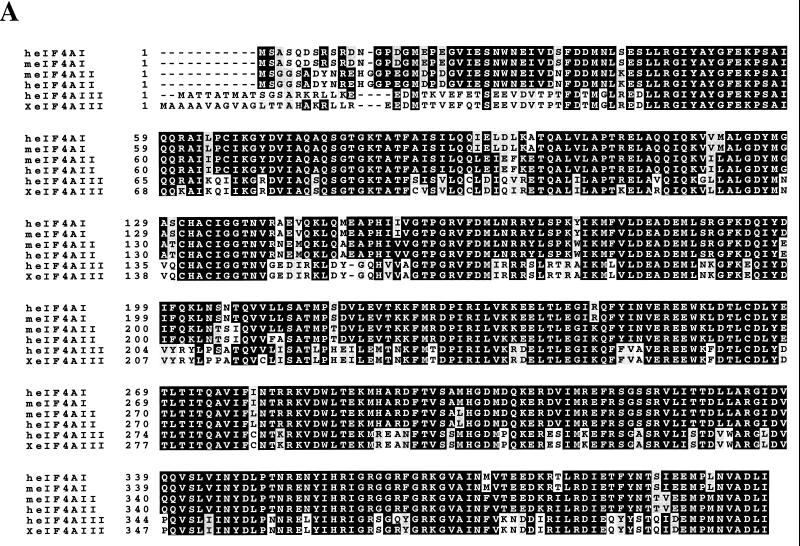
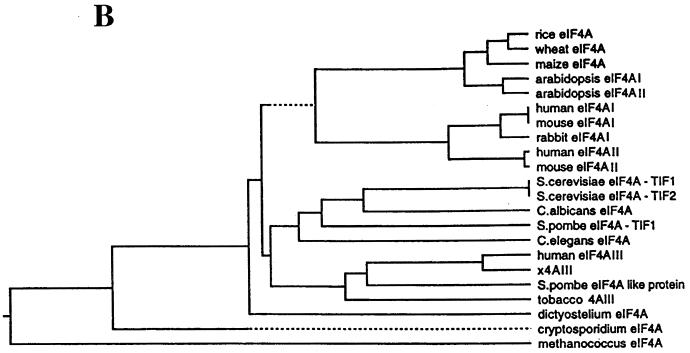
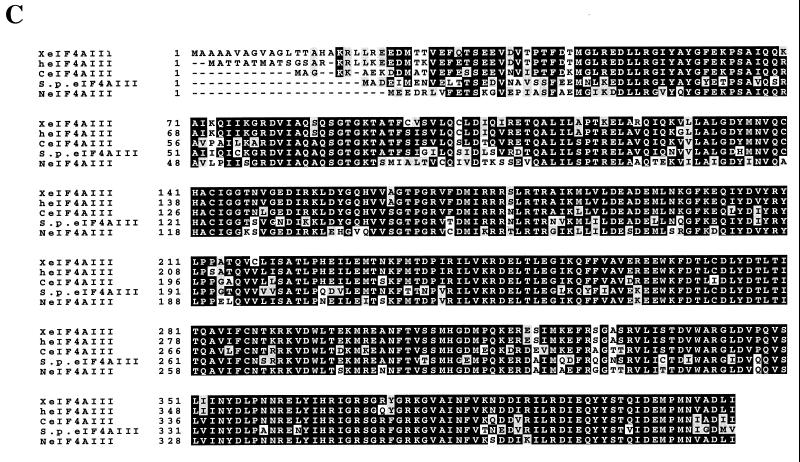
A cDNA clone (Nuk-34), isolated from an undifferentiated human keratinocyte cDNA library (a kind gift from Henrik Leffers), encodes a protein (411 aa) exhibiting high homology at the amino acid level to eIF4AI and eIF4AII (67 and 68% identity, respectively [Fig. 1A]), which is referred to as human eIF4AIII. A phylogenetic tree of eIF4A proteins (Fig. 1B) shows that Xenopus eIF4AIII is the closest homolog of human eIF4AIII (93% identity [Fig. 1C]), followed by the Schizosaccharomyces pombe eIF4A-like protein (75.3% identity) and tobacco eIF4AIII (74.5% identity). A striking feature of the eIF4AIII proteins is its strong phylogenetic conservation (75.3% identity and 88.5% homology between human and S. pombe). eIF4AIII is different from eIF4AI primarily in the N-terminal region, which diverged significantly from the other two eIF4A RNA helicases. When the N-terminal 40 aa of eIF4AIII are not taken into consideration, eIF4AIII is 73% identical to human eIF4AI.
FIG. 1.
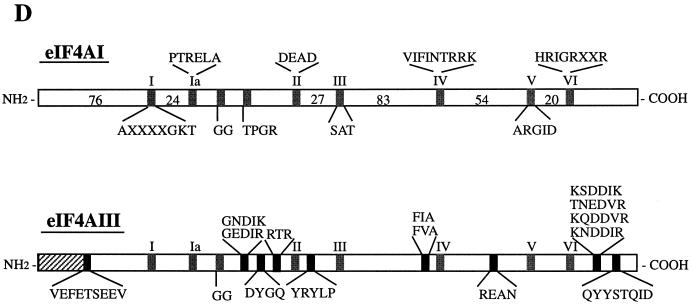
(A) Protein sequence alignment of eIF4A proteins. The pattern-induced multisequence alignment program was used to align the amino acid sequences of human (h), mouse (m), and Xenopus (X) eIF4A proteins. Identical and conserved amino acid residues are in black and shaded boxes, respectively. (B) Phylogenetic relationship among members of the eIF4A gene family based on aligned amino acid sequences, which was created by the PILEUP program. (C) Sequence alignment of human (h), Xenopus (X), S. pombe (S.p.), C. elegans (C), and Nicotiana plumbaginifolia (N) eIF4AIII. The accession numbers are as follows: heIF4AI (P04765), meIF4AI (S00986), heIF4AII (D30655), meIF4AII (S00985), XeIF4AIII (AAB71410), S.P.eIF4AIII (CAA92238), CeIF4AIII (AAB96704), and NeIF4AIII (P41380). (D) Amino acid motifs which are conserved in the eIF4A family and which are specific for eIF4AIII. The N-terminal divergent sequence in eIF4AIII is hatched.
eIF4AIII possesses all of the conserved motifs of the DEAD box protein family (Fig. 1D). The biochemical functions of DEAD box proteins can be attributed to these conserved motifs (50). Motif I (AXXGXGKT) is common to both DNA and RNA helicases and is responsible for ATP binding (49, 56); motif II (DEAD) is involved in ATP hydrolysis and couples ATP hydrolysis to helicase activity; motif III (SAT) is critical for RNA unwinding. Motif VI (HRIGRXXR) is unique to RNA helicases and is involved in the ATP hydrolysis-dependent RNA binding of eIF4A (37). eIF4AIIIs contain approximately nine short conserved sequences which eIF4AI and eIF4AII lack. They are dispersed across the entire length of the molecule among the conserved domains of the DEAD box family (Fig. 1D). For instance, instead of RA(E/D)VQ in eIF4AI, there is GEDIR in eIF4AIII.
Expression of eIF4AIII.
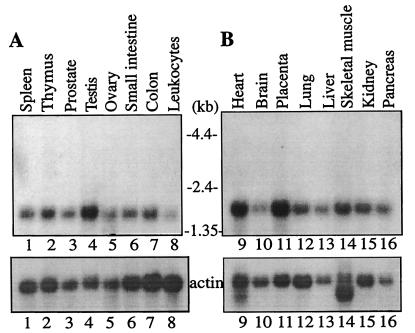
While eIF4AII mRNA levels vary among tissues, levels of eIF4AI mRNA are relatively uniform in all tissues examined (32, 53). The human tissue expression of eIF4AIII mRNA was examined by using a probe derived from the 3′ untranslated region, which exhibits no homology to human eIF4AI and eIF4AII. A single hybridizing band (1.5 kb) was observed in all tissues examined, demonstrating that eIF4AIII mRNA is ubiquitously expressed (Fig. 2). Relatively higher expression was observed in testis, heart, and placenta.
FIG. 2.
Northern blot analysis of human eIF4AIII mRNA expression in human tissues. A multiple-tissue Northern blot, MTN1 (A) and MTN2 (B) (CLONTECH), containing 2 μg of poly(A)+ RNA per lane from the indicated human tissues was hybridized with a 32P-labeled human eIF4AIII (Nuk34) cDNA probe (a 0.3-kb fragment from the 3′ untranslated sequence), as specified by the manufacturer. The filters were also hybridized with a [32P]-labeled actin probe. Taken from reference 43.
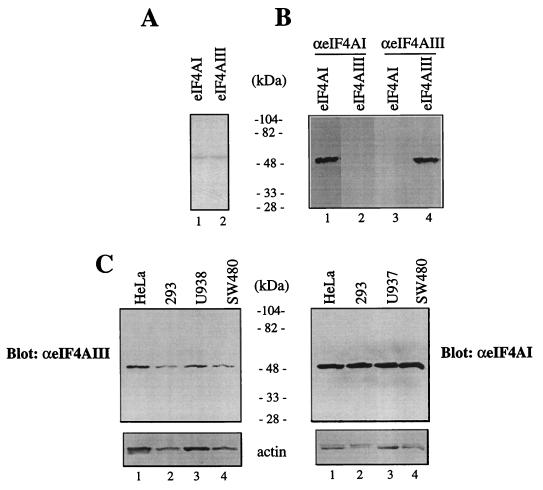
To determine the expression of eIF4AIII protein, a polyclonal antiserum was raised against a synthetic peptide corresponding to the N-terminal region of human eIF4AIII (aa 8 to 22), which exhibits no sequence similarity to eIF4AI or eIF4AII. Recombinant eIF4AI and eIF4AIII (Fig. 3A; Coomassie blue stain) were used to assess the specificity of the antibody. The anti-eIF4AIII serum is specific, since it detected recombinant eIF4AIII (Fig. 3B, lane 4) but not eIF4AI (lane 3), while the anti-eIF4AI antibody recognized recombinant eIF4AI (lane 1) but not eIF4AIII (lane 2). These results validate the use of the anti-eIF4AIII antiserum for quantitation of eIF4AIII. Extracts from four different cell lines were analyzed for eIF4AIII expression by Western blot analysis. eIF4AIII is present in approximately equal amounts in the cell lines examined (Fig. 3C, top).
FIG. 3.
Levels of eIF4AIII protein. (A) Coomassie blue staining. Recombinant eIF4AI and eIF4AIII (1 μg) were resolved by SDS-PAGE (10% polyacrylamide). Molecular masses (in kilodaltons) of protein standards are indicated. (B) Immunological identification of eIF4AI and eIF4AIII. Recombinant eIF4AI (1 μg; lanes 1 and 3) and eIF4AIII (1 μg; lanes 2 and 4) were subjected to SDS-PAGE (10% polyacrylamide), and proteins were transferred onto a nitrocellulose membrane, which was probed with anti-eIF4AI (lanes 1 and 2) or anti-eIF4AIII (lanes 3 and 4) antibodies. Protein bands were visualized on an X-ray film by an enhanced chemiluminescence detection system. (C) Cell extracts (10 μg) were resolved by SDS-PAGE (10% polyacrylamide). Western blotting was performed with anti-eIF4AI or anti-eIF4AIII. The same membrane was reprobed with anti-actin antibody (bottom).
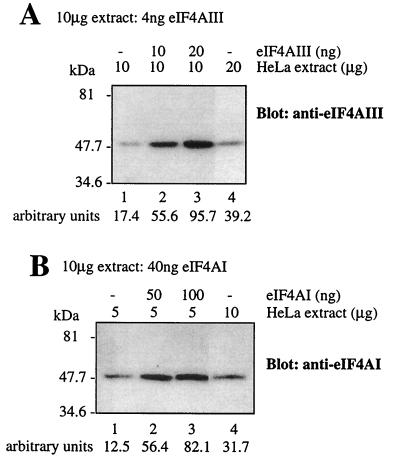
eIF4AI is the most abundant translation initiation factor. The factor/ribosome ratios of eIF4G, eIF4A, and eIF4E in HeLa cells are 0.52, 2.96, and 0.26, respectively (8). The proteins account for 0.33, 0.38, and 0.02% of the total cytoplasmic protein, respectively (8). The amount of eIF4AIII and eIF4AI in a cytoplasmic extract of HeLa cells was determined with recombinant proteins as standards in Western blotting. It was calculated that 10 μg of the cytoplasmic extract contains 4 ng of eIF4AIII and 40 ng of eIF4AI (Fig. 4). The concentration of eIF4AI obtained here was similar to that reported by Duncan et al. (8). eIF4AIII and eIF4AI were estimated to account for 0.04 and 0.4% of the total cytoplasmic protein, respectively. Thus, eIF4AIII is present at 1/10 of the concentration of eIF4AI in HeLa cells.
FIG. 4.
Quantitation of endogenous eIF4AIII and eIF4AI in HeLa cells. The indicated amounts of cell extract and amounts of eIF4AIII (A) or eIF4AI (B) were mixed and resolved by SDS-PAGE (10% polyacrylamide). Proteins were electroblotted onto a nitrocellulose membrane and probed with a polyclonal anti-eIF4AIII (A) or monoclonal eIF4AI (B) antibody. Signals were quantified on a phosphorimager, and the values obtained are indicated at the bottom of the figure.
eIF4AIII exhibits RNA-dependent ATPase activity and ATP-dependent helicase activity.
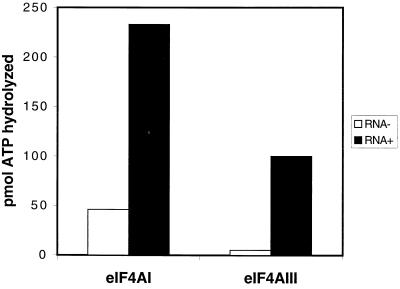
Next, we wished to study the function of eIF4AIII. An ATPase assay was performed in the presence of poly(A). The reaction involving eIF4AIII and no poly(A) showed a low level of ATPase activity (5 pmol of ATP hydrolyzed per reaction), while addition of poly(A) increased the ATPase activity 20-fold (Fig. 5; this is representative of two independent experiments with less than 10% variation). eIF4AI showed higher background ATPase activity than did eIF4AIII without RNA, and a fivefold increase in the presence of RNA. Thus, eIF4AIII possesses RNA-dependent ATPase activity.
FIG. 5.
eIF4AIII possesses an RNA-stimulated ATPase activity. ATPase assays were performed for 15 min with 100 μM ATP and 1 μg of recombinant eIF4AIII or recombinant eIF4AI in the absence or presence of 0.3 absorbance at 260 nm units of poly(A) RNA. The amount of inorganic phosphate released from a reaction with no eIF4A and no poly(A) RNA was subtracted. The values are the mean of three independent experiments.
To examine the helicase activity of eIF4AIII, an RNA helicase assay was performed with a 32P-labeled RNA substrate containing a 12-bp duplex with a 34-nucleotide 5′ single-strand extension (47) (Fig. 6A, top). The RNA migrated as a duplex in a nondenaturing polyacrylamide gel (Fig. 6A, lane 1). Heating the duplex RNA to 95°C caused its melting to generate single-stranded RNA (compare lanes 1 and 2). To examine factor-mediated unwinding, the RNA duplex was incubated with highly purified, nuclease-free reticulocyte eIF4AI or recombinant eIF4AIII. As previously demonstrated with this RNA duplex (47), eIF4AI exhibited significant ATP-dependent helicase activity (37% monomer; compare lanes 3 and 4), which was stimulated by the addition of eIF4B (84% monomer; compare lanes 4 and 8). Recombinant eIF4AIII also unwound the duplex to a significant extent in an ATP-dependent manner (22% monomer; compare lanes 9 and 10), and the helicase activity of eIF4AIII was stimulated by the addition of eIF4B (62% monomer; lanes 10 and 11). eIF4AIII in combination with eIF4B unwound the duplex RNA in a dose-dependent manner (Fig. 6B, lanes 4 to 7). Taken together, these data demonstrate that eIF4AIII possesses RNA-dependent ATPase and ATP-dependent RNA helicase activity which can be enhanced by eIF4B.
eIF4AIII fails to substitute for eIF4AI in a ribosome binding assay and inhibits translation.
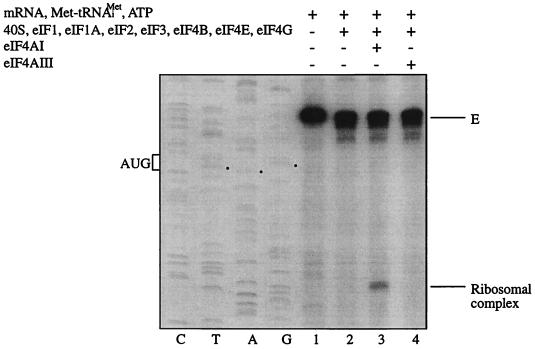
In light of the ATPase and helicase activities exhibited by eIF4AIII, it was pertinent to determine whether eIF4AIII could play a role similar to eIF4AI in translation. A ribosome binding-toeprinting assay was performed with purified translation factors and β-globin mRNA (40, 42). In the absence of translation factors (Fig. 7, lane 1) or eIF4AI (lane 2), there was no 48S ribosomal complex formation. Addition of eIF4AI resulted in formation of a 48S ribosomal complex, as evidenced by the toeprint band obtained (lane 3). However, no ribosomal complex formation was observed when eIF4AI was replaced by eIF4AIII (lane 4). This indicates that unlike eIF4AI, eIF4AIII cannot facilitate ribosome binding to β-globin mRNA, in spite of its activity as an RNA-dependent ATPase and an ATP-dependent RNA helicase.
FIG. 7.
eIF4AIII does not substitute for eIF4AI in the ribosome binding assay. The indicated translation components were incubated with β-globin mRNA. Formation of the 48S ribosomal complex was detected by toeprinting. Full-length cDNA is marked E. The cDNA product that maps to 15 to 17 nucleotides downstream of the initiation codon of β-globin mRNA is labeled Ribosomal complex. The position of the initiation codon is shown to the left of the reference lanes, which represent the β-globin sequence obtained with the same primer as for the toeprinting.
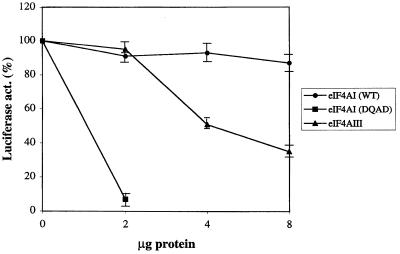
The finding that eIF4AIII could not replace eIF4AI in the ribosome binding assay raised the possibility that eIF4AIII functions as an inhibitor of translation. To address this, the effect of eIF4AIII on translation of luciferase mRNA in a reticulocyte lysate was examined (Fig. 8). Addition of wild-type eIF4AI had no significant effect on translation, whereas addition of 2 μg of a dominant negative mutant of eIF4AI (DEAD to DQAD) inhibited translation by 90%, as reported previously (38). Addition of 4 and 8 μg of eIF4AIII decreased translation by 50 and 65%, respectively, suggesting that eIF4AIII is a negative regulator of translation.
FIG. 8.
eIF4AIII inhibits translation. Increasing amounts of recombinant eIF4AI (2 to 8 μg), eIF4AI mutant (DQAD) (2 μg) (28), or eIF4AIII (2 to 8 μg) were preincubated in a rabbit reticulocyte lysate for 5 min at 30°C. Uncapped uciferase mRNA was added to the lysate, and translation was carried out at 37°C for 30 min. The luciferase activity of the lysate preincubated with buffer alone was set at 100%. Each point represents the mean of four experiments. WT, wild type.
eIF4AIII possesses only one binding site on eIF4G.
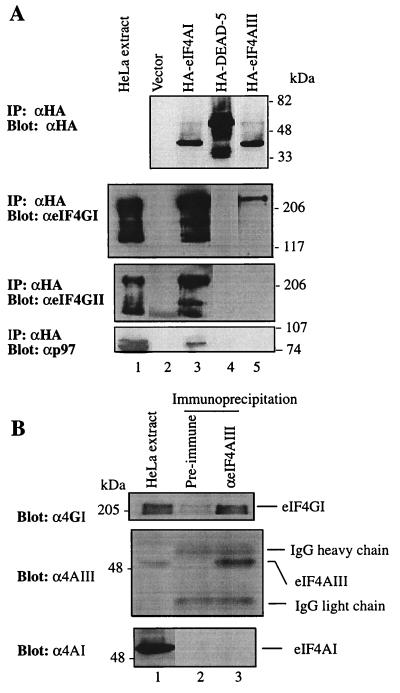
To investigate why eIF4AIII inhibits rather than stimulates translation, the binding of eIF4AIII to eIF4G was examined, since eIF4AI functions in translation through its binding to eIF4GI (10). Hemagglutinin (HA)-tagged eIF4AIII and eIF4AI were expressed in HeLa cells, and extracts were immunoprecipitated with anti-HA antibody. The immunoprecipitates were assayed by Western blotting for the presence of eIF4GI, eIF4GII (12), and p97, a homolog of eIF4G which also binds eIF4AI (19). An unrelated DEAD-box protein, DEAD-5 (9), whose yeast homolog functions in nucleocytoplasmic mRNA transport (51, 55), was used as a negative control. HA-tagged eIF4AI and eIF4AIII were expressed to approximately similar levels (Fig. 9A, top). eIF4AIII was coimmunoprecipitated with eIF4GI, but much less efficiently than was eIF4AI (second panel from top, compare lanes 2 and 4). eIF4GI did not coimmunoprecipitate with the negative control DEAD-5 (second panel, lane 3). No binding of eIF4AIII to eIF4AII or p97 was detected (bottom two panels, lane 4). Because the signal for eIF4GII and p97 was weaker than that of eIF4GI, it is likely that the interaction between eIF4GII and p97 with eIF4AIII was not sufficiently strong to be detected.
FIG. 9.
eIF4AIII interacts with eIF4G. (A). Expression plasmids pcDNA3-HA, pcDNA3-HA-eIF4AI, pcDNA3-HA-DEAD5, and pcDNA3-HA-eIF4AIII were transfected into HeLa cells after infection with vTF7-3, as described in Materials and Methods. Immunoprecipitates (IP) obtained with anti-HA antibody (αHA) were resolved by SDS-PAGE (10% polyacrylamide). Western blotting was performed with anti-HA (top), anti-eIF4GI (αeIF4GI) (second panel), anti-eIF4GII (αeIF4GII) (third panel), or anti-p97 antibody (αp97) (bottom). (B) HeLa cell extract (168 μg) was incubated with preimmune serum or anti-eIF4AIII serum. Following precipitation with protein G-Sepharose, bound proteins were resolved by SDS-PAGE (10% polyacrylamide). Western blotting was performed with anti-eIF4GI (top), anti-eIF4AIII (middle), or anti-eIF4AI antibody (bottom). HeLa cell extract (12 μg) was used for Western blotting (lane 1). IgG, immunoglobulin G.
To examine whether endogenous eIF4AIII is also associated with eIF4GI, HeLa extracts were immunoprecipitated with anti-eIF4AIII serum. The immunoprecipitates were subjected to Western blotting with an anti-eIF4GI, anti-eIF4AIII, or anti-eIF4AI antibody. eIF4GI coimmunoprecipitated with eIF4AIII, while preimmune serum failed to precipitate either protein (Fig. 9B, top). Thus, eIF4AIII interacts with eIF4GI in vivo. eIF4AI did not coprecipitate with eIF4AIII (bottom, lane 3), suggesting that eIF4AI and eIF4AIII do not bind simultaneously to the same molecule of eIF4GI.
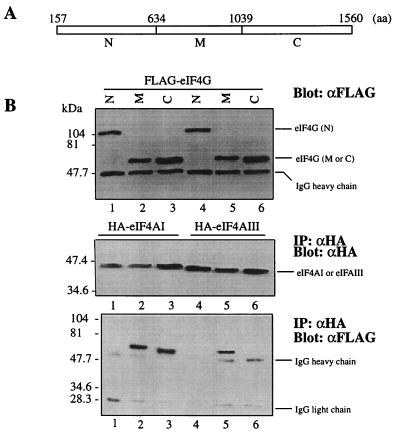
Human eIF4GI possesses two separate and independent binding sites for eIF4AI: one is located in the middle third of the protein (aa 634 to 1039), and the other is in the C-terminal third (aa 1040 to 1560) (20). The interaction of eIF4AIII with the eIF4GI middle and C-terminal regions was investigated by immunoprecipitation. HA-tagged eIF4AIII and FLAG-tagged N-terminal, middle or C-terminal regions of eIF4GI were coexpressed in HeLa cells. Immunoprecipitates obtained with an anti-HA antibody were analyzed by Western blotting with anti-FLAG and anti-HA antibodies. All of the proteins generated from the transfected plasmids were expressed at similar levels (Fig. 10B, top and middle). The migration of the N-terminal fragment is extremely anomalous, since its predicted molecular mass is 53 kDa but it migrates as a ∼105 kDa polypeptide (top, lanes 1 and 4). This anomalous migration has been observed in numerous previous studies, where an N-terminally cleaved product from poliovirus-infected cells migrated much slower than expected (see, for example, reference 16). The reason for the anomalous migration is most probably the presence of clusters of glutamines and prolines in the N-terminal region. As expected, eIF4AI bound to both the eIF4GI middle and C-terminal regions (20) (Fig. 10B, bottom, lanes 2 and 3). In contrast, eIF4AIII coprecipitated with the middle region of eIF4GI (bottom, lane 5), but failed to coprecipitate the C-terminal region of eIF4GI (bottom, lane 6). Neither eIF4AI nor eIF4AIII coprecipitated with the N-terminal domain (aa 157 to 613) of eIF4GI (bottom, lanes 1 and 4; an N-terminal fragment including the newly found sequence [aa 1 to 156] [18] was not used, because it was not well expressed). Taken together, these experiments show that eIF4AIII is remarkably different from eIF4AI in its eIF4GI binding properties.
FIG. 10.
eIF4AIII binds only to the middle portion of eIF4GI. (A) Schematic map of the three fragments of eIF4GI. (B) FLAG-eIF4GI N-terminal region (N) (lanes 1 and 4), middle region (M) (lanes 2 and 5), and C-terminal region (C) (lanes 3 and 6) expression plasmids were cotransfected into HeLa cells with pcDNA3-HA-eIF4AI (lanes 1 to 3) or pcDNA3-HA-eIF4AIII (lanes 4 to 6). An aliquot of the extract was removed for Western blotting with anti-FLAG (top). The remaining portion was used for immunoprecipitation (IP) with anti-HA antibody (αHA). Western blotting of immunoprecipitates was performed with anti-HA (αHA) (middle) or anti-FLAG (αFLAG) (bottom) antibody.
DISCUSSION
In this study, a third member of the eIF4A family, eIF4AIII, was characterized biochemically. eIF4AIII is the second closest relative of eIF4AI, after eIF4AII. eIF4AIII shares the same core region as eIF4AI and eIF4AII with members of the DEAD-box helicase family. However, while eIF4AII is 91% identical to eIF4AI at the amino acid level, eIF4AIII is only 65% identical to eIF4AI. The major differences reside in the unique N-terminal 30-aa region of eIF4AIII and in nine short sequence segments that are characteristic of the eIF4AIII homologs in different species (Fig. 1D). Consistent with the similarity between eIF4AIII and eIF4AI, eIF4AIII exhibits biochemical activities that had been demonstrated for eIF4AI: RNA-dependent ATPase, ATP-dependent helicase which is stimulated by eIF4B, and binding to eIF4G. Based on these properties, it is likely that eIF4AIII serves some function related to translation. Although the similarities between eIF4AI and eIF4AIII would suggest a functional redundancy, eIF4AIII fails to substitute for eIF4AI in a ribosome binding assay and inhibits translation in an in vitro reticulocyte lysate translation system.
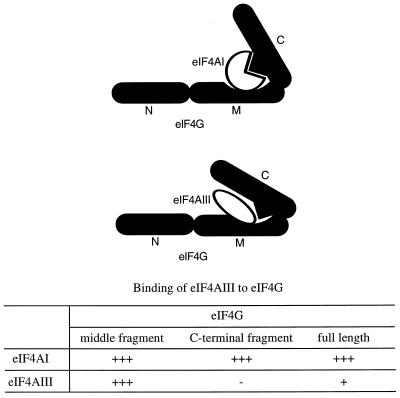
The mechanism underlying the translational inhibition by eIF4AIII might be explained by the finding that eIF4AIII binds eIF4GI only through the middle domain of eIF4G (Fig. 11). In spite of the robust binding of eIF4AIII to the middle fragment of eIF4G, its binding to the full-length eIF4GI is poor (Fig. 9 and 10). Although the C-terminal domain, which harbors another binding site for eIF4AI, is not essential for translation (31, 42), a point mutation in eIF4GI which abolishes the C-terminal eIF4A binding inhibited eIF4GI activity, even in the presence of excess eIF4AI (31). Thus, it is likely that for mammalian eIF4AI to function efficiently, binding of eIF4AI to the C-terminal domain, when present in eIF4G, is important. One plausible model is that the C-terminal third of eIF4GI folds over the middle domain to inhibit eIF4G function. The model in Fig. 11 depicts a scenario by which eIF4AI, through its simultaneous binding to the middle and carboxy regions of eIF4G, would cause a change in the conformation of eIF4G to alleviate inhibition. However, it should be emphasized that other models are possible, since two molecules of eIF4AI might bind to one molecule of eIF4G.
FIG. 11.
Model of binding of eIF4AI or eIF4AIII to eIF4GI. While eIF4AI binds to the middle and C-terminal regions of eIF4GI, eIF4AIII binds only to the middle region. It should be noted that there is a distinct possibility that two molecules of eIF4AI bind to one molecule of eIF4G. N, N-terminal fragment; M, middle fragment; C, C-terminal fragment.
Several natural proteinaceous inhibitors of translation initiation have been described. p97 is homologous to the C-terminal two-thirds of eIF4G and functions as a repressor of translation by forming translationally inactive complexes that include eIF4A and eIF3 but exclude eIF4E (19). The eIF4E binding proteins (4E-BPs) bind to eIF4E and prevent its association with eIF4G, because the 4E-BPs and eIF4G have a common site for eIF4E binding (15, 27). eIF4AIII may participate in a third novel inhibition pathway. It is possible that the different repressors have different mRNA specificities and thus affect disparate physiological responses. Upon treatment of cells with insulin and growth factors, 4E-BPs become phosphorylated. This leads to the dissociation of 4E-BPs from eIF4E and subsequent formation of the eIF4F complex and stimulation of translation (4, 23, 36). It remains to be determined whether the activity of eIF4AIII is modulated under different circumstances.
When and where does eIF4AIII function? The reticulocyte lysate, which was used in the in vitro translation experiments (9 μl) (Fig. 8), contains 1.5 μg of endogenous eIF4AI (38). Addition of eIF4AI (2 to 8 μg) did not affect translation, suggesting that the system is already saturated with eIF4AI. However, addition of eIF4AIII (4 μg) decreased translation by 50%. Thus, the amount of eIF4AIII which is required to inhibit translation significantly exceeds that of eIF4AI. This inhibition may not occur in HeLa cells, because the concentration of eIF4AIII with respect to eIF4AI is only 1/10 (Fig. 4). However, several reports suggest that eIF4AI exists in a limiting amount in embryonic cells (30, 57), and injection of purified eIF4AI into Xenopus oocytes stimulated translation (2). Overexpression of eIF4AII, which is functionally interchangeable with eIF4AI, in Xenopus animal caps induced neural fold genes (30). In fact, eIF4AII is expressed specifically in the prospective neurectoderm from stage 11.5 of the Xenopus embryo (30). Interestingly, eIF4AIII transcripts are elevated in Xenopus cells cultured in the presence of BMP-4, an epidermis inducer, and are also elevated in the ventral ectoderm (57). Overexpression of eIF4AIII inhibits neuralization and induces epidermis (57). It remains to be determined whether different mRNAs are translationally affected to the same extent by eIF4AIII.
A comparison between eIF4AI and eIF4AIII sequences should be valuable in mapping the eIF4G binding sites on eIF4A and determining their importance for translation. eIF4AI binds to eIF4G efficiently through both the middle and C-terminal regions of eIF4G, whereas eIF4AIII binds only to the middle region of eIF4G and thus binds to full-length eIF4G weakly. The functionally unrelated DEAD-box protein, DEAD-5 (9, 51, 55), does not bind to eIF4G. It is thus likely that the amino acids conserved between eIF4AI and eIF4AIII but not in DEAD-5 are involved in the binding to the middle region of eIF4G, while an eIF4AI-specific sequence is important in the binding to the C-terminal region of eIF4G and is not represented in eIF4AIII. Because the major sequence differences between eIF4AI and eIF4AIII are at the N terminus and might be involved in their specific biological function, swapping experiments of the N-terminal sequences between eIF4AI and eIF4AIII should be helpful in determining the importance of the N-terminal region.
Although it is likely that eIF4AIII functions in translation, it is possible that eIF4AIII has a function in some other pathway. The RNA helicases of the DEAD-box protein family play key roles in disparate aspects of RNA metabolism, including pre-mRNA splicing, rRNA processing, mRNA turnover, mRNA transport, and ribosome assembly, as well as in oogenesis, spermatogenesis, cell growth, and division (26).
Finally, the interesting possibility that eIF4AIII functions to stimulate rather than inhibit translation under certain circumstances cannot be ruled out. For example, during apoptosis eIF4G is cleaved into three fragments, resulting in the separation of the middle and carboxy regions (7, 28). This could then result in the efficient binding of eIF4AIII to the eIF4G middle fragment and subsequent enhancement of translation of mRNAs harboring an internal ribosomal entry site (IRES), because the middle fragment of eIF4GI is sufficient to mediate IRES-dependent translation (42). Interestingly, IRES-dependent translation of XIAP mRNA encoding an inhibitor of apoptosis has been recently detected during apoptosis (17). Thus, eIF4AIII could potentially play a positive role in translation initiation of XIAP mRNA or other IRES-dependent mRNAs during apoptosis.
ACKNOWLEDGMENTS
We thank Henrik Leffers for human eIF4AIII (Nuk-34) cDNA, D. C. Weinstein and A. Hemmati-Brivanlou for Xenopus eIF4AIII, Colin Lister for help with the phylogenetic tree program, Hans Trachsel for antibody against eIF4AI, Alessandra Gradi for anti-eIF4GII antibody, Francis Poulin for the actin Northern blot in Fig. 2, and Anne-Claude Gingras and Brian Raught for critical reading of the manuscript.
This work was supported by a grant from the Medical Research Council of Canada to N.S. N.S. is a Medical Research Council of Canada distinguished scientist and a Howard Hughes Medical Institute International Scholar.
REFERENCES
- 1.Allen M L, Metz A M, Timmer A T, Rhoads R E, Browning K S. Isolation and sequence of the cDNAs encoding the subunits of the isozyme form of wheat protein synthesis initiation factor 4F. J Biol Chem. 1992;267:23232–23236. [PubMed] [Google Scholar]
- 2.Audet R G, Goodchild J, Richter J D. Eukaryotic initiation factor 4A stimulates translation in microinjected Xenopus oocyte. Dev Biol. 1987;121:58–68. doi: 10.1016/0012-1606(87)90138-2. [DOI] [PubMed] [Google Scholar]
- 3.Belsham G J, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beretta L, Gingras A C, Svitkin Y V, Hall M N, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 5.Blum S, Schmid S R, Pause A, Buser P, Linder P, Sonenberg N, Trachsel H. ATP hydrolysis by initiation factor 4A is required for translation initiation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1992;89:7664–7668. doi: 10.1073/pnas.89.16.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browning K S, Webster C, Roberts J K, Ravel J M. Identification of an isozyme form of protein synthesis initiation factor 4F in plants. J Biol Chem. 1992;267:10096–10100. [PubMed] [Google Scholar]
- 7.Clemens M J, Bushell M, Morley S J. Degradation of eukaryotic polypeptide chain initiation factor (eIF) 4G in response to induction of apoptosis in human lymphoma cell lines. Oncogene. 1998;17:2921–2931. doi: 10.1038/sj.onc.1202227. [DOI] [PubMed] [Google Scholar]
- 8.Duncan R, Milburn S C, Hershey J W. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem. 1987;262:380–388. [PubMed] [Google Scholar]
- 9.Gee S L, Conboy J G. Mouse erythroid cells express multiple putative RNA helicase genes exhibiting high sequence conservation from yeast to mammals. Gene. 1994;140:171–177. doi: 10.1016/0378-1119(94)90541-x. [DOI] [PubMed] [Google Scholar]
- 10.Gingras A-C, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 11.Goyer C, Altmann M, Lee H S, Blanc A, Deshmukh M, Woolford J L, Jr, Trachsel H, Sonenberg N. TIF4631 and TIF4632: two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol Cell Biol. 1993;13:4860–4874. doi: 10.1128/mcb.13.8.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gradi A, Svitkin Y V, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grifo J A, Tahara S M, Morgan M A, Shatkin A J, Merrick W C. New initiation factor activity required for globin mRNA translation. J Biol Chem. 1983;258:5804–5810. [PubMed] [Google Scholar]
- 15.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haghighat A, Svitkin Y, Novoa I, Kuechler E, Skern T, Sonenberg N. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J Virol. 1996;70:8444–8450. doi: 10.1128/jvi.70.12.8444-8450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk R G. A novel internal ribosome entry site (IRES) motif potentiates XIAP mediated cytoprotection under cellular stress. Nat Cell Biol. 1999;1:190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- 18.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imataka H, Olsen H S, Sonenberg N. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 1997;16:817–825. doi: 10.1093/emboj/16.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson R J, Howell M T, Kaminski A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem Sci. 1990;15:477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- 22.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 23.Lin T A, Kong X, Haystead T A, Pause A, Belsham G, Sonenberg N, Lawrence J C., Jr PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 24.Linder P, Lasko P F, Ashburner M, Leroy P, Nielsen P J, Nishi K, Schnier J, Slonimski P P. Birth of the D-E-A-D box. Nature. 1989;337:121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- 25.Linder P, Slonimski P P. Sequence of the genes TIF1 and TIF2 from Saccharomyces cerevisiae coding for a translation initiation factor. Nucleic Acids Res. 1988;16:10359. doi: 10.1093/nar/16.21.10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luking A, Stahl U, Schmidt U. The protein family of RNA helicases. Crit Rev Biochem Mol Biol. 1998;33:259–296. doi: 10.1080/10409239891204233. [DOI] [PubMed] [Google Scholar]
- 27.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marissen W E, Lloyd R E. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol Cell Biol. 1998;18:7565–7574. doi: 10.1128/mcb.18.12.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merrick W C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 31–70. [Google Scholar]
- 30.Morgan R, Sargent M G. The role in neural patterning of translation initiation factor eIF4AII; induction of neural fold genes. Development. 1997;124:2751–2760. doi: 10.1242/dev.124.14.2751. [DOI] [PubMed] [Google Scholar]
- 31.Morino, S., H. Imataka, Y. V. Svitkin, T. V. Pestova, and N. Sonenberg. Unpublished data. [DOI] [PMC free article] [PubMed]
- 32.Nielsen P J, McMaster G K, Trachsel H. Cloning of eukaryotic protein synthesis initiation factor genes: isolation and characterization of cDNA clones encoding factor eIF-4A. Nucleic Acids Res. 1985;13:6867–6880. doi: 10.1093/nar/13.19.6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen P J, Trachsel H. The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. EMBO J. 1988;7:2097–2105. doi: 10.1002/j.1460-2075.1988.tb03049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owttrim G W, Hofmann S, Kuhlemeier C. Divergent genes for translation initiation factor eIF-4A are coordinately expressed in tobacco. Nucleic Acids Res. 1991;19:5491–5496. doi: 10.1093/nar/19.20.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pain V M. Initiation of protein synthesis in eukaryotic cells. Eur J Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 36.Pause A, Belsham G J, Gingras A C, Donze O, Lin T A, Lawrence J C, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 37.Pause A, Methot N, Sonenberg N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol Cell Biol. 1993;13:6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pause A, Methot N, Svitkin Y, Merrick W C, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pestova T V, Borukhov S I, Hellen C U. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 41.Pestova T V, Hellen C U, Shatsky I N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pestova T V, Shatsky I N, Hellen C U. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poulin F, Gingras A C, Olsen H, Chevalier S, Sonenberg N. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J Biol Chem. 1998;273:14002–14007. doi: 10.1074/jbc.273.22.14002. [DOI] [PubMed] [Google Scholar]
- 44.Pyronnet S, Imataka H, Gingras A C, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ray B K, Lawson T G, Kramer J C, Cladaras M H, Grifo J A, Abramson R D, Merrick W C, Thach R E. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985;260:7651–7658. [PubMed] [Google Scholar]
- 46.Richter-Cook N J, Dever T E, Hensold J O, Merrick W C. Purification and characterization of a new eukaryotic protein translation factor. Eukaryotic initiation factor 4H. J Biol Chem. 1998;273:7579–7587. doi: 10.1074/jbc.273.13.7579. [DOI] [PubMed] [Google Scholar]
- 47.Rogers G W, Richter-Cook N J, Merrick W C. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 48.Rozen F, Edery I, Meerovitch K, Dever T E, Merrick W C, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saraste M, Sibbald P R, Wittinghofer A. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 50.Schmid S R, Linder P. D-E-A-D protein family of putative RNA helicases. Mol Microbiol. 1992;6:283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 51.Snay-Hodge C A, Colot H V, Goldstein A L, Cole C N. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sonenberg N. mRNA 5′ cap-binding protein eIF4E and control of cell growth. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 271–294. [Google Scholar]
- 53.Sudo K, Takahashi E, Nakamura Y. Isolation and mapping of the human eIF4A2 gene homologous to the murine protein synthesis initiation factor 4A-II gene eIF4A2. Cytogenet Cell Genet. 1995;71:385–388. doi: 10.1159/000134145. [DOI] [PubMed] [Google Scholar]
- 54.Svitkin Y V, Gradi A, Imataka H, Morino S, Sonenberg N. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J Virol. 1999;73:3467–3472. doi: 10.1128/jvi.73.4.3467-3472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tseng S S, Weaver P L, Liu Y, Hitomi M, Tartakoff A M, Chang T H. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 1998;17:2651–2662. doi: 10.1093/emboj/17.9.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinstein D C, Honore E, Hemmati-Brivanlou A. Epidermal induction and inhibition of neural fate by translation initiation factor 4AIII. Development. 1997;124:4235–4242. doi: 10.1242/dev.124.21.4235. [DOI] [PubMed] [Google Scholar]
- 58.Yan R, Rychlik W, Etchison D, Rhoads R E. Amino acid sequence of the human protein synthesis initiation factor eIF-4 gamma. J Biol Chem. 1992;267:23226–23231. [PubMed] [Google Scholar]
- 59.Yoder-Hill J, Pause A, Sonenberg N, Merrick W C. The p46 subunit of eukaryotic initiation factor (eIF)-4F exchanges with eIF-4A. J Biol Chem. 1993;268:5566–5573. [PubMed] [Google Scholar]