Stimulation of Homologous Recombination through Targeted Cleavage by Chimeric Nucleases (original) (raw)
Abstract
Chimeric nucleases that are hybrids between a nonspecific DNA cleavage domain and a zinc finger DNA recognition domain were tested for their ability to find and cleave their target sites in living cells. Both engineered DNA substrates and the nucleases were injected into Xenopus laevis oocyte nuclei, in which DNA cleavage and subsequent homologous recombination were observed. Specific cleavage required two inverted copies of the zinc finger recognition site in close proximity, reflecting the need for dimerization of the cleavage domain. Cleaved DNA molecules were activated for homologous recombination; in optimum conditions, essentially 100% of the substrate recombined, even though the DNA was assembled into chromatin. The original nuclease has an 18-amino-acid linker between the zinc finger and cleavage domains, and this enzyme cleaved in oocytes at paired sites separated by spacers in the range of 6 to 18 bp, with a rather sharp optimum at 8 bp. By shortening the linker, we found that the range of effective site separations could be narrowed significantly. With no intentional linker between the binding and cleavage domains, only binding sites exactly 6 bp apart supported efficient cleavage in oocytes. We also showed that two chimeric enzymes with different binding specificities could collaborate to stimulate recombination when their individual sites were appropriately placed. Because the recognition specificity of zinc fingers can be altered experimentally, this approach holds great promise for inducing targeted recombination in a variety of organisms.
Procedures and reagents that allow the directed alteration of genes in situ constitute a powerful toolbox for experimental genetics and potentially for agricultural and therapeutic applications. In many organisms, however, and particularly in higher eukaryotes, the efficiency of recombination between an introduced DNA and the homologous chromosomal target is discouragingly low. For example, such events typically occur in mammalian cells at a frequency of only about 1 for each 106 cells treated (3, 31). We are interested in developing procedures that would substantially improve the frequency of gene targeting.
A major impediment to efficient gene replacement is the status of the chromosomal target. Increasing the number of target sequences has little or no effect on targeting efficiency (54, 60). In contrast, making an intentional double-strand break (DSB) in the target DNA increases the yield of specific homologous recombination events up to 1,000-fold or more (10, 11, 14, 44, 46). It is believed that exonucleases act at broken ends to generate single-stranded tails that are recombinagenic in any of several pathways. In particular, the single-strand annealing mechanism (33), by which homologous recombination involving exogenous DNA usually occurs in higher eukaryotes (53), cannot proceed unless both the donor and target have ends (5, 48).
Whatever the mechanism of recombination, it is clear that the frequency of targeted recombination can be substantially improved by introducing a targeted DSB. The feasibility of this approach has been demonstrated by directing cleavage with meganucleases, like I-_Sce_I (20); however, the utility of such enzymes is limited by the need to introduce the corresponding recognition site by a traditional, low-efficiency process before it can be cleaved. More useful would be cleavage reagents that either inherently possess or can be designed to have affinity for natural chromosomal sequences. If the recognition specificity of such reagents could be manipulated to attack different targets in different circumstances, this would be most powerful.
In the present study we investigate the potential of a class of chimeric nucleases for DNA cleavage and initiation of recombination in living cells. These enzymes are hybrids between the nonspecific cleavage domain of the type IIs restriction endonuclease _Fok_I and a DNA-binding domain made up of three Cys2His2 zinc fingers (Fig. 1A) (7, 17, 24, 26–30, 37, 51). Recognition of DNA by zinc fingers is modular: each finger contacts primarily three consecutive base pairs in the target, and a few key residues in the protein mediate recognition. These features have encouraged attempts to develop zinc finger combinations with novel specificities, which have proved quite successful (8, 12, 13, 16, 18, 19, 43, 49, 58, 59). In fact, randomization of the codons for the recognition residues allows the selection of new fingers that have high affinity for arbitrarily chosen DNA sequences. Furthermore, zinc fingers are natural DNA-binding modules, and engineered fingers have been shown to act on their designed targets in living cells (1, 9, 25, 34). Thus, nucleases based on zinc fingers should be targetable to specific but arbitrary recognition sites.
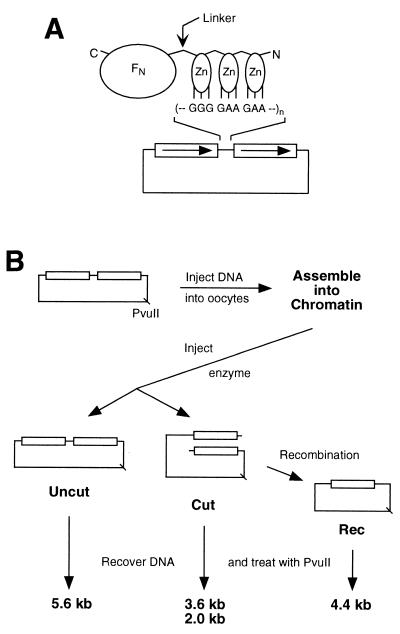
FIG. 1.
(A) Schematic diagram of a chimeric nuclease and DNA substrate. The nuclease consists of three zinc fingers (Zn) connected to the cleavage domain of _Fok_I (FN) by a flexible peptide linker. The N and C termini of the protein are indicated. Each finger makes contact with three consecutive base pairs in the recognition sequence. In the DNA substrates, the canonical binding site for QQR, 5′-GGG GAA GAA, was inserted, in various numbers and orientations, between the 1.25-kb direct repeats (boxes with arrows) of plasmid pRW4. (B) Scheme for the oocyte injection experiments. The DNA substrate is diagrammed at the top left, and the position of the unique _Pvu_II site is shown. For each sample the DNA was injected into the nuclei of 20 to 40 oocytes; they were incubated for 3 to 4 h to allow chromatin assembly, and then QQR was injected into the nuclei. After various lengths of time, DNA was recovered from the oocytes, digested with _Pvu_II, and analyzed by Southern blot hybridization. This distinguishes DNA molecules not cleaved by QQR (Uncut) from cleaved DNA (Cut) and from cleaved molecules that have undergone homologous recombination (Rec). The locations of the _Pvu_II sites and the sizes of the resulting _Pvu_II fragments are shown.
Here we characterize the cleavage abilities of the chimeric nucleases in Xenopus laevis oocytes. These enormous cells have a large capacity for homologous recombination that is readily accessed by microinjection of appropriate substrates (5) and that proceeds by the same single-strand annealing mechanism that is the principal pathway available to exogenous DNAs in cultured mammalian cells (5, 53). Injected linear DNAs undergo efficient recombination if they carry appropriately placed homologous sequences, and a single oocyte can process more than 109 molecules into completed recombination products in a few hours (6, 35, 36). Injected circular DNAs are assembled into apparently normal chromatin and are inert for recombination, but they can be induced to interact with a homologous partner, if they are cleaved (48). A circular DNA thus serves as an effective model for an inactive chromosomal target.
We report that chimeric nucleases based on zinc fingers are capable of finding their recognition sites in oocytes, directing specific cleavage, and stimulating local homologous recombination. The substrate requirements for cleavage in living cells are described, and future applications are discussed.
MATERIALS AND METHODS
Enzymes.
Zif-QQR-FN (29) and Zif-ΔQNK-FN (51) were purified from overproducing bacteria as previously described (51, 52). We refer to them by the three-letter designations QQR and QNK, respectively. Coding sequences for enzyme variants with altered linkers were created from the original QQR clone in the pET15b vector by PCR using primers carrying the desired alterations. The resulting plasmids were verified by sequence analysis and transformed into Escherichia coli BLR(DE3)/pLysS; induction, lysis, and enrichment of the enzymes were carried out as described for QQR (51, 52). In vitro reactions were performed in 20 μl containing 10 mM Tris (pH 8.5), 50 mM NaCl, 1 mM dithiothreitol, 100 μM ZnCl2, 50 μg of bovine serum albumin per ml, 100 μg of tRNA per ml, and 50 ng of substrate DNA (final concentration, about 0.7 nM). After addition of various amounts of enzyme, the mixture was incubated for 30 min at room temperature. MgCl2 was added to a final concentration of 10 mM, and incubation was continued for 1 h at room temperature. Cleavage was monitored by electrophoresis in 1% agarose gels.
DNA substrates.
The parent plasmid, pRW4, consists of pBR322 sequences with a direct duplication of 1.25 kb and a unique _Xho_I site between the repeats (36). Substrate DNAs were constructed by insertion of oligonucleotide duplexes containing the recognition site for QQR (5′-GGG GAA GAA) and/or QNK (5′-GGG GCG GAA), in various arrangements, at the _Xho_I site. Sequences of the inserts were verified experimentally in all cases, and they are reported explicitly elsewhere (52).
Oocyte injections.
Injections, DNA recovery, and analysis were performed as described previously (4, 36). A mixture containing 1 ng of substrate DNA (about 0.3 fmol) and 1 ng of the recovery control plasmid pHSS6 (22) was injected into each oocyte in a volume of 20 nl. After incubation for 3 to 4 h to allow chromatin assembly, the chimeric nuclease in 10% glycerol was delivered to the nuclei in a volume of 2.5 to 15 nl. Two different QQR solutions were used for injections: one with an estimated concentration of 3 fmol/nl, and the other with 7 fmol/nl. The linker variant enzymes were approximately 7 fmol/nl, and the QNK stock was approximately 3 fmol/nl. Because glycerol can be toxic to oocytes, the injection volume was limited. Some aliquots of enzyme tended to lose activity in the injection needles, so care was taken to load a fresh needle for each small batch of oocytes and to use it within about 20 min. Samples taken from the needle before and after oocyte injections were assayed to ensure that the enzyme remained active, and only examples that retained activity were processed further.
After incubation and recovery of the DNA, it was digested with _Pvu_II and analyzed by Southern blot hybridization with radioactive pBR322 as a probe. The radioactivity in each band was quantitated with a Molecular Dynamics model 400E PhosphorImager using ImageQuant software. Reported percentages of recombination product were calculated as R/(R + U) × 100, where R is the number of counts in the recombinant band and U is the counts in the uncut substrate. The intensity of the pHSS6 control band (C) was used to determine the recovery of substrate DNA by comparing (R + U)/C in the recovered DNA to U/C in an uninjected sample. While there appeared to be some loss of cleaved DNA in some cases, the total recovery was essentially always above 80%.
RESULTS
Experimental design.
The general designs of the enzymes and DNA substrates used in this study are shown in Fig. 1A. The chimeric nucleases are composed of three zinc fingers at the N terminus, connected by a flexible linker to the cleavage domain of _Fok_I (FN) at the C terminus. Each finger contacts primarily three consecutive base pairs of DNA. Each plasmid DNA carries one or more copies of the recognition sequence for the chimeric nuclease between 1.25-kb direct repeats.
The protocol for injection into Xenopus oocytes is illustrated in Fig. 1B. For each combination of experimental parameters, the substrate DNA was injected into the nuclei of 20 to 40 oocytes and allowed 3 to 4 h to assemble into chromatin (15, 48). We have verified chromatin assembly in our conditions by showing that only about one-quarter of restriction sites in the plasmids are susceptible to cleavage by enzymes injected into the oocytes and that recovered DNA is supercoiled after deproteinization (data not shown). The chimeric nuclease was then injected, again directly into the oocyte nucleus. DNA was recovered after various times of incubation and assayed for cleavage and recombination by digestion with _Pvu_II, gel electrophoresis, and Southern blot hybridization. Substrate molecules that were not cut by the chimeric nuclease, cut molecules, and molecules that had undergone homologous recombination all yield characteristic fragments in this analysis (Fig. 1B). Although recombination products may be formed from these substrates largely by intramolecular events, we have shown previously that intermolecular reactions, very similar in principle to a gene-targeting setup, are stimulated to a comparable extent by directed DSBs in oocytes (5, 48).
Requirements for cleavage and recombination in oocytes.
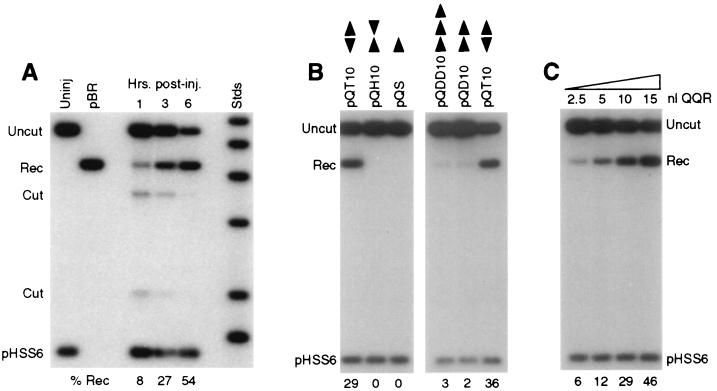
Figure 2A shows a time course experiment in which a plasmid DNA with paired inverted recognition sites for QQR (pQT10) was injected into oocytes and then isolated at various times after enzyme injection. Products of cleavage at the expected site (Cut) were observed within 1 h after enzyme injection. There are two cut bands because the linear DNA was digested with _Pvu_II for analysis. A band corresponding to recombination products (Rec) was also visible, and in longer exposures a faint trailing smear representing recombination intermediates (35) was seen. Both cleavage and recombination proceeded through the 3-h time point, and the process was essentially complete by 6 h. This corresponds well to time courses of recombination of linear DNAs in oocytes determined previously (5, 35). In this experiment, the final level of recombination was 54%; both cleavage and recombination products were absent if the nuclease was not injected (6, 48) (data not shown). The total recovery of injected substrates was always good, as indicated by comparison to the circular recovery control plasmid pHSS6.
FIG. 2.
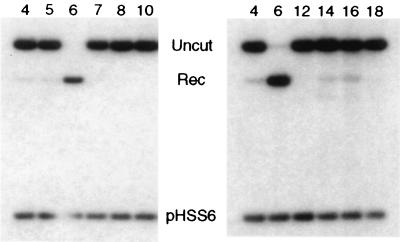
Cleavage and recombination in oocytes. (A) Time course of cleavage and recombination in Xenopus oocytes after injection of QQR. Circular pQT10 DNA (0.3 fmol; this corresponds to 0.6 fmol of binding sites) was injected into oocyte nuclei, following the scheme shown in Fig. 1B; 10 nl (30 fmol) of a solution of QQR was delivered to each oocyte, and DNA was recovered at the indicated times after enzyme injection. Recovered DNA was analyzed as described in the legend to Fig. 1B; the positions of the expected fragments (Uncut, Cut, and Rec) are indicated. pHSS6 is a circular plasmid that was included as a recovery control (22). Uninjected pQT10 (Uninj) is shown in the first lane, and uninjected pBR322 (pBR) serves as a marker for the position of recombination products. Stds, linear size standards. The percentage of the recovered DNA that was in the product band (Rec) is indicated below each lane. (B) Effect of recognition site disposition on cleavage and recombination. The number and orientation of sites are indicated by the arrowheads above each lane, with the point designating the A end of the binding site. pQS has a single recognition site; pQT10 has two sites in tail-to-tail inverted orientation; the two sites in pQH10 are in head-to-head orientation; pQD10 has two sites in direct repeat orientation; and pQDD10 has three direct repeats. Each pair of neighboring sites is separated by 10 bp. DNA and enzyme concentrations were as in panel A; incubation was done overnight. (C) Cleavage and recombination in oocytes after injection of various amounts of QQR (3 fmol/nl), as indicated. The DNA substrate was pQT10, and incubation was overnight.
Studies of the chimeric nucleases in vitro demonstrated that the enzymes require two recognition sites in close proximity to effect double-strand cleavage (52). We tested the ability of QQR to cleave substrates with various site configurations in oocytes (Fig. 2B). Incubation continued overnight, so only completed recombination products were scored. A pair of tail-to-tail inverted repeats (pQT10) provided an effective substrate for cleavage and recombination. A single copy of the zinc finger binding site (pQS in Fig. 2B) did not support cleavage by QQR; this substrate was recovered entirely as intact circles (Uncut). Similarly, a DNA with head-to-head inverted repeats (pQH10) was completely ineffective as a substrate in oocytes. Substrates with two (pQD10 in Fig. 2B) or three (pQDD10) direct repeats yielded only low levels of recombination products after injection of QQR. The requirements for cleavage in oocytes are thus somewhat stricter than those in vitro, where direct repeats were readily cleaved and head-to-head repeats showed some susceptibility (52).
The level of cleavage and recombination was governed at least partly by the amount of enzyme activity delivered to the oocytes, since injection of increasing volumes of enzyme solution led to increased product yields (Fig. 2C). At the highest level of enzyme shown, 46% of pQT10 ultimately recombined, but even higher levels were achieved with this substrate using more concentrated enzyme stocks (see Fig. 3). Although the molar amount of QQR injected greatly exceeded that of the DNA substrate in these experiments, the effective concentration could be substantially lower, since we do not know how much of the enzyme remained intact, active, and nuclear during the incubation.
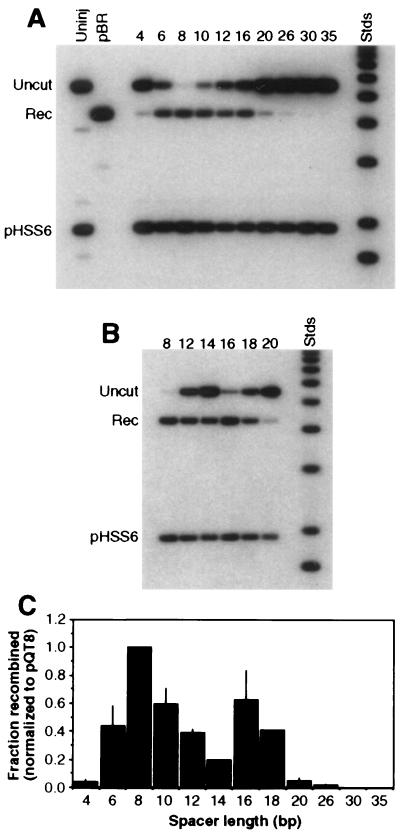
FIG. 3.
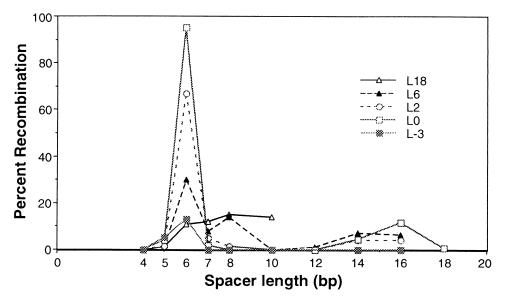
Effect of spacer length on efficiency of cleavage and recombination. All substrates carried two recognition sites in tail-to-tail orientation, separated by the number of base pairs given above the lanes. Labels and markers are as in Fig. 2. Two independent experiments are shown, one (A) covering a broad range of recognition site separations and the other (B) focusing on the range from 12 to 20 bp. About 0.3 fmol of DNA and 70 fmol of QQR were injected in each sample. (C) Histogram summarizing the results of several independent oocyte injection experiments. Recombination yields are plotted against the distance in base pairs between inverted sites. Values were normalized to the recombination fraction measured for pQT8 in each experiment; thin lines represent standard deviations from three or four independent experiments. The values for spacers of 14 and 18 bp are based on a single experiment but were confirmed qualitatively by two additional observations.
These results demonstrate that (i) the chimeric nuclease QQR can locate its target and produce recombinagenic DSBs in living cells even when the target is incorporated into chromatin; (ii) paired, inverted recognition sites are required to effect efficient cleavage; and (iii) a large fraction of the substrate can be converted to recombination products.
Influence of recognition site separation on efficiency.
A series of substrates carrying paired inverted repeats of the recognition site separated by various lengths of spacer DNA were tested for the ability of QQR to stimulate recombination in Xenopus oocytes. Two different experiments are shown in Fig. 3, one that includes a broad range of site separations (4 to 35 bp; Fig. 3A), and the other that concentrates on the range between 12 and 20 bp (Fig. 3B). Effective cleavage and recombination occurred with several substrates, but there was a rather sharp dependence on the length of the spacer. Little product was formed when the separation was 4 bp; this substrate was also not cleaved effectively in vitro, probably due to steric clash between enzyme molecules that attempt to bind to the two sites (52). The yield was better with a separation of 6 bp, and a spacer of 8 bp consistently gave the largest yield. In the experiment shown in Fig. 3A, 94% of this substrate was converted to recombination product; in Fig. 3B the yield was 95%. A secondary maximum was often seen with a 16-bp spacer (Fig. 3B). The efficiency dropped at larger separations, and with distances of 20 bp and greater, very little or no product was formed. (In a separate experiment [not shown], a separation of 5 bp gave no recombination, and the yield with 7 bp was intermediate between those for 6 and 8 bp.)
Quantitated results from several experiments are summarized in Fig. 3C. These data have been normalized to the yield obtained with a separation of 8 bp because the level of enzyme activity delivered to the oocytes could not be precisely controlled between experiments, and thus the absolute yields are not strictly comparable. The oocyte results contrast with those obtained in vitro (52) in several respects. First, the range of effective site separations was narrower in cells: all substrates with spacers of between 6 and 35 bp were cleaved to completion with a modest excess of enzyme in vitro, while the effective range in oocytes was 6 to 18 bp. Second, the dependence of cleavage efficiency on distance is sharper in cells, with a decided maximum at 8 bp, while the efficiency in vitro was roughly constant throughout the effective range. Third, no nicking by the nuclease was observed in cells (not shown). This may be a reflection of the DNA repair capabilities of the oocytes rather than a difference in inherent nuclease activity.
Linker variants in vitro.
The specificity of the chimeric nuclease is determined by how many related sites are recognized and cleaved. This is governed largely by the discrimination achieved by the zinc fingers, but it also reflects how many different configurations of the binding sites can be effectively recognized. As illustrated in Fig. 4, when the binding sites are moved farther apart, the linker must extend a longer distance. For a given linker length, there will be a limit to the distance between recognition sites that is consistent with both binding and dimerization. When the linker is shortened, we expect this limit to be reduced. We therefore attempted to limit the range of competent targets by shortening the flexible peptide linker between the binding and cleavage domains of QQR.
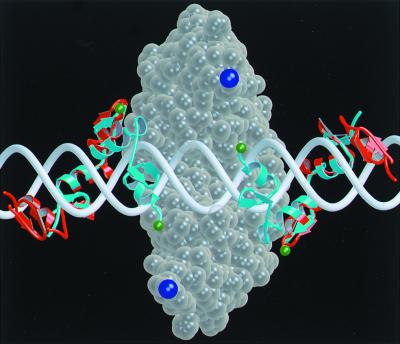
FIG. 4.
Molecular model of the domains of the chimeric nuclease on DNA. The cleavage domain dimer (ball representation in transparent wheat) sits largely behind the DNA (white) in this view and reaches around the duplex at the top and bottom. The zinc finger domains wind through the major groove and are shown in ribbon representation centered around the cleavage dimer: cyan for binding sites separated by 6 bp and red for a 10-bp separation. The residues that must be connected by the flexible linker are colored green on the zinc finger domains and dark blue on the cleavage domains. In moving from the 6-bp to the 10-bp separation, the attachment sites for the linker have retracted both axially and into the plane of the picture. The distance that the linker must extend to join the binding and cleavage domains has gone from about 20 Å for the 6-bp spacer to >30 Å for the 10-bp case. If the linker cannot reach this distance once the zinc finger domains are bound, the cleavage domain cannot dimerize. This model (52) was produced with the program O, and the figure was generated with MolScript.
The original QQR construct has 18 amino acids inserted between domains obtained from a zinc finger protein and _Fok_I (26, 52). We prepared derivatives in which this number was reduced to 6, 2, and 0 amino acids, and we further encroached on the cleavage domain by deleting an additional 3 or 6 residues from its N terminus. In the shorthand we have chosen for these proteins, QQR is designated L18, and the derivatives are L6, L2, L0, L−3, and L−6, respectively. These enzymes were used to treat the range of substrates shown in Fig. 3 both in vitro and in oocytes. (L−6 was not studied in oocytes due to the relative ineffectiveness of L−3.)
As noted earlier, L18 is capable in vitro of cleaving to completion all substrates with 6 to 35 bp between recognition sites, while spacers of 4, 5, and 40 bp do not support cleavage. Examining the altered enzymes, we found a progressive decrease in cleavage of some DNAs with decreasing linker length (Table 1). The first to show reduced cleavage were spacers of 10, 20, and 30 bp, which were essentially resistant to L6 and all smaller linkers. Next, distances of 8, 18, and 35 became resistant, and finally cleavage of the 7- and 12-bp spacers disappeared. Concomitantly, DNAs with spacers of 4 and 5 bp became sensitive as the linker was shortened. The 5-bp construct was cleaved moderately well by L6 and very effectively by all shorter linkers, while the 4-bp target showed weak cleavage by L2 and L0 and strong cleavage by L−3 and L−6.
TABLE 1.
In vitro digestion by QQR linker variants
| Substrate | Cleavagea by enzyme: | |||||
|---|---|---|---|---|---|---|
| L18 | L6 | L2 | L0 | L−3 | L−6 | |
| pQT4 | − | − | ± | ± | +++ | ++ |
| pQT5 | − | ++ | +++ | +++ | +++ | +++ |
| pQT6 | +++ | ++ | ++ | +++ | +++ | +++ |
| pQT7 | +++ | ++ | ++ | ++ | ± | − |
| pQT8 | +++ | ++ | + | ± | − | − |
| pQT10 | +++ | + | − | − | − | − |
| pQT12 | +++ | +++ | ++ | + | ± | − |
| pQT14 | +++ | +++ | ++ | +++ | +++ | +++ |
| pQT16 | +++ | +++ | ++ | +++ | +++ | ++ |
| pQT18 | +++ | +++ | ± | − | − | − |
| pQT20 | ++ | − | − | − | − | − |
| pQT26 | +++ | +++ | ++ | ++ | + | + |
| pQT30 | ++ | − | − | − | − | − |
| pQT35 | +++ | + | ± | − | − | − |
| pQT40 | − |
The substrates that retained the greatest ability to be cleaved with shorter linkers had spacers of 5, 6, 14, and 16 bp, with lesser cleavage at 7 and 26 bp. Molecular modeling (52) showed that these separations allow the linker peptide to lie entirely on one side of the DNA duplex, which appears to be a favorable situation. The steric constraints that prevent cleavage of the 4- and 5-bp separations by L18 are relieved by deletion of linker residues and particularly by deletion into the cleavage domain (L−3 and L−6).
Linker variants in oocytes.
We tested the capabilities of the same enzyme variants (except L−6) to cut substrates with different spacer lengths in oocytes. The basic observations were that the range of susceptible targets became restricted as the linker was shortened; and for each enzyme, as with QQR (L18), the range was narrower in cells than in vitro. Some examples are shown for L0 in Fig. 5. At moderate concentrations of the nuclease, essentially the only substrate cleaved was that with a 6-bp spacer. Neither the 5-bp nor the 7-bp construct showed any recombination, and those with 14-bp and 16-bp spacers gave very weak product bands. At higher enzyme inputs (not shown), greater cleavage of the 14- and 16-bp substrates could be forced, but there was still no cleavage of any shorter spacer except 6 bp.
FIG. 5.
Activity of the L0 linker variant. Two separate experiments showing cleavage and recombination of substrates with various spacer lengths (indicated above each lane) after injection of L0 nuclease. DNA (0.3 fmol) was injected, followed by 5 fmol of enzyme in A and 10 fmol of enzyme in B. Other conditions are as in Fig. 2.
The linkerless L0 protein not only exhibited the most restricted substrate preference, it also had the greatest activity in the injection experiments. L2 had similar substrate selectivity but somewhat less activity. This is shown graphically, along with data for the other variants, in Fig. 6. For these experiments the input quantities were adjusted so that the physical amounts of nuclease protein (judged by Coomassie blue staining) and the in vitro cleavage activities were matched. One aspect to note is that QQR (L18) was used at a much lower concentration than shown in earlier figures, and the extent of recombination was substantially reduced as a consequence.
FIG. 6.
Summary of the activities of all linker variants in oocytes. Results of experiments like those in Fig. 3 and 6 were quantitated, and the percent recombination is plotted against spacer length (in base pairs) for each enzyme. In all experiments 0.3 fmol of DNA and approximately 10 fmol of nuclease were injected. For L18 (QQR), this is considerably less enzyme than was used in the experiments shown in Fig. 2.
Cleavage of paired nonidentical recognition sites.
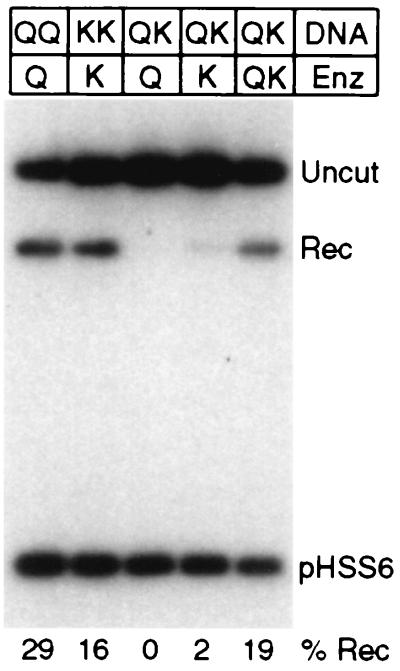
When cleavage of a chromosomal target is desired, it is very unlikely that exact inverted repeats of a 9-bp sequence will be located in favorable positions. Thus, it will be necessary to devise nucleases with two different sets of zinc fingers designed to bind two different 9-mers. We tested the feasibility of this scheme using two chimeric nucleases that recognize related but different sites. The preferred site for QQR is 5′-GGG GAA GAA (29, 50), while that for QNK is 5′-GGG GCG GAA (23, 51). Three substrates were prepared: one with two sites for QQR (QQ), one with two sites for QNK (KK), and one with one site of each type (QK). An 8-bp spacer was chosen because it was optimal for cleavage and recombination in oocytes (see Fig. 3). Earlier experiments had shown that similar substrates with 14-bp spacers were cleaved in vitro by a combination of the two enzymes (52).
When the hybrid substrate was injected into Xenopus oocytes, it was activated effectively for recombination only when both enzymes were injected together (Fig. 7). Both single-enzyme controls were positive on their own substrates, but cross-cleavage was not observed. Injection of QQR stimulated recombination of the QQ substrate, and the yield of product was 29%. Injection of QNK led to cleavage and recombination of KK, with a yield of 16%. With the combined substrate (QK), essentially no recombination was observed when QQR was injected alone; with QNK a low level of recombination was seen (yield, 2%), reflecting the lower selectivity of this enzyme. When a mixture of the two enzymes was injected, a level of recombination was observed (yield, 19%) that was comparable to that obtained with the single-enzyme substrates. Thus, two enzymes directed to nonidentical sites can collaborate to produce a recombinagenic DSB in cells.
FIG. 7.
Cleavage and recombination with paired nonidentical sites. The DNA substrates carried two sites for QQR (QQ), two sites for QNK (KK), or one each for QQR and QNK (QK). They were injected into oocytes followed by no enzyme (−), with a single enzyme (Q or K), or with a mixture of the two enzymes (QK). In each case, 0.3 fmol of DNA and 15 fmol of each enzyme were injected.
DISCUSSION
Requirements of the chimeric nucleases.
Our long-term goal is to provide reagents that will substantially improve the frequency of targeted homologous recombination. Because DSBs stimulate recombination dramatically in their vicinity in a wide variety of organisms (10, 11, 14, 38, 39, 41, 44, 46–48), we have focused our attention on chimeric nucleases that have the potential for making targeted breaks at arbitrarily selected DNA sequences.
The enzymes of particular interest carry a nonspecific DNA cleavage domain linked to a DNA-binding domain comprised of Cys2His2 zinc fingers. We have shown that these chimeras are capable of locating their target sequences in chromatin, cleaving with good efficiency, and thereby stimulating homologous recombination. Although the substrates we used were engineered plasmid DNAs and the cellular milieu was that of the Xenopus oocyte, our findings should be applicable to mammalian somatic cells and to many other cells and organisms that do not have the ability to initiate recombination efficiently at unbroken chromosomal targets. This might include organisms popular with geneticists, like Drosophila melanogaster and nematodes. In the former case, target cleavage might be combined with the recently described procedures for producing linear donor DNA in vivo (45) to achieve maximum efficiency.
Our results define the requirements for cleavage by the chimeric nucleases in living cells. Because dimerization of the cleavage domain is required for nuclease activity (2, 52), two recognition sites for the zinc fingers must occur in close proximity. These sites must be in the inverted repeat orientation that directs the two cleavage domains toward the space between the sites. Furthermore, only a limited range of separations between binding sites is tolerated. With the original 18-amino-acid linker between the binding and cleavage domains of QQR, the range was 6 to 18 bp. When the linker was shortened, the range of effective spacer lengths was correspondingly constrained; and with no intentional linker between the domains, a separation of 6 bp was the only one cleaved efficiently.
When the above criteria were met and sufficient enzyme was provided, essentially all of the substrate molecules were cleaved in oocytes. This indicates that assembly into chromatin does not prevent access of the nucleases to their targets. There is reason to be optimistic that this will be true with other chimeras based on zinc fingers, since these domains are derived from transcription factors that are capable of locating their recognition sequences in the normal nuclear environment. Furthermore, other engineered zinc fingers have been shown to modulate transcription at their specific targets in mammalian cells (1, 9, 25, 34).
Interpretation of linker variants.
Our earlier molecular modeling of the complexes of the chimeric nucleases with DNA (52) provides a basis for evaluating the results with interdomain linkers of different lengths (see Fig. 4). Several conclusions were drawn. First, it was certainly anticipated that the upper limit on spacers that allow effective cleavage would be reduced as the linker was trimmed, since the linker must extend to allow dimerization of the cleavage domains attached to the two separately bound recognition domains.
Second, short spacers that did not allow cleavage by the original QQR (L18) were cleaved when the linker was reduced. Although the linker does not form part of the interface that causes steric interference between binding and cleavage domains, shortening it may allow adjustments in other parts of the cleavage domains to allow closer approach of the binding domains.
Third, the secondary maximum in cleavage efficiency at separations of 14 to 16 bp reflects the helical nature of DNA. This phenomenon is observed both in vitro (Table 1) and in oocytes (Fig. 3 and 6). The modeling and cleavage mapping results show that when the spacer between sites is 16 bp, the cleavage dimer sits off-center, 3 bp from one site and 13 bp from the other, and both linkers lie on the same helical face (52). When the separation between sites is 8, 10, or 12 bp, all possible domain arrangements require the linker to traverse around the helix to some extent. This extends the distance the linker must stretch, and it also seems that there is some inherent preference to keep the linker on one side of the DNA (52).
Fourth, it is still puzzling that enzymes with very short linkers cleave substrates with sites separated as widely as we observed. The 6-bp spacer requires the linker to stretch 18 to 20 Å by our estimate (52) (Fig. 4). In the L0 construct, there are three residues in each binding domain and three in each cleavage domain that are disordered in the crystal structures; these six residues could easily reach the necessary distance. This same enzyme also cuts the 16-bp substrate efficiently, however, and the required extension in that case is about 40 Å for one of the linkers. Furthermore, the L−3 and L−6 constructs, in which more of the cleavage domain has been deleted, also cut both the 6-bp and 16-bp substrates in vitro. It appears that some of the cleavage domain must become unstructured in order to accommodate these distances. The first regular feature at the N terminus of the cleavage domain is an α-helix that may be stabilized in native _Fok_I by interaction with elements of the binding domain (55, 56). In the chimeric nucleases, it is conceivable that this helix may be unfolded with modest energy input. In addition, the actual distance between the binding and cleavage domains could be reduced by a change in the DNA structure, for example, a bend or kink at the site of cleavage.
Cleavage parameters in oocytes.
Cleavage by the chimeric nucleases in oocytes shows stricter limits on effective recognition site separations with all linker deletions. While a site separation of 8 bp is optimal for the L18 enzyme, the optimum shifts to 6 bp as the linker is trimmed. The secondary optimum with spacers of 14 and 16 bp is maintained in oocytes except for the L−3 enzyme, presumably reflecting the preference for the linker to remain on one side of the DNA. With the L0 nuclease there is a very strong preference for sites exactly 6 bp apart, and this fact should enhance the specificity of cleavage.
We have never seen substrates with a single recognition site cleaved in oocytes, even though such a configuration can be cleaved in vitro at high enzyme concentrations (52). One difference between these situations is that oocytes, like other living cells, have the ability to repair single-strand breaks. If the cuts in the two strands are not made in a concerted fashion by the nuclease, particularly in less favorable situations, it is possible that the DNA repair machinery fixes one break before a second can be made.
The modeling indicates that binding and cleavage by two enzyme molecules require exposure of a fairly extensive region of the major groove. It seems unlikely that this could occur on DNA segments that remain closely associated with a nucleosome core. Either natural nucleosome dynamics expose all possible sequences in linker DNA during the course of an experiment, or binding of one zinc finger domain could facilitate the release of nearby segments of DNA (57). The different requirements for cleavage in vitro and in oocytes could reflect the structure of DNA exposed in cells, or the environment provided by the oocyte may impose structural constraints on the protein. For example, the interdomain linker may adopt a folded structure that restricts its extensibility in the cell nucleus.
The efficiency of cleavage in oocytes, even of the best substrates, also varied with the size of the linker. A substantially larger amount of the L18 nuclease than of the L0 construct had to be injected to achieve optimal cleavage. Since the binding and cleavage domains are unaltered by the linker manipulations, it seems unlikely that binding affinities differ in these cases. An alternative explanation is that the stability of the protein is greater in the L0 nuclease, perhaps because the L18 linker provides an unstructured target for oocyte proteases.
Applications to gene targeting.
A chromosomal gene targeting experiment utilizing the chimeric nucleases would presumably proceed as follows. A target site would be chosen within a gene of interest. Zinc finger combinations would be derived that bind inverted sites separated by 6 bp at the target locus. These sites need not be identical, as we have demonstrated that two chimeras with different DNA-binding domains are capable of collaborating to achieve cleavage. The zinc finger domains would be linked to the DNA cleavage domain and tested in vitro for specificity to ensure that the zinc fingers recognize the desired sites. For maximum specificity, linkerless (L0) constructs would be made; but if suitable sites spaced by exactly 6 bp could not be found, longer linkers could be incorporated to accommodate greater separations. The two new chimeric nucleases would then be delivered to cells along with a linear donor DNA molecule carrying the desired sequence alteration. The method of delivery would depend on the organism, cell type, and other experimental conditions.
Will the chimeric nucleases have sufficient specificity to attack the desired target without introducing breaks at many other chromosomal locations? The requirement for dimerization of the cleavage domain enforces a high level of sequence specificity, as long as the zinc fingers show good discrimination against related sites. Since each set of three fingers binds nine consecutive base pairs, two chimeric nucleases effectively demand an 18-bp target if each zinc finger domain has perfect specificity. Any given sequence of this length is predicted to be unique in a DNA as complex as the human genome (3 × 109 bp), since there are 418 (6.9 × 1010) different 18-mers. Furthermore, it has been shown that additional fingers provide enhanced specificity (1, 25, 34), so the number of zinc fingers in each DNA-binding domain could be increased.
What are the prospects for deriving zinc finger combinations that can recognize any desired 9-bp sequence? Fingers with new binding specificities have been produced by randomizing coding sequences for key residues that contact DNA, then selecting by phage display for combinations that bind the desired target most avidly (8, 12, 13, 19, 43, 59). In compelling demonstrations of the power of this approach, Greisman and Pabo (16) evolved zinc fingers that recognized completely new 9-bp sites by selecting sequentially for one finger at a time, and Segal et al. (49) systematically derived fingers that recognize the complete subset of GNN triplets. It is not known what limitations might exist on the ability of zinc fingers to bind the full spectrum of possible target sequences, but it is clear that the accessible range is large (18, 58).
Several additional issues remain to be addressed to confirm the utility of chimeric nucleases as tools for gene targeting. Among these are demonstrating discrimination against related sequences; proving the efficacy of zinc fingers designed to bind arbitrarily chosen sequences; and testing the cleavage of genuine chromosomal targets. The question of discrimination among potential binding sites is a particularly critical one. In this regard, neither QQR nor QNK is the ideal model enzyme, since both can bind alternative sites (23, 29, 51). Impressive zinc finger binding selectivity has been achieved recently with the assistance of negative selection against closely related base triplets (49). An additional concern is the existence of nonhomologous recombination pathways, which will compete with homologous recombination to repair the broken target. It may be possible to take advantage of differences in the genetic requirements of these processes (21, 32, 40) to tip the balance in favor of homologous events.
Assuming that these issues can be resolved satisfactorily, the use of chimeric nucleases for targeted gene manipulation should be applicable to a wide variety of organisms and experimental purposes. At present the effort required to produce zinc finger combinations with novel binding specificities will likely restrict application of the chimeric nucleases to situations in which the same site is targeted repeatedly. As experience accumulates, methods of producing new specificities will be improved, and even single-use applications may become feasible.
ACKNOWLEDGMENTS
This project is an equal collaboration between the Carroll and Chandrasegaran labs.
We thank Frank Whitby for providing Fig. 4. We are grateful to Jeremy Berg for advice on zinc finger recognition, to H. O. Smith for continuing interest in this project, and to Tim Formosa, Wes Sundquist, and Mario Capecchi for comments on various versions of the manuscript.
This work was supported in part by grants from the National Institutes of Health (GM50739 and GM58504) and the University of Utah to D.C. and by grants from the National Institutes of Health (GM53923) and the National Science Foundation (MCB 9415861) to S.C. Assistance was also provided by the Markey Center for Protein Biophysics, the Huntsman Cancer Institute at the University of Utah, and the Environmental Health Sciences Core Facility at Johns Hopkins University. S.C. is a member of the Scientific Advisory Board of Sangamo Biosciences, Inc.
REFERENCES
- 1.Beerli R R, Segal D J, Dreier B, Barbas C F., III Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci USA. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bitinaite J, Wah D A, Aggarwal A K, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capecchi M R. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 4.Carroll D. DNA recombination and repair in Xenopus oocytes and eggs: substrate design, direct microinjection, and extract preparation. In: Richter J D, editor. A comparative methods approach to the study of oocytes and embryos. New York, N.Y: Oxford University Press; 1999. pp. 173–195. [Google Scholar]
- 5.Carroll D. Homologous genetic recombination in Xenopus: mechanism and implications for gene manipulation. Prog Nucleic Acid Res Mol Biol. 1996;54:101–125. doi: 10.1016/s0079-6603(08)60361-x. [DOI] [PubMed] [Google Scholar]
- 6.Carroll D, Wright S H, Wolff R K, Grzesiuk E, Maryon E B. Efficient homologous recombination of linear DNA substrates after injection into Xenopus laevis oocytes. Mol Cell Biol. 1986;6:2053–2061. doi: 10.1128/mcb.6.6.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandrasegaran S, Smith J. Chimeric restriction enzymes: what is next? Biol Chem. 1999;380:841–848. doi: 10.1515/BC.1999.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choo Y, Klug A. Toward a code for the interactions of zinc fingers with DNA: selection of randomized fingers displayed on phage. Proc Natl Acad Sci USA. 1994;91:11163–11167. doi: 10.1073/pnas.91.23.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choo Y, Sanchez-Garcia I, Klug A. In vivo repression by a site-specific DNA-binding protein designed against an oncogene sequence. Nature. 1994;372:642–645. doi: 10.1038/372642a0. [DOI] [PubMed] [Google Scholar]
- 10.Choulika A, Perrin A, Dujon B, Nicolas J-F. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen-Tannoudji M, Robine S, Choulika A, Pinto D, El Marjou F, Babinet C, Louvard D, Jaisser F. I-SceI-induced gene replacement at a natural locus in embryonic stem cells. Mol Cell Biol. 1998;18:1444–1448. doi: 10.1128/mcb.18.3.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desjarlais J R, Berg J M. Toward rules relating zinc finger protein sequences and DNA binding site preferences. Proc Natl Acad Sci USA. 1992;89:7345–7349. doi: 10.1073/pnas.89.16.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desjarlais J R, Berg J M. Use of a zinc finger consensus sequence framework and specificity rules to design specific DNA binding proteins. Proc Natl Acad Sci USA. 1993;90:2256–2260. doi: 10.1073/pnas.90.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott B, Richardson C, Winderbaum J, Nickoloff J A, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol Cell Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gargiulo G, Worcel A. Analysis of the chromatin assembled in germinal vesicles of Xenopus oocytes. J Mol Biol. 1983;170:699–722. doi: 10.1016/s0022-2836(83)80128-4. [DOI] [PubMed] [Google Scholar]
- 16.Greisman H A, Pabo C O. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 17.Huang B, Schaeffer C J, Li Q, Tsai M-D. Sp1ase: a new class IIS zinc-finger restriction endonuclease with specificity for Sp1 binding sites. J Protein Chem. 1996;15:481–489. doi: 10.1007/BF01886856. [DOI] [PubMed] [Google Scholar]
- 18.Isalan M, Klug A, Choo Y. Comprehensive DNA recognition through concerted interactions from adjacent zinc fingers. Biochemistry. 1998;37:12026–12033. doi: 10.1021/bi981358z. [DOI] [PubMed] [Google Scholar]
- 19.Jamieson A C, Kim S-H, Wells J A. In vitro selection of zinc fingers with altered DNA-binding specificity. Biochemistry. 1994;33:5689–5695. doi: 10.1021/bi00185a004. [DOI] [PubMed] [Google Scholar]
- 20.Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- 21.Jeggo P A. DNA breakage and repair. Adv Genet. 1998;38:185–218. doi: 10.1016/s0065-2660(08)60144-3. [DOI] [PubMed] [Google Scholar]
- 22.Jeong-Yu S, Carroll D. Effect of terminal nonhomologies on homologous recombination in Xenopus laevis oocytes. Mol Cell Biol. 1992;12:5426–5437. doi: 10.1128/mcb.12.12.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim C A, Berg J M. A 2.2 Å resolution crystal structure of a designed zinc finger protein bound to DNA. Nat Struct Biol. 1996;3:940–945. doi: 10.1038/nsb1196-940. [DOI] [PubMed] [Google Scholar]
- 24.Kim J-S, Kim J, Cepek K L, Sharp P A, Pabo C O. Design of TATA box-binding protein/zinc finger fusions for targeted regulation of gene expression. Proc Natl Acad Sci USA. 1997;94:3616–3620. doi: 10.1073/pnas.94.8.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J-S, Pabo C O. Getting a handhold on DNA: Design of poly-zinc finger proteins with femtomolar dissociation constants. Proc Natl Acad Sci USA. 1998;95:2812–2817. doi: 10.1073/pnas.95.6.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim Y-G, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to FokI cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim Y-G, Chandrasegaran S. Chimeric restriction endonuclease. Proc Natl Acad Sci USA. 1994;91:883–887. doi: 10.1073/pnas.91.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y-G, Kim P S, Herbert A, Rich A. Construction of a Z-DNA-specific restriction endonuclease. Proc Natl Acad Sci USA. 1997;94:12875–12879. doi: 10.1073/pnas.94.24.12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y-G, Shi Y, Berg J M, Chandrasegaran S. Site-specific cleavage of DNA-RNA hybrids by zinc finger-FokI cleavage domain fusions. Gene. 1997;203:43–49. doi: 10.1016/s0378-1119(97)00489-7. [DOI] [PubMed] [Google Scholar]
- 30.Kim Y-G, Smith J, Durgesha M, Chandrasegaran S. Chimeric restriction enzyme: Gal4 fusion to FokI cleavage domain. Biol Chem. 1998;379:489–495. doi: 10.1515/bchm.1998.379.4-5.489. [DOI] [PubMed] [Google Scholar]
- 31.Koller B H, Smithies O. Altering genes in animals by gene targeting. Annu Rev Immunol. 1992;10:705–730. doi: 10.1146/annurev.iy.10.040192.003421. [DOI] [PubMed] [Google Scholar]
- 32.Lieber M R. The biochemistry and biological significance of nonhomologous DNA end joining: an essential repair process in multicellular organisms. Genes Cells. 1999;4:77–85. doi: 10.1046/j.1365-2443.1999.00245.x. [DOI] [PubMed] [Google Scholar]
- 33.Lin F-L, Sperle K, Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for the ends in the recombination process. Mol Cell Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Q, Segal D J, Ghiara J B, Barbas C F., III Design of polydactyl zinc-finger proteins for unique addressing within complex genomes. Proc Natl Acad Sci USA. 1997;94:5525–5530. doi: 10.1073/pnas.94.11.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maryon E, Carroll D. Characterization of recombination intermediates from DNA injected into Xenopus laevis oocytes: evidence for a nonconservative mechanism of homologous recombination. Mol Cell Biol. 1991;11:3278–3287. doi: 10.1128/mcb.11.6.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maryon E, Carroll D. Degradation of linear DNA by a strand-specific exonuclease activity in Xenopus laevis oocytes. Mol Cell Biol. 1989;9:4862–4871. doi: 10.1128/mcb.9.11.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nahon E, Raveh D. Targeting a truncated Ho-endonuclease of yeast to novel DNA sites with foreign zinc fingers. Nucleic Acids Res. 1998;26:1233–1239. doi: 10.1093/nar/26.5.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osman F, Subramani S. Double-strand break-induced recombination in eukaryotes. Prog Nucleic Acid Res Mol Biol. 1998;58:263–299. doi: 10.1016/s0079-6603(08)60039-2. [DOI] [PubMed] [Google Scholar]
- 39.Ozenberger B, Roeder G S. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol Cell Biol. 1991;11:1222–1231. doi: 10.1128/mcb.11.3.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pâques F, Haber J E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plessis A, Perrin A, Haber J E, Dujon B. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics. 1992;130:451–460. doi: 10.1093/genetics/130.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puchta H, Dujon B, Hohn B. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 1993;21:5034–5040. doi: 10.1093/nar/21.22.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rebar E J, Pabo C O. Zinc finger phage: affinity selection of fingers with new DNA-binding specificities. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 44.Richardson C, Moynahan M E, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rong Y S, Golic K G. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- 46.Rouet P, Smith F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudin N, Haber J E. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol Cell Biol. 1988;8:3918–3928. doi: 10.1128/mcb.8.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segal D J, Carroll D. Endonuclease-induced, targeted homologous extrachromosomal recombination in Xenopus oocytes. Proc Natl Acad Sci USA. 1995;92:806–810. doi: 10.1073/pnas.92.3.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Segal D J, Dreier B, Beerli R R, Barbas C F., III Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN′-3′ DNA target sequences. Proc Natl Acad Sci USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi Y, Berg J M. Specific DNA-RNA hybrid binding by zinc finger proteins. Science. 1995;268:282–284. doi: 10.1126/science.7536342. [DOI] [PubMed] [Google Scholar]
- 51.Smith J, Berg J M, Chandrasegaran S. A detailed study of the substrate specificity of a chimeric restriction enzyme. Nucleic Acids Res. 1999;27:674–681. doi: 10.1093/nar/27.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith J, Bibikova M, Whitby F G, Reddy A R, Chandrasegaran S, Carroll D. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramani S, Seaton B L. Homologous recombination in mitotically dividing mammalian cells. In: Kucherlapati R, Smith G R, editors. Genetic recombination. Washington, D.C.: American Society for Microbiology; 1988. pp. 549–574. [Google Scholar]
- 54.Thomas K R, Folger K R, Capecchi M R. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986;44:419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- 55.Wah D A, Bitinaite J, Schildkraut I, Aggarwal A K. Structure of FokI has implications for DNA cleavage. Proc Natl Acad Sci USA. 1998;95:10564–10569. doi: 10.1073/pnas.95.18.10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wah D A, Hirsch J A, Dorner L F, Schildkraut I, Aggarwal A K. Structure of the multimodal endonuclease FokI bound to DNA. Nature. 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 57.Widom J. Structure, dynamics, and function of chromatin in vitro. Annu Rev Biophys Biomol Struct. 1998;27:285–327. doi: 10.1146/annurev.biophys.27.1.285. [DOI] [PubMed] [Google Scholar]
- 58.Wolfe S A, Greisman H A, Ramm E I, Pabo C O. Analysis of zinc fingers optimized via phage display: evaluating the utility of a recognition code. J Mol Biol. 1999;285:1917–1934. doi: 10.1006/jmbi.1998.2421. [DOI] [PubMed] [Google Scholar]
- 59.Wu H, Yang W-P, Barbas C F., III Building zinc fingers by selection: toward a therapeutic application. Proc Natl Acad Sci USA. 1995;92:344–348. doi: 10.1073/pnas.92.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng H, Wilson J H. Gene targeting in normal and amplified cell lines. Nature. 1990;344:170–173. doi: 10.1038/344170a0. [DOI] [PubMed] [Google Scholar]