PCR Bias in Ecological Analysis: a Case Study for Quantitative Taq Nuclease Assays in Analyses of Microbial Communities (original) (raw)
Abstract
Succession of ecotypes, physiologically diverse strains with negligible rRNA sequence divergence, may explain the dominance of small, red-pigmented (phycoerythrin-rich) cyanobacteria in the autotrophic picoplankton of deep lakes (C. Postius and A. Ernst, Arch. Microbiol. 172:69–75, 1999). In order to test this hypothesis, it is necessary to determine the abundance of specific ecotypes or genotypes in a mixed background of phylogenetically similar organisms. In this study, we examined the performance of Taq nuclease assays (TNAs), PCR-based assays in which the amount of an amplicon is monitored by hydrolysis of a labeled oligonucleotide (TaqMan probe) when hybridized to the amplicon. High accuracy and a 7-order detection range made the real-time TNA superior to the corresponding end point technique. However, in samples containing mixtures of homologous target sequences, quantification can be biased due to limited specificity of PCR primers and probe oligonucleotides and due to accumulation of amplicons that are not detected by the TaqMan probe. A decrease in reaction efficiency, which can be recognized by direct monitoring of amplification, provides experimental evidence for the presence of such a problem and emphasizes the need for real-time technology in quantitative PCR. Use of specific primers and probes and control of amplification efficiency allow correct quantification of target DNA in the presence of an up to 104-fold excess of phylogenetically similar DNA and of an up to 107-fold excess of dissimilar DNA.
The introduction of techniques in microbial ecology that allow sensitive detection and identification of DNA sequences has revealed a much higher diversity in natural microbial communities than the classical approach based on strain isolation. The greatest resolution is achieved using PCR-based identification techniques in which two oligonucleotides and the resulting PCR fragment of 102 to 103 nucleotides are used to characterize a genotype. PCR-based techniques are not bound to analysis of ribosomal DNA (rDNA). For example, the filamentous cyanobacterium Planktothrix rubescens in Lake Zurich forms a population consisting of strains that differ in size and gas vesicle pressure resistance (5) but not in rDNA sequence (4). Genetic variability was noticed in an operon coding for gas vesicle proteins. A PCR-based assay was developed that allowed discrimination between different alleles of the operon on the basis of the length and restriction polymorphism of the amplicons. In these species, PCR can be performed with single filaments. This allows quantification of subpopulations with different resistance to hydrostatic pressure, a property that is thought to be relevant in strain selection during winter circulation and in the appearance of blooms during the following growth period (4).
In many studies, PCR itself is treated as a semiquantitative detection method. It is assumed that the relative abundance of amplicons generated during a fixed number of PCR cycles provides a measure of the gene dose and gene ratio in the starting mixture. However, a number of factors are known to bias amplification and jeopardize this assumption. Amplification may be hampered by suboptimal reaction conditions, including lack of primer specificity or formation of secondary structures of the template (21, 26). Moreover, the misincorporation rate of Taq polymerase (7) or the formation of chimeric molecules during PCR (37) and heterogeneity of 16S rDNA sequences (38) were reported to bias PCR product formation. In particular PCRs, a bias toward a 1:1 product formation ratio was observed irrespective of the initial template ratio (34). In many reactions, however, amplicons generated from two templates seem to compete in such a way that amplification of the minor template appears to be suppressed by the amplification of the more abundant template. This observation led to the development of quantitative competitive PCR. In this approach, a fixed amount of a competitor DNA which is amplified by the same set of primers as the target DNA is added in a known concentration to a serial dilution of a template with an unknown concentration (12, 22, 24, 36). The target and the competitor are distinguished in post-PCR analysis, either by a difference in size or by the presence of a unique restriction site, and concentration is determined by comparison of the intensity of a stained amplicon. A theoretical basis for this quantification method was provided by Schnell and Mendoza (31).
Quantitative competitive PCR requires time- and resource-consuming post-PCR analyses. An alternative are single-step quantification techniques in which Taq polymerase activity or the accumulation of the amplicon is directly monitored during or after the PCR. Accumulation of PCR products can be monitored using a fluorescent dye, for example, SYBR Green, that forms fluorescent adducts with double-stranded DNA without compromising the polymerization reaction (14, 39). A different approach makes use of the 5′→3′ exonuclease activity of Taq DNA polymerase (15, 16). In a Taq nuclease assay (TNA; also called a 5′ nuclease assay), this activity is used to cleave a labeled oligonucleotide, the TaqMan probe, that hybridizes to the PCR template but is 3′ phosphorylated to prevent polymerization by Taq polymerase (18). The TaqMan probe is labeled at the 5′ end with a fluorescein derivative whose fluorescence is quenched by a rhodamine derivative linked either a few bases downstream of the 5′ end or at the 3′ end of the probe (18, 19). Hydrolysis of the probe by Taq nuclease activity releases the fluorescence of fluorescein, and the shortened probe will be displaced from the target sequence without disturbing polymerase activity. As the probe is hydrolyzed only when bound to the target DNA, TaqMan PCR reports the activity of Taq polymerase with respect to a particular target DNA (11). The concentration of the reported amplicon is proportional to the fluorescence of the reporter dye. The reaction is calibrated by amplification of known amounts (concentrations) of the target sequence and by measuring the fluorescence before and after a fixed number of PCR cycles (end point determination) or by monitoring the increase in fluorescence cycle by cycle (real-time monitoring of PCR). The latter allows calibration by the threshold cycle method (13). Quantification is based on the number of cycles required to reach a certain concentration of amplicons rather than on the concentration reached after a fixed number of cycles. The threshold cycle CT is defined as the PCR cycle at which a fluorescence signal, developed by a dye-DNA complex or by the fluorescence of the TaqMan reporter dye, passes a preset value. This value corresponds to an amount of amplicons which was generated in a few cycles if a large amount of templates was present initially or after many cycles if the PCR started with few templates. Real-time TNA was reported to match the sensitivity of protocols for post-PCR analysis by DNA-binding dyes (1), radiolabeled primers (6), qualitative nested PCR (25, 29) or hybridization techniques (30).
The ability to quantify a particular amplicon in a single step in the presence of homologous DNA that may be coamplified led us to examine the feasibility of TNA, calibrated as end point PCR or via the threshold value method, for the quantification of a specific ecotype in a highly diversified population of phycoerythrin-rich Synechococcus spp. These small, unicellular cyanobacteria dominate the autotrophic picoplankton of Lake Constance (9, 27). For this case study, one isolated strain, Synechococcus sp. strain BO 8807, was selected as the target strain. This strain exhibits a unique glycosylated surface layer (10) and, perhaps due to this surface structure, decreased attractiveness to predators (3, 23). All other isolates of this population lack the surface layer. The genome of these _Synechococcus_-type cyanobacterial strains contains two ribosomal operons (A. Ernst, unpublished data). They exhibit less than 1% 16S rDNA sequence divergence, but the target strain BO 8807 differs in approximately 8% of the nucleotides of the first internal transcribed spacer (ITS-1) of the ribosomal operon (27; Ernst, unpublished). Problems in a PCR-based quantification of subspecies may be caused by the presence of a high background level of homologous and heterologous DNAs, by the lack of hybridization specificity of oligonucleotides used as primers and probes in a TNA, and by a quantification bias caused by the accumulation of amplicons not detected by the TaqMan probe. We examined these problems in end point and continuously monitored TNAs.
MATERIALS AND METHODS
Organisms and culture conditions.
Pelagic Synechococcus sp. strains BO 8805, BO 8807, BO 8808, BO 8809, and BO 9404 and Synechocystis sp. strain BO 8402 isolated from the pelagic zone of Lake Constance (Bodensee) (8, 28) were cultured in 40 ml of the mineral liquid medium BG11 (32) under low light intensity (5 to 10 microeinsteins m−2s−1). Microcystis sp., an isolate originating from Lake Constance, was received from H. Lampert, MPI Limnologie, Plön, Germany. This isolate and Anabaena sp. strain PCC 7120, Anabaena variabilis strain ATCC 29413, and Anacystis nidulans (also known as Synechococcus leopoliensis strain SAG 1402-1 or Synechococcus sp. strain PCC 6301) were cultivated accordingly.
Isolation of genomic DNA.
Cultures of cyanobacteria were harvested by centrifugation for 10 min at 2,000 × g. The pellet was resuspended in a protein digestion buffer containing 5 mM Tris/HCl, 50 mM NaCl, and 5 mM EDTA, pH 8.5. The phenol-chloroform method, performed in accordance with the instructions of Wood and Townsend (40), was used to extract genomic DNA (28). After precipitation with isopropanol, the DNA was dissolved either in sterile distilled water or in TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0). The concentration and purity of genomic DNA were determined by measuring the _A_260/_A_280 ratio. Genomic DNA from Escherichia coli strain K-12 was a gift of W. Boos, University of Constance, Constance, Germany; herring sperm DNA was obtained from Boehringer, Mannheim, Germany. For the estimation of genome copy numbers, we assumed a genome size of 3 Mbp for pelagic Synechococcus spp. and 2.69 Mbp for A. nidulans (33). Using an approximate molecular mass for a base pair of 650 Da, 1 ng of genomic DNA represented 3 × 105 and 3.3 × 105 genome copies of Synechococcus spp. and A. nidulans, respectively.
PCR and end point TNA.
PCR primers and labeled probes (Table 1) were designed for pelagic freshwater Synechococcus spp. using PCRplan from PCGene (version 6.7; IntelliGenetics, Mountain View, Calif.) and Primer Express primer design software (version 1.0; PE Biosystems, Foster City, Calif.). The PCR mixtures contained (in 25 μl) 50 to 800 nM primers (from PE Biosystems, Weiterstadt, Germany, or Interactiva, Ulm, Germany), 1.5 to 7.5 mM Mg2+, 2.5 μl of 10× reaction buffer without Mg2+, and 0.625 U of Taq polymerase from either Qiagen, Hilden, Germany, or Sigma, Deisenhofen, Germany. Genomic target DNA was added as indicated in the figure legends. For TNA, PCR was performed in the presence of 20 or 50 nM double-labeled oligonucleotide probe synthesized by PE Biosystems, Weiterstadt, Germany. The probes were labeled with FAM as the reporter and TAMRA as the quencher and were 3′ phosphorylated to prevent polymerization by Taq polymerase. As the TaqMan probe is hydrolyzed only when bound to the target DNA, annealing of primers and probe and the polymerization by Taq polymerase were performed in a combined annealing-extension step in a two-step cyclic protocol in a PTC-100 thermal cycler (MJ Research, Inc.). After initial denaturation at 95°C (3 min), the program comprised 30 cycles of 1.5 to 3 min of annealing-extension at various temperatures and denaturation (40 s) at 94°C. All reactions were terminated by polymerization at 70°C for 5 min.
TABLE 1.
Primers and probes used in TNAs
| Primer or probea | Sequence (5′→3′)b | Tm (°C)c |
|---|---|---|
| Primers | ||
| PITSANF | CGTAACAAGGTAGCCGTAC | 50 |
| PITSEND | CTCTGTGTGCCAAGGTATC | 50 |
| P8807AP | CATTCTTGACAAGTTAACCAGTTAGCTG | 60 |
| P8807AM | CAAGGTTCTGCTGACATTCAAACA | 60 |
| P8807PE | GTGATCTTGACTGTTGTTCGCTGA | 60 |
| P100PA | GGTTTAGCTCAGTTGGTAGAGCGC | 61 |
| P3 | TTGGATGGAGGTTAGCGGACT | 60 |
| Probes | ||
| S8807 | R–ATTTGTGCT–QCTCGGCTTTACCCGC | 67 |
| S8807A | R–TCTCCAGGGCAGCATTGAATCCAG–Q | 67 |
| S100A | R–CTTTGCAAGCAGGATGTCAGCGGTT–Q | 68 |
Analysis of PCR products.
After 30 cycles, PCR products (2 μl) were analyzed by agarose gel electrophoresis (1% agarose, 1× TAE buffer containing 40 mM Tris acetate and 2 mM EDTA). λ DNA restricted with Pst_I served as molecular size markers. The gel was recorded and stored with a gel reading program (ImageQuant by Molecular Dynamics). Relative amounts of amplified DNA were estimated from the intensities integrated over the area covered by an ethidium bromide-stained PCR product. For measurement of the fluorescence of probes and their hydrolyzed products, 20 μl of the TNA was diluted 10-fold with distilled water and excited at 480 nm. Fluorescence spectra between 515 and 585 nm were recorded with a fluorescence spectrophotometer (F-2000; Hitachi, Colora, Germany) using a correction function to adjust for decreased detector sensitivity at long wavelengths (8). We measured the fluorescence emission of the reporter (R) FAM at 521 nm and that of the quencher (Q) TAMRA at 580 nm. The R fluorescence was normalized to the Q signal. Normalization was validated using a Gaussian peak-fitting program (Origin 5.0; Microcal Software Inc., Northampton, Mass.). In accordance with Livak et al. (19), the normalized fluorescence signal of the TNA was calculated as Δ_RQ = (_R_DNA/_Q_DNA) − (_R_0/_Q_0).
Real-time monitoring of TNA.
TNA mixtures contained (in 25 μl) 300 nM primers, 12.5 μl of TaqMan universal PCR master mix (PE Biosystems, Foster City, Calif.; including Mg2+ at a final concentration of 5 mM, ROX as an internal fluorescence reference, a deoxynucleoside triphosphate (except dTTP), dUTP, AmpliTaq Gold DNA polymerase for hot-start PCR, and AmpErase UNG), and various amounts of genomic DNA. For real-time monitoring of TNA, 50 nM fluorescent probe was added. The assay mixture was pretreated for 2 min at 50°C and for 10 min at 95°C before 45 cycles of a two-step cycling program (annealing and extension at 60°C for 60 s and 15 s of denaturation at 95°C) was completed. Fluorescence was measured at the end of the annealing-extension phase of each cycle using the GeneAmp 5700 Sequence Detection System (PE Biosystems, Foster City, Calif.). Data were exported in Origin format and converted to graphics in Origin, version 5. A threshold value for the fluorescence of all samples was set manually in accordance with the instruction manual of the GeneAmp 5700 Sequence Detection System. The reaction cycle at which the TNA exceeded this fluorescence threshold was identified as threshold cycle CT (13) and was used for construction of standard curves for quantitative PCR.
RESULTS
Specificity of TaqMan probes S8807 and S8807A.
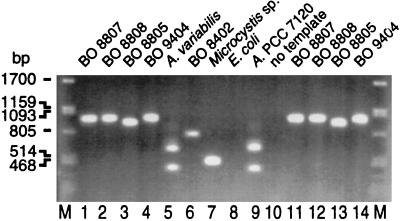
As a first step in our investigation, we compared two TaqMan probes constructed for the specific detection of Synechococcus sp. strain BO 8807 in a PCR performed with primers suitable for all Synechococcus spp. isolated from Lake Constance. The PCR primers (PITSANF and PITSEND [Table 1]) targeted sequences at the 3′ end of 16S rDNA and the 5′ end of 23S rDNA, and the probes targeted sequences in noncoding sections of the ITS-1 unique to strain BO 8807. In probe S8807, the quencher dye (TAMRA) was located at nucleotide 9, near the center of the oligonucleotide. Mismatches with other phycoerythrin-rich Synechococcus isolates were located close to the 5′ end of the oligonucleotide between the reporter (FAM) at the 5′ terminal and the quencher (Fig. 1A). This probe was constructed to perform allelic discrimination as suggested by Lee et al. (18). In probe S8807A, the fluorescent dyes were located at the 5′ (FAM) and 3′ (TAMRA) ends of the oligonucleotide, respectively, and a single central mismatch was anticipated for strains other than target strain BO 8807 (Fig. 1B). This construction followed instructions for optimum target specificity described by Livak et al. (20). End point TNA performed with the primers and probes yielded single amplicons comprising the ribosomal ITS-1 from Synechococcus spp. (Fig. 2, lanes 1 to 4 and 11 to 14), Synechocystis sp. strain BO 8402 (lane 6), and Microcystis sp. (lane 7). Multiple ribosomal operons of Anabaena spp. were amplified in some reactions by PITSANF and PITSEND, but this did not occur consistently under our assay conditions (Fig. 2, compare lanes 5 and 9). E. coli DNA (lane 8) was not amplified with these primers. The amount of amplicon produced after 30 cycles and detected by agarose gel electrophoresis was compared with the normalized fluorescence signal Δ_RQ_ produced by the hydrolysis of labeled TaqMan probes (Table 2). In PCR mixtures containing DNAs from organisms other than phycoerythrin-rich Synechococcus spp., no significant Δ_RQ_ signal was measured; i.e., probes S8807 and S8807A were not hydrolyzed. In particular, the probes discriminated against phycocyanin-rich Synechococcus strain BO 8805 from Lake Constance, although the ITS-1 of this strain was amplified during the PCR. However, among phycoerythrin-rich Synechococcus spp., probe S8807 exhibited significant cross-reactivity with templates from strains other than BO 8807: a fraction of probe S8807 was hydrolyzed (Δ_RQ_, >0.05) when DNA from either strain BO 8808 or BO 9404 was amplified. Both strains exhibit two mismatched nucleotides in the target area of the probe (Fig. 1A). In contrast, probe S8807A, constructed with a single central mismatch for nontarget DNA, exhibited no significant hydrolysis in the presence of DNA of strains other than BO 8807 in end point determinations.
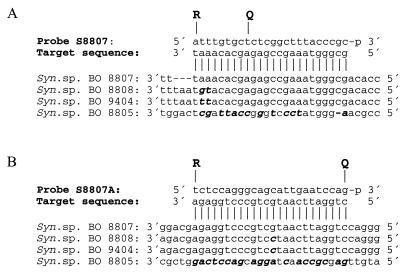
FIG. 1.
TaqMan probes used in this study. Shown are the positions of reporter (R) and quencher (Q) of probes S8807 (A) and S8807A (B) and alignment with complementary strands of target sequences in Synechococcus (Syn.) sp. strains BO 8807, BO 8808, BO 9404, and BO 8805. Mismatches are in boldface and in italics.
FIG. 2.
Ethidium bromide-stained PCR products from end point TNAs. Two-microliter volumes were analyzed in a 1% agarose gel. Lanes: M, λ DNA digested with _Pst_I; 1 to 4, DNAs from Synechococcus sp. strains BO 8807, BO 8808, BO 8805, and BO 9404; 5, A. variabilis strain ATCC 29413; 6, Synechocystis sp. strain BO 8402; 7, Microcystis sp.; 8, E. coli strain K-12; 9, Anabaena sp. strain PCC 7120; 10, no-template control; 11 to 14, Synechococcus spp. Assay mixtures (25 μl) contained 0.625 U of Taq polymerase and primers PITSANF and PITSEND (200 nM each), amplifying the ITS-1 in the ribosomal operon of many cyanobacteria. The assay mixtures analyzed in lanes 1 to 9 contained 10 ng of DNA, 1.5 mM Mg2+, and 20 nM probe S8807. The assay mixtures analyzed in lanes 11 to 14 contained 1 ng of DNA, 2.5 mM Mg2+, and 50 nM probe S8807A. Templates were amplified in 30 cycles using a two-step PCR program (3 min of annealing and polymerization at 59°C, denaturation at 94°C).
TABLE 2.
Comparison of the amount of amplified DNA and the fluorescence signal in end point TNAsa with probes S8807 and S8807A
| Expt and templateb | Amplified DNA | Probed | Fluorescence signal | ||
|---|---|---|---|---|---|
| Arbitrary areac | % | Avg arbitrary Δ_RQ_ ± SD | % | ||
| A | |||||
| BO 8807 | 7.1 | 92.2 | S8807 | 1.02 ± 0.17 | 100 |
| BO 8808 | 7.3 | 94.8 | S8807 | 0.22 ± 0.06 | 21.6 |
| BO 8805 | 7.1 | 92.2 | S8807 | 0.02 ± 0.04 | 2.0 |
| BO 9404 | 7.3 | 94.8 | S8807 | 0.17 ± 0.04 | 16.7 |
| A. variabilis | NDe | ND | S8807 | 0.03 ± 0.02 | 2.9 |
| BO 8402 | 4.6 | 59.7 | S8807 | 0.01 ± 0.03 | 1.0 |
| Microcystis sp. | 7.7 | 100 | S8807 | 0 ± 0.04 | 0 |
| E. coli | 0 | 0 | S8807 | 0 ± 0.08 | 0 |
| Anabaena strain PCC 7120 | ND | ND | S8807 | 0.01 ± 0.03 | 1.0 |
| None | 0 | 0 | S8807 | 0 | |
| B | |||||
| BO 8807 | 6.2 | 88.6 | S8807A | 0.8 ± 0.02 | 100 |
| BO 8808 | 5.9 | 84.3 | S8807A | −0.01 ± 0.04 | |
| BO 8805 | 5.4 | 77.1 | S8807A | −0.03 ± 0.04 | |
| BO 9404 | 7.0 | 100 | S8807A | 0.02 ± 0.03 | 2.5 |
Melting behavior of probes S8807 and S8807A.
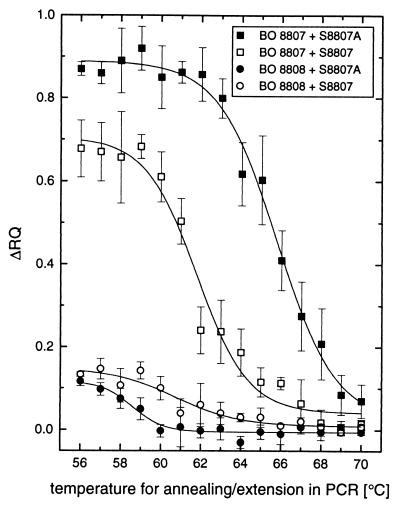
In order to understand the different specificities of probes S8807 and S8807A, the melting behavior of the two probes was evaluated. End point TNAs were conducted at different annealing-polymerization temperatures with DNAs from strains BO 8807 (perfect match) and BO 8808 (two and one mismatches with S8807 and S8807A, respectively). Amplification of the ITS-1 at various annealing and extension temperatures in a two-step PCR program was evaluated by agarose gel analysis. The PCR primer concentration was increased at annealing temperatures higher than 65°C in order to yield identical amounts of PCR products as judged from ethidium bromide-stained gels (data not shown). The Δ_RQ_ signal, representing the amount of TaqMan probe that hybridized to the target DNA during annealing-extension in the PCR was plotted against the temperature. In the presence of target DNA from strain BO 8807, the hydrolysis of the probes by Taq nuclease activity was strongly affected by the annealing-polymerization temperature, generating sigmoidal melting curves with _Tm_s of 61 and 65°C for probes S8807 and S8807A, respectively (Fig. 3). Below 60°C, probe S8807A exhibited partial annealing to a mismatched target sequence in Synechococcus sp. strain BO 8808 (Fig. 1), leading to a weak fluorescence signal, but no signal was observed at 60°C or higher temperatures. In contrast, probe S8807 exhibited significant hydrolysis in the presence of DNA from strain BO 8808 between 56 and 70°C. This temperature range overlaps the temperature required for efficient probe annealing to DNA of BO 8807 and, hence, leads to the observed lack of specificity of probe S8807 (Table 2).
FIG. 3.
Melting curves of probes S8807 and S8807A. The melting behavior of two TaqMan probes hybridizing to template DNAs from Synechococcus spp. strains BO 8807 and BO 8808 was analyzed by end point TNA. PCR conditions: 1 ng of template DNA, 50 nM probe (S8807 or S8807A), 2.5 mM Mg2+, 0.625 U of Taq polymerase, 50 nM primers PITSANF and PITSEND at annealing-extension temperatures of 56 to 65°C, 200 nM at 66 and 67°C, 400 nM at 68 and 69°C, and 800 nM at 70°C. Cycling conditions: 1.5 min of annealing-polymerization, 30 cycles.
Detection ranges of end point TNA and real-time PCR.
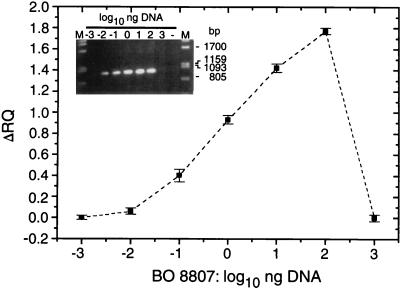
The use of TNA offers the advantage of direct quantification of the amplicon concentration by measurement of the fluorescence of the hydrolyzed TaqMan probe. We examined the detection range in end point TNA and real-time PCR. End point TNA was set up for 10−3 to 103 ng of genomic DNA from Synechococcus sp. strain BO 8807, which represented 3 × 102 to 3 × 108 copies of the genome, assuming a genome size of approximately 3 Mbp. In a 30-cycle PCR, 10−2 ng of DNA, corresponding to 3 × 103 copies of the genome, represented the lower limit of detection on agarose gel, as well as in TNA (Fig. 4). Partial inhibition of the TNA, detected as a deviation of signal size from the expected log-linear increase, occurred when 102 ng of DNA was added to the assay mixture. The greatest amount of phenol-extracted DNA used in these experiments, 1 μg, apparently introduced PCR inhibitors that led to elimination of amplification and a strong decrease in Δ_RQ_, although the reaction was performed with a higher Mg2+ concentration (2.5 mM) than usually suggested (Fig. 4). The result showed a log-linear correlation between Δ_RQ_ and the DNA amount in the initial assay of less than 3 orders of magnitude. Thus, in a 30-cycle protocol, quantification was possible for 3 × 104 to 3 × 106 genomes per assay. Higher sensitivity can be achieved by increasing the number of PCR cycles, but in this case, PCR will reach a plateau in which the correlation between a high initial template number and the amount of product is lost (22). Thus, the detection range of end point PCR cannot be extended by increasing the PCR cycle number.
FIG. 4.
Quantification of DNA from Synechococcus sp. strain BO 8807 by end point TNA. Twenty-five-microliter Taq nuclease assay mixtures contained 10−3 to 103 ng of DNA, representing approximately 3 × 102 to 3 × 108 genome copies of Synechococcus sp. strain BO 8807; 50 nM primers PITSANF and PITSEND; 50 nM probe S8807A; 2.5 mM Mg2+; and 0.625 U of Taq polymerase. PCR conditions: annealing-polymerization at 60°C for 1.5 min, 30 cycles. The insert shows a 1% agarose gel containing 2 μl of each assay mixture per lane. PCR products were stained with ethidium bromide. Lanes M, λ DNA digested with _Pst_I.
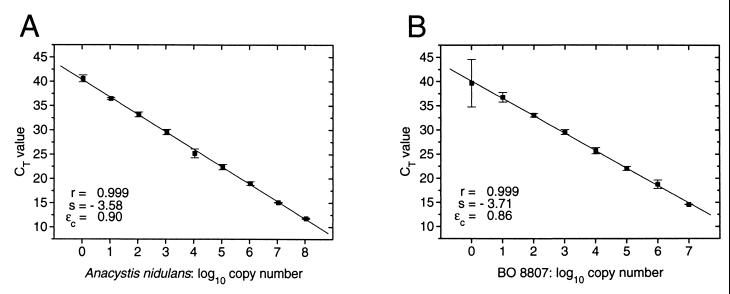
The application of the threshold cycle method in real-time PCR (14) allows calibration in a wide dynamic range in which the number of cycles can be adjusted to the signal size. We tested serial dilutions of genomic DNA from the _Synechococcus_-type cyanobacteria used in this study and obtained similar log-linear standard curves of up to 8 orders of magnitude (Fig. 5; data not shown). For each template DNA, different sets of primers and probe were used to obtain a short amplicon of 75 to 107 bp. Template numbers lower than 10 genomes per assay were detectable, as demonstrated with DNA from A. nidulans (Fig. 5A), a strain with a genome size of 2.69 Mbp and two identical ribosomal operons per genome (33, 35). The calculation of the copy number of Synechococcus sp. strain BO 8807 (Fig. 5B), which also contains two ribosomal operons (Ernst, unpublished), represents an assumption based on an arbitrarily chosen genome size of 3 Mbp.
FIG. 5.
Standard curves obtained by the CT method in real-time PCR. For TNA, 25-μl assay mixtures contained approximately 100 to 107 (108) copies of genomes, 12.5 μl of TaqMan universal PCR master mix (5 mM [final concentration] Mg2+), 300 nM primers, and 50 nM probe. Target DNA, primers (P), and probes (S) in panel A: A. nidulans, P100PA and P3, S100A. Target DNA, primers, and probes in panel B, Synechococcus sp. strain BO 8807, P8807AP and P8807AM, S8807A. The PCR comprised 45 cycles with 1 min at 60°C for annealing-polymerization and 15 s of denaturation at 95°C. Fluorescence threshold (Δ_RQ_) = 0.04; s = slope. Amplification efficiency was calculated as follows: ɛc = 10−1/s − 1.
TNA in the presence of heterologous and homologous background DNAs.
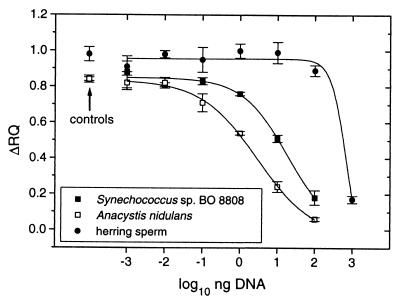
Using the calibration curves depicted in Fig. 4 for end point TNA or in Fig. 5 for real-time PCR and the highly specific TaqMan probe S8807A, quantification of amplicons from Synechococcus sp. strain BO 8807 was expected to be possible in the presence of a background of heterologous and homologous DNAs. Addition of herring sperm DNA, which yielded no detectable PCR product with the primers used (data not shown), to an assay containing 1 ng of DNA of strain BO 8807 yielded a Δ_RQ_ of approximately 1 after 30 cycles, as expected from the standard curve for end point TNA (Fig. 4). However, the signal vanished when the herring sperm DNA was added in 1,000-fold surplus (103 ng; Fig. 6). In assays containing a background of DNA from Synechococcus sp. strain BO 8808 or A. nidulans, both of which contain targets of PCR primers but are not detected by the TaqMan probe, the fluorescent Δ_RQ_ signal produced by hydrolysis of probe S8807A already decreased with much smaller additions (Fig. 6). We investigated if the primer concentration may limit the PCR in mixed assays. A fourfold higher primer concentration (200 nM) eliminated the inhibition of TNA in assays with herring sperm DNA. However, no compensation was seen in assays performed with a background of DNA from Synechococcus sp. strain BO 8808 or A. nidulans, even if the primer concentration was increased to 1 μM (data not shown).
FIG. 6.
Competitive end point TNA. Assay mixtures contained 1 ng of genomic DNA from Synechococcus sp. strain BO 8807 and 10−3 to 103 ng of DNA from Synechococcus sp. strain BO 8808, A. nidulans, or herring sperm. The PCR assay conditions comprised 30 cycles with annealing and extension at 60°C (1.5 min), 50 nM primers PITSANF and PITSEND was used for amplification of ITS-1, and 50 nM probe S8807A was used to detect strain BO 8807. Controls: Synechococcus sp. strain BO 8807 only.
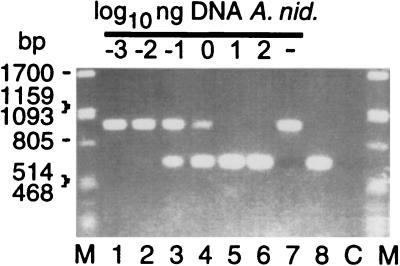
It is known that in mixtures of homologous DNAs, product formation can become biased by the competition of amplicons with PCR primers for template annealing (31, 34). The ITS-1 of A. nidulans and Synechococcus strains isolated from Lake Constance can be amplified with primers PITSANF and PITSEND (Fig. 2 and 7). As the amplicons of the phycoerythrin-rich Synechococcus spp. have sizes of 1,001 to 1,004 bp, the ITS-1 of A. nidulans (amplicon size, 637 bp) can easily be distinguished on agarose gels. Figure 7 shows that the amplicon amount produced from 1 ng of BO 8807 DNA was reduced in the presence of 1 ng of A. nidulans DNA. Detection of strain BO 8807 by PCR failed if it comprised less than 10% of the initial total amount of DNA (Fig. 7, lane 6). The experiment showed that 1 ng of A. nidulans DNA alone produced a more intense band than the amplicon obtained from 1 ng of Synechococcus sp. strain BO 8807 DNA (Fig. 7, lanes 8 and 7, respectively). During simultaneous amplification, this difference was enhanced (Fig. 7, lane 4), probably because of a higher efficiency of amplification of the shorter amplicon. This control on the gel showed that we had indeed encountered a problem caused by conditions of competitive PCR. In this case, end point TNA (Fig. 6), just as the intensity of stained fragments (Fig. 7), led to underestimation of the number of targets in the initial assay.
FIG. 7.
For quantitative PCR, a 25-μl TNA mixture contained 1 ng of genomic DNA from Synechococcus sp. strain BO 8807 and 10−3 to 102 ng of DNA from A. nidulans (lanes 1 to 6) or 1 ng of DNA from Synechococcus sp. strain BO 8807 and A. nidulans alone (lanes 7 and 8, respectively). The assay conditions are described in the legend to Fig. 6. A 2-μl volume of each assay mixture was analyzed on a 1% agarose gel and stained with ethidium bromide. λ DNA digested with _Pst_I was used as molecular size markers (lanes M). Lane C, no-template control.
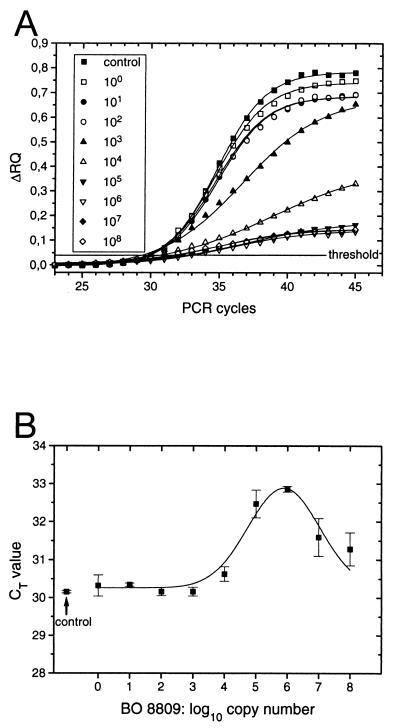
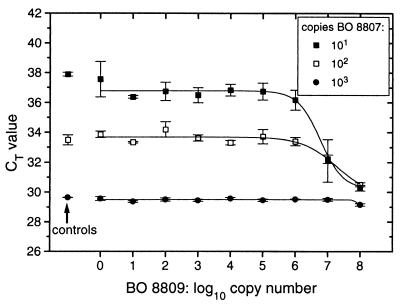
Real-time analysis of TNA showed that the problem caused by competitive PCR conditions is observed not only in end point determinations but also when the threshold cycle method is used for quantification. It furthermore showed that the problem persists even when significantly smaller amplicons are used for quantification. Figure 8A shows the amplification of a 148-bp amplicon in the ITS-1 of BO 8807 monitored by hydrolysis of the same TaqMan probe, S8807A, as used in the experiment whose results are shown in Fig. 6. Under conditions of competitive PCR, the Δ_RQ_ fluorescence signal derived from hydrolysis of probe S8807A decreased once the initial copy number of the competitor, Synechococcus sp. strain BO 8809 (with the same amplicon size of 148 bp), was similar to or higher than that of BO 8807. Figure 8B shows that, as in end point TNA, the CT value leads to underestimation of the initial target concentration. Surprisingly, at competitor copy numbers in excess of 106 the CT values appeared to decline again (Fig. 8B). However, this artifact was caused by the inability of the TaqMan technique to resolve differences in Δ_RQ_ at the threshold level in strongly inhibited reactions. Note that due to the higher sensitivity of TNA calibrated by the threshold value method, these assay mixtures contained a 300-fold lower initial concentration of BO 8807 as template than the assay mixtures used in the experiment whose results are depicted in Fig. 6.
FIG. 8.
Real-time competitive PCR (A) and corresponding CT values (B). Assay mixtures contained approximately 103 copies of Synechococcus sp. strain BO 8807 DNA (control) and approximately 100 to 108 copies of Synechococcus sp. strain BO 8809 as a competitor, 300 nM primers P8807AM and P8807PE, 50 nM probe S8807A, and 12.5 μl of TaqMan universal PCR master mix (5 mM [final concentration] Mg2+). The assays were run for 45 cycles with 1 min of annealing and extension at 60°C and 15 s of denaturation at 95°C. The threshold was set to Δ_RQ_ = 0.04. The control was Synechococcus sp. strain BO 8807 only.
Real-time quantitative PCR with specific primers.
In order to avoid competitive assay conditions, a new PCR primer, P8807AP, was designed. This primer exhibited a minimum of three mismatches with homologous DNAs from phylogenetically closely related Synechococcus isolates from Lake Constance. Using this primer, correct quantification of 10 copies of BO 8807 in assays containing up to 105 copies of Synechococcus sp. strain BO 8809 was achieved (Fig. 9). The same template-to-background ratio of 1:104 was observed in assay mixtures containing 102 or 103 copies of Synechococcus sp. strain BO 8807. At higher ratios, the CT value decreased, falsely indicating higher initial template numbers. In a background of the phylogenetically more distant strain A. nidulans (from Waller Creek, Tex.) and Synechococcus sp. strain BO 8805 (a phycocyanin-rich strain isolated from Lake Constance), both lacking a sequence complementary to that of the probe in the amplified fragment, 10 copies of Synechococcus sp. strain BO 8807 were detected at target/background ratios of 1:107 and 1:105, respectively (data not shown).
FIG. 9.
Detection of Synechococcus sp. strain BO 8807 DNA in the presence of similar DNA (7% sequence divergence in the ITS-1) using PCR primer P8807AP. Approximately 100 to 108 copies of Synechococcus sp. strain BO 8809 DNA were added to approximately 101 (■), 102 (□), or 103 (●) genome copies of Synechococcus sp. strain BO 8807. The control was Synechococcus sp. strain BO 8807 only. A 25-μl TNA mixture contained the DNA, 300 nM primers P8807AP and P8807AM, 50 nM probe S8807A, and 12.5 μl of TaqMan universal PCR master mix (5 mM [final concentration] Mg2+). The PCR comprised 45 cycles of 1 min of annealing-polymerization at 60°C and 15 s of denaturation at 95°C. The threshold value Δ_RQ_ was 0.04.
DISCUSSION
In a TNA, hydrolysis of a labeled oligonucleotide is used to monitor the amplification of PCR products to which this oligonucleotide is bound during polymerization by Taq polymerase. This method seemed to be suitable for analysis of the abundance of specific genotypes in natural populations or communities that contain numerous homologous sequences of phylogenetically related organisms.
In a case study using Synechococcus strains isolated from the autotrophic picoplankton of Lake Constance, we intended to quantify a particular strain in a background of phylogenetically closely related strains. Comparison of ribosomal sequences of isolated strains provided information required to design specific PCR primers and TaqMan probes targeting variable regions in the ITS-1.
Comparison of the two TaqMan probes, S8807 and S8807A, revealed surprisingly dissimilar capacities for allelic discrimination. The two probes were selected to have the same Tm (67 or 64°C when calculated using the program Primer Express or PCR Plan from PCGene; see Materials and Methods). Probe S8807, exhibiting two mismatches with target DNA of phycoerythrin-rich Synechococcus strains other than BO 8807, was less specific than probe S8807A, with only one mismatch with these strains. In probe S8807, the mismatches were located in an AT-rich environment close to the 5′ end (Fig. 1A). The calculated Tm for a shortened 18-bp oligonucleotide comprising the GC-rich part of the probe is 59°C (PCGene), close to the measured Tm of this probe of 61°C (Fig. 3). Possibly, the asymmetric distribution of AT- and GC-rich domains caused not only significant deviation of the calculated and measured _Tm_s (Table 1 and Fig. 3) but also broadening of the temperature range for partial hybridization with mismatched target DNA (Fig. 3) and, hence, lack of specificity in a TNA (Table 2). In probe S8807A, the mismatched base divides the probe into two parts with significantly lower _Tm_s than the matching probe, a characteristic that was also found to provide high specificity in fluorescence in situ hybridization technology (2). These results indicate that the probe oligonucleotide should not target a sequence delimited by AT-rich sections because a significant fraction of the probe is able to bind to target sequences that exhibit mismatches in these sections.
We demonstrated that in a TNA, short and long DNA fragments can be used to establish calibration curves for quantitative PCR (Fig. 4 and 5). Our experiments showed that an end point TNA is not more sensitive than gel-based PCR product analysis (Fig. 4). However, the comparison of the end point TNA with continuously monitored real-time PCR demonstrated that the latter allows calibration over a much wider range of initial template concentrations. The log-linear range of end point PCR is limited to 2 to 3 orders of magnitude due to a saturation-like plateau reached by PCR products after a number of cycles (also Fig. 8A). This plateau obliterates the semiquantitative correlation between the initial template concentration and the final amplicon concentration observed in earlier cycles (22). The problem caused by the plateau is avoided by calibrating PCRs using the threshold cycle method (13). At the fluorescence threshold, the TNA contains similar concentrations of the reported amplicons, irrespective of the initial template concentration. In PCR assays, in which a unique template is amplified, template numbers ranging over 7 orders of magnitude can be measured in a 25-μl assay with a lower limit of less than 10 genomic copies per assay (Fig. 5). In samples that contain DNAs from different organisms, PCR-based quantification techniques can become severely biased (22, 34). This is also true for quantitative PCR using a TNA.
We encountered three sources of error: (i) loss of signal due to a large surplus of complex DNA; (ii) false-positive signals and, hence, overestimation of the target DNA due to limited probe specificity; and (iii) problems caused by competitive PCR conditions.
Problem i, loss of a signal due to the presence of a large surplus of highly complex DNA (Fig. 6), was solved by increasing the primer concentration. This indicates that the problem was caused by primers binding to the background DNA without producing significant amounts of amplicons. Note that in the absence of a sufficient number of targets, the increase in the PCR primer concentration will lead to increased production of primer artifacts, the so-called primer dimers. Therefore, it is not recommended to increase primer concentrations for standard conditions.
Problem ii, wrong TNA signals caused by hybridization of the TaqMan probe to sequences that exhibit one or two mismatches, was demonstrated in Table 2 for a TaqMan probe that lacked specificity but also in Fig. 9 using a highly specific probe. In principle, the sensitivity of PCR in combination with the wide detection range of the threshold cycle calibration method allows detection of very small fractions of total DNA (Fig. 9). However, if the background contains a large excess of very similar target sequences, i.e., a 105-fold excess of DNA exhibiting three mismatches in the forward primer and one mismatch in the TaqMan probe, a small fraction of oligonucleotides will show mismatched binding and contribute to the signal generated by the specific target. The result, a decrease in the threshold cycle number corresponding to an increased number of copies of target DNA of strain BO 8807 (Fig. 9, compare calibration in Fig. 5B), produces overestimation of the target strain in the assay. The problem caused by the finite specificity of primer and probe oligonucleotides is probably enhanced by the high concentration of Mg2+ (5 mM) that is required in a TNA to improve quenching of reporter fluorescence in the intact probe (19).
Problem iii was caused by accumulation of amplicons not detected by the TaqMan probe under competitive PCR conditions. Competition occurs if different strains have identical target sequences for PCR primers, for example, if the PCR primers target highly conserved sequences in the rDNA. Competitive conditions caused an apparent inhibition of amplification (lowered band intensity in Fig. 7), lower Δ_RQ_ in end point TNA (Fig. 6, compare Fig. 4 for calibration), and a CT increase in a real-time PCR (Fig. 8B, compare Fig. 5 for calibration). In most assays, this led to a significant underestimation of the copy number (concentration) of target DNA. Real-time analysis showed that the apparent decrease in the signal is caused by a decrease in amplification efficiency (Fig. 8A). This invalidates the use of the calibration shown in Fig. 5B.
Amplification efficiency can be calculated by two methods derived from different PCR models. Model 1 assumes a constant amplification efficiency, ɛc (0 < ɛc <1), for all PCR cycles up to the threshold cycle. The efficiency of a particular primer-probe-template combination (ɛc) can be calculated from a standard curve by transforming the equation Tn = T0(1 + ɛc)n describing the amount of template (Tn) reached after n PCR cycles (17). This equation can be solved to ɛc = 10−1/s − 1 using the slope (s) of the standard curves depicted in Fig. 5. The constant efficiency (ɛc) varied for different primer pairs and templates (Fig. 5 and data not shown).
More realistic than model 1 is the supposition that competition between primers and amplicons for template binding can be neglected during initial cycles but increases during a PCR due to the accumulation of amplicons. Schnell and Mendoza (31) compared the competition between PCR primers and single-stranded amplicons during the annealing phase of a PCR with the competition of substrates and products for a binding site of an enzyme. In both cases, the competition decreases the efficiency as the reaction reaches a plateau (equilibrium). Using the formalism of Michaelis-Menten kinetics, Schnell and Mendoza showed that the efficiency of a reaction in the _i_th cycle (ɛi) depends on a rate constant (Km) and the template concentration [_Ti_], as shown by the relationship ɛi ≅ Km ([_Ti_] + Km)−1. We estimated Km (Ti is equivalent to Δ_RQ_i; Δ_RQ_0 = 10−10) using a nonlinear least-squares fit of the equation T(i+1) = Ti (1 + Km ([Ti_] + Km)−1) between cycles 1 and 35 and calculated the amplification efficiency (ɛ0.04) at Δ_RQ = 0.04, the fluorescence value used to determine the CT. Reaction efficiencies calculated by this method (Table 3) were lower than those calculated from the standard curve (Fig. 5B), even if no competitor was present, indicating that the PCR had already started to deviated from 100% efficiency at CT. Any further decrease in ɛ indicates a bias that can be caused by the presence of competing templates not recognized by the probe (Fig. 8A) or the presence of polymerase inhibitors (data not shown). In either case, the standard curve is not valid for quantification and, hence, the initial template concentration cannot be determined.
TABLE 3.
Decrease in amplification efficiency caused by coamplified competitor DNA in a real-time competitive PCRa
| Competitor DNA copy no.b | Avg Kmc ± SD | Avg ɛd ± SD |
|---|---|---|
| 0 | 0.1012 ± 0.0018 | 0.7166 ± 0.0036 |
| 100 | 0.0926 ± 0.0031 | 0.6983 ± 0.0071 |
| 101 | 0.0849 ± 0.0055 | 0.6793 ± 0.0140 |
| 102 | 0.0901 ± 0.0033 | 0.6925 ± 0.0076 |
| 103 | 0.0543 ± 0.0021 | 0.5757 ± 0.0095 |
| 104 | 0.0183 ± 0.0003 | 0.3143 ± 0.0036 |
| 105 | 0.0094 ± 0.0004 | 0.1896 ± 0.0068 |
| 106 | 0.0084 ± 0.0000 | 0.1736 ± 0.0004 |
| 107 | 0.0124 ± 0.0011 | 0.2364 ± 0.0162 |
| 108 | 0.0120 ± 0.0012 | 0.2300 ± 0.0181 |
In analysis of microbial communities, a large detection range and high specificity are essential prerequisites. Although these two criteria are met by real-time PCR, we showed that when TaqMan technology is used, different biases may obscure quantification. Under conditions leading to decreased reaction efficiency, caused by amplicons not detected by the TaqMan probe but recognized by the PCR primers, the concentration of a target genome will be underestimated (Fig. 8B). On the other hand, the finite specificity of probes and primers will lead to overestimation of the target genome in the presence of a large surplus of DNA from phylogenetically very similar organisms (Table 2 and Fig. 9). Nevertheless, real-time PCR performed with primers exhibiting specificity identical to or higher than that of the probes used to monitor the amplification currently represents the only method allowing quantification of genomes in a complex and highly variable background such as that anticipated for microbial communities.
ACKNOWLEDGMENTS
We are grateful to PE Biosystems, Weiterstadt, Germany, for enabling the utilization of the GeneAmp 5700 Sequence Detection System and thank P. Herman, Yerseke, The Netherlands, for help in data evaluation.
This work was supported by the Deutsche Forschungsgemeinschaft through Sonderforschungsbereich 454, Bodenseelitoral.
Footnotes
†
Publication 2667 of the NIOO-Centre for Estuarine and Coastal Ecology.
REFERENCES
- 1.Albis-Camps M, Blasczyk R. Fluorotyping of HLA-DRB by sequence-specific priming and fluorogenic probing. Tissue Antigens. 1999;53:301–307. doi: 10.1034/j.1399-0039.1999.530312.x. [DOI] [PubMed] [Google Scholar]
- 2.Amann R, Snaidr J, Wagner M, Ludwig W, Schleifer K-H. In situ visualization of high genetic diversity in a natural microbial community. J Bacteriol. 1996;178:3496–3500. doi: 10.1128/jb.178.12.3496-3500.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Assmann D. Nahrungsselektion und Nahrungsverwertung chroococcaler Cyanobakterien durch heterotrophe Nanoflagellaten. Ph.D. thesis. Constance, Germany: Universität Konstanz; 1998. [Google Scholar]
- 4.Beard S J, Handley B A, Hayes P K, Walsby A E. The diversity of gas vesicle genes in Planktothrix rubescens from Lake Zurich. Microbiology. 1999;145:2757–2768. doi: 10.1099/00221287-145-10-2757. [DOI] [PubMed] [Google Scholar]
- 5.Bright D I, Walsby A E. The relationship between critical pressure and width of gas vesicles in isolates of Planktothrix rubescens from Lake Zurich. Microbiology. 1999;145:2769–2775. doi: 10.1099/00221287-145-10-2769. [DOI] [PubMed] [Google Scholar]
- 6.Desjardin L E, Chen Y, Perkins M D, Teixeira L, Cave M D, Eisenach K D. Comparison of the ABI 7700 system (TaqMan) and competitive PCR for quantification of IS6110 DNA in sputum during treatment of tuberculosis. J Clin Microbiol. 1998;36:1964–1968. doi: 10.1128/jcm.36.7.1964-1968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckert K A, Kunkel T A. DNA polymerase fidelity and the polymerase chain reaction. PCR Methods Appl. 1991;1:17–24. doi: 10.1101/gr.1.1.17. [DOI] [PubMed] [Google Scholar]
- 8.Ernst A. Cyanobacterial picoplankton from Lake Constance. I. Isolation by fluorescence characteristics. J Plankton Res. 1991;13:1307–1312. [Google Scholar]
- 9.Ernst A, Becker S, Hennes K, Postius C. Is there a succession in the autotrophic picoplankton of temperate zone lakes? In: Bell C R, Brylinski M, Johnson-Green P, editors. Microbial biosystems: new frontiers. Proceedings of the 8th International Symposium on Microbial Ecology. Halifax, Nova Scotia, Canada: Atlantic Canada Society for Microbial Ecology; 2000. pp. 623–629. [Google Scholar]
- 10.Ernst A, Postius C, Böger P. Glycosylated surface proteins reflect genetic diversity among Synechococcus species of Lake Constance. Arch Hydrobiol Spec Issues Adv Limnol. 1996;48:1–6. [Google Scholar]
- 11.Gelmini S, Orlando C, Sestini R, Vona G, Pinzani P, Ruocco L, Pazzagli M. Quantitative polymerase chain reaction-based homogeneous assay with fluorogenic probes to measure c-erbB-2 oncogene amplification. Clin Chem. 1997;43:752–758. [PubMed] [Google Scholar]
- 12.Gilliland G, Perrin S, Blanchard K, Bunn H F. Analysis of cytokine mRNA and DNA: detection and quantitation by competitive polymerase chain reaction. Proc Natl Acad Sci USA. 1990;87:2725–2729. doi: 10.1073/pnas.87.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heid C A, Stevens J, Livak K J, Williams P M. Real-time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 14.Higuchi R, Dollinger G, Walsh P S, Griffith R. Simultaneous amplification and detection of specific DNA sequences. BioTechnology. 1992;10:413–417. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- 15.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′→3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Innis M A, Myambo K B, Gelfand D H, Brow M D. DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc Natl Acad Sci USA. 1988;85:9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein D, Janda P, Steinborn R, Müller M, Salmons B, Günzburg W H. Proviral load determination of different feline immunodeficiency virus isolates using real-time polymerase chain reaction: influence of mismatches on quantification. Electrophoresis. 1999;20:291–299. doi: 10.1002/(SICI)1522-2683(19990201)20:2<291::AID-ELPS291>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 18.Lee L G, Connel C R, Bloch W. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res. 1993;21:3761–3766. doi: 10.1093/nar/21.16.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak K J, Flood S J A, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 20.Livak K J, Marmaro J, Todd J A. Towards fully automated genome-wide polymorphism screening. Nat Genet. 1995;9:341–342. doi: 10.1038/ng0495-341. [DOI] [PubMed] [Google Scholar]
- 21.Meyerhans A, Vartanian J-P, Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;18:1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrison C, Gannon F. The impact of the PCR plateau phase on quantitative PCR. Biochim Biophys Acta. 1994;1219:493–498. doi: 10.1016/0167-4781(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 23.Müller H. Selective feeding of a freshwater chrysomonad, Paraphysomonas sp., on chroococcoid cyanobacteria and nanoflagellates. Arch Hydrobiol Spec Issues Adv Limnol. 1996;48:63–71. [Google Scholar]
- 24.Orlando C, Pinzani P, Pazzagli M. Developments in quantitative PCR. Clin Chem Lab Med. 1998;36:255–269. doi: 10.1515/CCLM.1998.045. [DOI] [PubMed] [Google Scholar]
- 25.Pahl A, Kühlbrandt U, Brune K, Röllinghoff M, Gessner A. Quantitative detection of Borrelia burgdorferi by real-time PCR. J Clin Microbiol. 1999;37:1958–1963. doi: 10.1128/jcm.37.6.1958-1963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pallansch L, Beswick H, Talian J, Zelenka P. Use of an RNA folding algorithm to choose regions for amplification by the polymerase chain reaction. Anal Biochem. 1990;185:57–62. doi: 10.1016/0003-2697(90)90254-7. [DOI] [PubMed] [Google Scholar]
- 27.Postius C, Ernst A. Mechanisms of dominance: coexistence of picocyanobacterial genotypes in a freshwater ecosystem. Arch Microbiol. 1999;172:69–75. doi: 10.1007/s002030050742. [DOI] [PubMed] [Google Scholar]
- 28.Postius C, Ernst A, Kenter U, Böger P. Persistence and genetic diversity among strains of phycoerythrin-rich cyanobacteria from the picoplankton of Lake Constance. J Plankton Res. 1996;18:1159–1166. [Google Scholar]
- 29.Pusterla N, Huder J B, Leutenegger C M, Braun U, Madigan J E, Lutz H. Quantitative real-time PCR for detection of members of the Ehrlichia phagocytophila genogroup in host animals and Ixodes ricinus ticks. J Clin Microbiol. 1999;37:1329–1331. doi: 10.1128/jcm.37.5.1329-1331.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryncarz A J, Goddard J, Wald A, Huang M-L, Roizman B, Corey L. Development of a high-throughput quantitative assay for detecting herpes simplex virus DNA in clinical samples. J Clin Microbiol. 1999;37:1941–1947. doi: 10.1128/jcm.37.6.1941-1947.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnell S, Mendoza C. Enzymological considerations for a theoretical description of the quantitative competitive polymerase chain reaction (QC-PCR) J Theor Biol. 1997;184:433–440. doi: 10.1006/jtbi.1996.0283. [DOI] [PubMed] [Google Scholar]
- 32.Stanier R H, Kunisawa R, Mandel M, Cohen-Baziere G. Purification and properties of unicellular blue-green algae (order Chroococcales) Bacteriol Rev. 1971;35:171–205. doi: 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugiura M. From plastid to cyanobacterial genomes. In: Peschek G A, Löffelhardt W, Schmetterer G, editors. The phototrophic prokaryotes. New York, N.Y: Kluwer Academic Press; 1999. pp. 411–420. [Google Scholar]
- 34.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomioka N, Shinozaki K, Sugiura M. Molecular cloning and characterization of ribosomal RNA genes from a blue-green alga, Anacystis nidulans. Mol Gen Genet. 1981;184:259–363. [Google Scholar]
- 36.Wang A M, Doyle M V, Mark D F. Quantitation of mRNA by the polymerase chain reaction. Proc Natl Acad Sci USA. 1989;86:9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang G C-Y, Wang Y. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl Environ Microbiol. 1997;63:4645–4650. doi: 10.1128/aem.63.12.4645-4650.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Wintzingerode F, Göbel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 39.Wittwer C T, Herrmann M G, Moss A A, Rasmussen R P. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques. 1997;22:130–138. doi: 10.2144/97221bi01. [DOI] [PubMed] [Google Scholar]
- 40.Wood A M, Townsend D. DNA polymorphism within the WH7803 serogroup of marine Synechococcus spp. (cyanobacteria) J Phycol. 1990;26:576–585. [Google Scholar]