Antimicrobial resistance genes aph(3′)-III, erm(B), sul2 and tet(W) abundance in animal faeces, meat, production environments and human faeces in Europe (original) (raw)
Abstract
Background
Real-time quantitative PCR (qPCR) is an affordable method to quantify antimicrobial resistance gene (ARG) targets, allowing comparisons of ARG abundance along animal production chains.
Objectives
We present a comparison of ARG abundance across various animal species, production environments and humans in Europe. AMR variation sources were quantified. The correlation of ARG abundance between qPCR data and previously published metagenomic data was assessed.
Methods
A cross-sectional study was conducted in nine European countries, comprising 9572 samples. qPCR was used to quantify abundance of ARGs [aph(3′)-III, erm(B), sul2, tet(W)] and 16S rRNA. Variance component analysis was conducted to explore AMR variation sources. Spearman’s rank correlation of ARG abundance values was evaluated between pooled qPCR data and earlier published pooled metagenomic data.
Results
ARG abundance varied strongly among animal species, environments and humans. This variation was dominated by between-farm variation (pigs) or within-farm variation (broilers, veal calves and turkeys). A decrease in ARG abundance along pig and broiler production chains (‘farm to fork’) was observed. ARG abundance was higher in farmers than in slaughterhouse workers, and lowest in control subjects. ARG abundance showed a high correlation (Spearman’s ρ > 0.7) between qPCR data and metagenomic data of pooled samples.
Conclusions
qPCR analysis is a valuable tool to assess ARG abundance in a large collection of livestock-associated samples. The between-country and between-farm variation of ARG abundance could partially be explained by antimicrobial use and farm biosecurity levels. ARG abundance in human faeces was related to livestock antimicrobial resistance exposure.
Introduction
Antimicrobial resistance (AMR) poses a threat not only to humans but also to animals worldwide.1 High antimicrobial use (AMU) leads to the selection of resistant bacteria, which limits therapeutic options in animals and humans.
A variety of methods have been carried out to quantify AMR levels in faeces, such as conventional testing of phenotypes of antimicrobial susceptibility of selected organisms.2,3 Besides, next-generation sequencing (NGS) is emerging as a new method to detect genetic determinants conferring AMR in selected isolates or metagenomically in DNA of bacterial communities.4–6 However, due to the high costs and technological constraints of NGS methods, the number of samples per study is often limited, or animal samples are pooled together (e.g. at farm or herd level).4 Consequently, in pooled samples, within-farm variation of antimicrobial resistance gene (ARG) abundance cannot be determined. Compared with NGS, real-time quantitative PCR (qPCR) is an affordable and widely applied method that can also provide precise quantification of certain ARG targets,4 allowing comparisons of ARG abundance between and within sampling sites.
Direct and indirect exposure of humans to livestock, companion animals or animal products are known risk factors for AMR acquisition.7–9 Intensity and frequency of contact with animals have been shown to represent risk factors for carrying resistant strains such as livestock-associated MRSA (LA-MRSA) or ESBL-producing bacteria.10,11 Although the correlation of qPCR-quantified ARG abundance between animals and humans has been studied,12–14 the insights are limited by geographical distribution or sample type variation.
As part of the Ecology from Farm to Fork Of microbial drug Resistance and Transmission (EFFORT) project, we used qPCR to quantify four frequently occurring ARGs [aph(3′)-III, erm(B), sul2 and tet(W)] in a total of 9572 samples collected in nine European countries. We analysed samples from animal faeces, meat, production environments and human faeces. The objectives of the current study were: (1) to describe ARG abundance among different sample types and countries; (2) to quantify AMR variation sources (between countries, between and within farms, and determinants) based on variance component analysis (VCA); and (3) to determine the correlation between ARG abundance quantified by qPCR and metagenomics.
Materials and methods
Study population and sampling procedure
Between 2014 and 2017, we collected a large set of samples (9572 samples) from various sources. The sampling procedures have been partially described before.5,6,13,15–23 Faecal samples were taken from farm animals [pigs,5,13,23 broilers,6,23 veal calves,15 turkeys,16 fish (intestines were collected)], companion animals (cats, dogs)17 and wild boars. Carcass samples were taken from pigs13 and broilers at slaughterhouses. Raw meat (pork,13 chicken, turkey, veal and trout) samples were purchased at food stores. We also collected environmental samples [electrostatic dustfall collector (EDC)19,20 and gloves of slaughterhouse workers13] and human faeces (humans occupationally exposed to pigs or broilers at Dutch and German farms or slaughterhouses, and control subjects from the Dutch ‘Lifelines’ cohort).13,18,21,22 Animal samples were collected from nine European countries (Belgium, Bulgaria, Germany, Denmark, Spain, France, Italy, the Netherlands and Poland). More details about faecal sampling are described in the Supplementary methods, available as Supplementary data at JAC Online (Sampling procedure of faeces).
Before DNA extraction, animal faecal samples were stored at 4°C, transported to the laboratory within 24 h and stored at −80°C.4–6,15,16,23 Exposed EDC cloths were put into a small resealable bag with sterile tweezers and frozen at −80°C.19,20 Carcass, meat and glove samples were collected in a stomacher bag (INTERSCIENCE, 400 mL, UK), transported and stored at 4°C, followed by further preparations.13 Human faecal samples were refrigerated directly after collection, transported and stored on dry ice.13,18,21,22
ARG target selection
Within the EFFORT project, the ARG target selection was based on several criteria, including: (1) sufficient abundance to be measured in >25% of the samples estimated from the prevalence and relative abundance of ARG targets in previous metagenomic analyses of pig and broiler faeces, in order to enable large-scale statistical analyses of risk factors such as AMU;4 (2) the inclusion of targets of unrelated antimicrobial classes; (3) limited correlation of the chosen gene concentrations to avoid redundancy; and (4) a PCR protocol should either exist or be achievable. More details about the selection process are described in the Supplementary methods (ARG targets selection process). Although genes of particular clinical relevance (such as genes encoding resistance mechanisms in WHO priority-resistant pathogens, i.e. ESBL, carbapenemase and/or vancomycin resistance genes) were initially prioritized, they were estimated to be detectable only in <10% of samples from at least one species (based on the strength of the respective metagenomic signal). These genes were deselected as such low prevalence would limit the power of the planned statistical analyses. The resulting genes represent resistance to the antimicrobial classes aminoglycosides, macrolides, sulphonamides and tetracyclines, all of which are used in animal practice to a varying extent.
DNA extraction, qPCR and sequencing
DNA of animal and human faeces was extracted using the modified QIAamp Fast DNA Stool Mini Kit (Cat. No. 51604; QIAGEN, The Netherlands) as described before.13,15,23 DNA of EDC and gloves was respectively extracted using the modified NucleoSpin® 8 Plant II Kit and NucleoSpin® 96 Food Kit (MACHEREY-NAGEL, Germany), while DNA of meat was extracted using the modified NucleoSpin® Food Kit (MACHEREY-NAGEL).13,20 Following DNA extraction, qPCR was conducted to quantify the abundance of four ARGs [aph(3′)-III, erm(B), sul2, tet(W)] along with the 16S rRNA gene as a measure of total bacterial DNA (Table S1). More details on the qPCR process have been described in previous papers13,15,23 and the Supplementary methods (qPCR process). qPCR quality control, comprising a number of elements, is described in the Supplementary methods (Quality control and quantification of qPCR results).
ARG copy number per unit of the sample was log10 transformed. Subsequently, relative ARG abundance was calculated using 16S rRNA as a general bacterial molecular marker to normalize the bacterial community size in samples.
DNA of pooled faecal samples from pigs and broilers was extracted at the Technical University of Denmark (DTU) with the same extraction method and shipped on dry ice for shotgun metagenomic sequencing at the Oklahoma Medical Research Foundation (OMRF; Oklahoma City, OK, USA). In total, pooled faecal DNA collected from 181 pig farms and 178 broiler farms were shotgun sequenced on the HiSeq 3000 platform (Illumina), resulting in >18 billion paired-end reads. More details on the subsequent processing of the metagenomic data were described in our previous study.4–6
Comparison of ARG abundance across sample types, farms and countries
We compared ARG loads between samples from nine countries, between samples collected along Dutch pig and German broiler production lines, and between different human populations.
Principle component analyses (PCA) were performed to evaluate the similarities and differences in the distributions of relative ARG abundance across sample sources and species. The package vegan24 was used on R version 4.0.3.25 Data were log10 transformed before PCA due to the right-skewed distribution.26
Except for PCA results, all comparisons of ARG abundance in this study were conducted using a classic or Welch’s analysis of variance (ANOVA), depending on the variance homogeneity.27,28 In the case of a significant difference in ANOVA (P < 0.05), post hoc tests [Games–Howell _post hoc_ test29 or Tukey’s honest significant difference test (Tukey HSD)30] were carried out to test differences between groups. Unless otherwise specified, appropriate post hoc test P values are reported.
VCA
VCA was conducted per ARG target with a null model (AMR ∼ country + farm) to evaluate the between-country, between-farm and within-farm variation of relative ARG abundance in faecal samples of four animal species (pigs, broilers, veal calves and turkeys) using the package VCA31 on R version 4.0.3.25 In addition, to be consistent with our study in pigs and broilers,23 we also determined the contribution of farm characteristics (e.g. AMU, biosecurity measures) in veal calves and turkeys to the variance by adjusting these factors into the null model. More details are described in the Supplementary methods (Variance component analyses in veal calves and turkeys).
Correlation between pooled qPCR and pooled metagenomic data
As data were not normally distributed, Spearman’s rank correlation of relative ARG abundance was evaluated between pooled qPCR data and earlier published pooled metagenomic data.4 To match the ARG targets of qPCR, all downstream gene abundance values for aph(3′)-III, erm(B), sul2 and tet(W) were collected from the metagenomic data [fragments per kb reference per million bacterial fragments (FPKM)] and summed per gene target. FPKM was log10 transformed after adding a pseudocount of 1.
Ethics
‘ Lifelines’ research was conducted according to the protocols approved by the Medical Ethics Review Board of the Medical Center Groningen (NL) (Protocol METc2007/152). Written consent was received from all participants. The Medical Ethical Committee of the University Medical Centre Utrecht (NL) confirmed that the Dutch ‘Medical Research Involving Human Subjects Act’ did not apply for the study of the EFFORT research (Protocols 14–346/C, 14–403/C). Participants in EFFORT were compensated financially (100 Euros per farm family, 25 Euros per slaughterhouse employee).4,21
Results
Table 1 describes the 9572 samples analysed by qPCR. Human stool samples were collected in the Netherlands and Germany, including pig and broiler farmworkers and their families (n = 127), pig and broiler slaughterhouse workers (n = 669) and a healthy control population (n = 46) (Table 1 and Table S2). After a quality check involving the limit of detection (LOD) and limit of quantification (LOQ), 7084 (74%) samples had detectable gene levels for erm(B) and 6700 (70%) for tet(W), while 4892 (80%) samples for aph(3′)-III and 4543 (74%) samples for sul2 could be detected (Table 1). Only a small number of ARG targets (generally less than 10% of the total sample size) were detected in fish faeces.
Table 1.
Overview of all sample types and numbers of samples with detectable qPCR targets
| Description | Final sample country | Final sample number, n (%) | Total sample number | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 16S | aph(3′)-III | erm(B) | sul2 | tet(W) | 16S | aph(3′)-III | erm(B) | sul2 | tet(W) | ||
| Pig faeces (farms) | All nine | All nine | All nine | All nine | All nine | 1230 (97) | 1084 (86) | 1259 (100) | 1042 (82) | 1262 (100) | 1265 |
| Broiler faeces (farms) | All nine | All nine | All nine | All nine | All nine | 884 (97) | 847 (93) | 902 (99) | 840 (93) | 898 (99) | 908 |
| EDC (pig farms) | All nine | All nine | All nine | All nine | All nine | 462 (92) | 431 (86) | 477 (95) | 436 (87) | 475 (94) | 503 |
| EDC (broiler farms) | All nine | All nine | All nine | All nine | All nine | 501 (90) | 430 (77) | 530 (95) | 341 (61) | 526 (94) | 559 |
| Pooled pig faeces (farms) | All nine | All nine | All nine | All nine | All nine | 165 (92) | 168 (94) | 176 (98) | 168 (94) | 175 (98) | 179 |
| Pooled broiler faeces (farms) | All nine | All nine | All nine | All nine | All nine | 169 (95) | 170 (96) | 177 (99) | 170 (96) | 176 (99) | 178 |
| Retail pork | All nine | — | All nine | — | BG, DK, ES, PL | 848 (98) | — | 140 (16) | — | 8 (1) | 862 |
| Retail chicken | All nine | — | All nine | — | All nine | 875 (99) | — | 592 (67) | — | 383 (43) | 882 |
| Turkey faeces (farms) | DE, ES, FR | DE, ES, FR | DE, ES, FR | DE, ES, FR | DE, ES, FR | 290 (95) | 263 (87) | 299 (98) | 264 (87) | 301 (99) | 304 |
| Veal calf faeces (farms) | DE, FR, NL | DE, FR, NL | DE, FR, NL | DE, FR, NL | DE, FR, NL | 405 (96) | 339 (81) | 415 (99) | 387 (92) | 420 (100) | 420 |
| Fish faeces (farms) | ES, FR, PL | FR, PL | ES, FR, PL | ES, FR, PL | ES, PL | 270 (77) | 5 (1) | 27 (8) | 140 (40) | 6 (2) | 352 |
| Wild boar faeces | IT, PL | IT, PL | IT, PL | IT, PL | IT, PL | 156 (78) | 43 (22) | 83 (42) | 30 (15) | 197 (98) | 200 |
| Cat faeces | BE, IT, NL | BE, IT, NL | BE, IT, NL | BE, IT, NL | BE, IT, NL | 141 (95) | 117 (79) | 121 (81) | 43 (29) | 148 (99) | 149 |
| Dog faeces | BE, IT, NL | BE, IT, NL | BE, IT, NL | BE, IT, NL | BE, IT, NL | 148 (99) | 104 (69) | 92 (61) | 82 (55) | 146 (97) | 150 |
| Retail turkey | DE, ES, FR | — | DE, ES, FR | — | DE, ES, FR | 143 (99) | — | 112 (77) | — | 59 (41) | 145 |
| Retail veal | DE, ES, FR | — | DE, ES, FR | — | ES | 130 (100) | — | 38 (29) | — | 2 (2) | 130 |
| Retail fish | FR, NL, PL | — | FP, PL | — | FR | 122 (99) | — | 6 (5) | — | 1 (1) | 123 |
| Human faeces (pig farms) | NL, DE | NL, DE | NL, DE | NL, DE | NL | 93 (99) | 90 (96) | 93 (99) | 57 (61) | 93 (99) | 94 |
| Human faeces (poultry farms) | NL, DE | NL, DE | NL, DE | NL, DE | DE | 33 (100) | 33 (100) | 32 (97) | 26 (79) | 33 (100) | 33 |
| Pig faeces (slaughterhouses) | NL | NL | NL | NL | NL | 59 (98) | 58 (97) | 60 (100) | 20 (33) | 60 (100) | 60 |
| Pig carcass (slaughterhouses) | NL | — | NL | — | NL | 166 (84) | — | 28 (14) | — | 21 (11) | 198 |
| Gloves (pig slaughterhouses) | NL | — | NL | — | NL | 310 (96) | — | 178 (55) | — | 148 (46) | 324 |
| Pig meat (slaughterhouses) | NL | — | . | — | . | 190 (96) | — | — | — | — | 198 |
| Human faeces (pig slaughterhouses) | NL | NL | NL | NL | NL | 479 (99) | 441 (91) | 480 (99) | 296 (61) | 480 (99) | 483 |
| Human faeces (control) | NL | NL | NL | NL | NL | 46 (100) | 46 (100) | 46 (100) | 17 (37) | 46 (100) | 46 |
| Broiler faeces (slaughterhouses) | DE | DE | DE | DE | DE | 60 (97) | 47 (76) | 62 (100) | 55 (89) | 62 (100) | 62 |
| Broiler carcass (slaughterhouses) | DE | — | DE | — | DE | 198 (98) | — | 159 (79) | — | 146 (72) | 202 |
| Gloves (broiler slaughterhouses) | DE | — | DE | — | DE | 231 (94) | — | 208 (84) | — | 190 (77) | 247 |
| Broiler meat (slaughterhouses) | DE | — | DE | — | DE | 127 (98) | — | 109 (84) | — | 54 (42) | 130 |
| Human faeces (broiler slaughterhouses) | DE | DE | DE | DE | DE | 180 (97) | 176 (95) | 183 (98) | 129 (69) | 184 (99) | 186 |
| Overall | All nine | All nine | All nine | All nine | All nine | 9111 (95) | 4892 (80) | 7084 (74) | 4543 (74) | 6700 (70) | 9572 |
Comparison of relative ARG abundance across sample types and countries
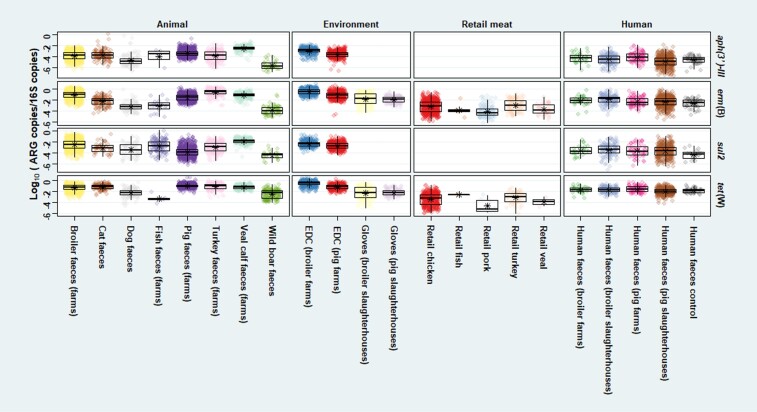
Among animal faeces, the highest mean relative abundance of tet(W) was generally seen in farm animals, especially in pigs (Figure 1). Among all sample types (Figure S1 and Table S3), the mean relative aph(3′)-III abundance was highest in broiler slaughterhouse faeces and lowest in wild boar faeces. For sul2, the mean relative abundance was lowest in wild boar faeces. The mean relative sul2 abundance was highest in veal calf faeces, which was significantly higher than all the other sample types (P < 0.01) (Figure S1 and Table S3). For _erm_(B) and _tet_(W), the mean relative abundance was highest in broiler farm dust samples and lowest in retail pork. The mean relative abundance of _tet_(W) in broiler farm dust samples was significantly higher than _tet_(W) abundance in all the other sample types (_P_ < 0.05). For all ARGs, cats showed higher mean relative abundance than dogs, but differences were not statistically significant (_P_ > 0.05) (Figure S1 and Table S3).
Figure 1.
Relative abundance of four targets [aph(3′)-III, erm(B), sul2, tet(W)] in all samples. Relative abundance of gene target was calculated by log10 (gene copies/16S copies). Asterisk shows the mean by sample type. Pooled faeces, slaughterhouse pig and broiler faeces, and slaughterhouse carcass samples were not included in this figure.
Between-country variation of relative ARG abundance in dust samples and in pig and broiler faeces is described elsewhere.20,23 For other animal species (cat, dog, veal calves), the Netherlands generally showed the lowest relative abundance of all ARGs (Figure S2). For veal calf faecal samples, we observed significantly lower relative abundance of aph(3′)-III (P < 0.05) and tet(W) (P < 0.01) in the Netherlands than in all the other countries (Figure S2).
Relative ARG abundance across the pig and broiler production chain
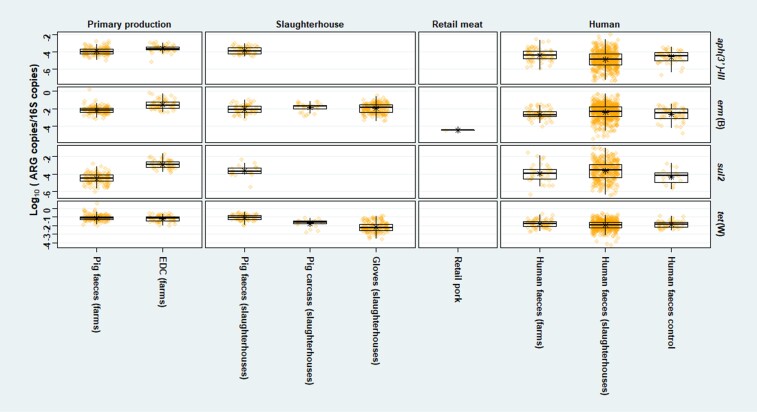
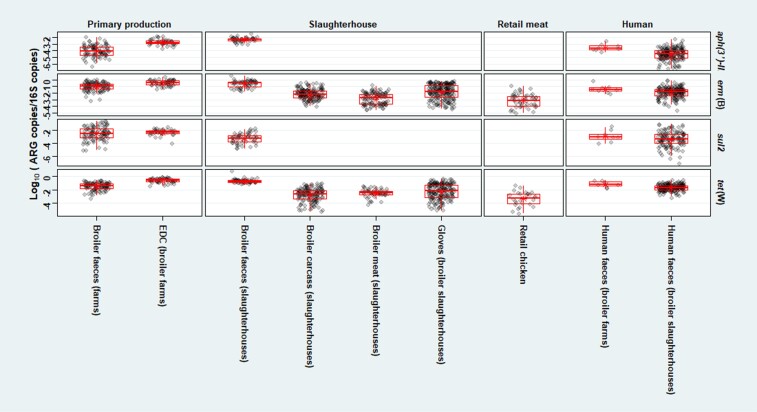
We found an overall decreasing trend of the mean relative ARG abundances along the production chain of pigs in the Netherlands and broilers in Germany, i.e. from primary production to slaughterhouse to retail meat (Figures 2 and 3). Farm dust showed higher (P < 0.01) mean relative ARG abundances than faeces recovered from the same farms, except for tet(W) in pigs. In addition, we found a significantly higher (P < 0.01) mean relative aph(3′)-III abundance in faeces of pig farmers than in faeces of pig slaughterhouse employees. A significantly higher (P < 0.05) mean relative sul2 abundance was observed in faeces of pig slaughterhouse employees than in faeces of control subjects (Figure 2). Meanwhile, faeces from broiler farmers showed a higher mean relative abundance of all ARGs than faeces from broiler slaughterhouse employees, which was significant for aph(3′)-III (P < 0.01) (Figure 3). Furthermore, we found higher mean relative erm(B) and tet(W) abundance in pig/broiler slaughterhouse carcasses than in slaughterhouse meat and retail meat, which was significant for erm(B) (P < 0.01) (Figures 2 and 3).
Figure 2.
Relative abundance of four targets [aph(3′)-III, erm(B), sul2, tet(W)] in samples related to pig production in the Netherlands. Relative abundance of gene target was calculated by log10 (gene copies/16S copies). Only pig farms and slaughterhouses in the Netherlands were involved. Human faeces of control subjects were collected from the ‘Lifelines’ cohort in the Netherlands.18 Asterisk shows the mean by sample type in the Netherlands.
Figure 3.
Relative abundance of four targets [aph(3′)-III, erm(B), sul2, tet(W)] in samples related to broiler production in Germany. Relative abundance of gene target was calculated by log10 (gene copies/16S copies). Only broiler farms and slaughterhouses in Germany were involved. Asterisk shows the mean by sample type in Germany.
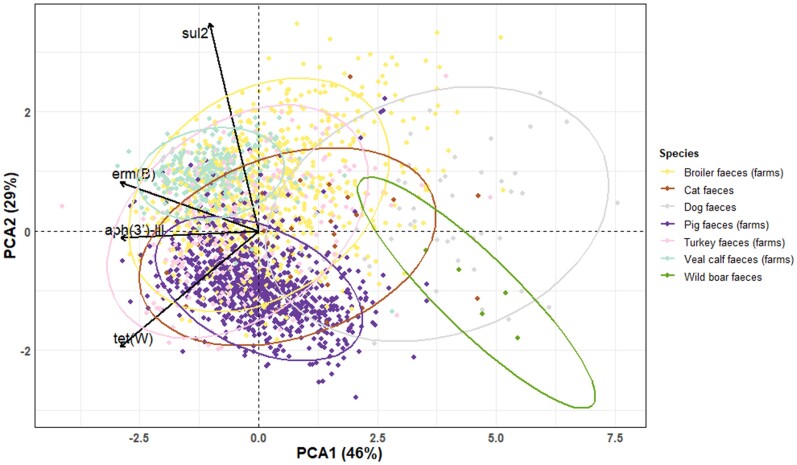
Determination of differences in ARG patterns by animal species
The faecal resistome, here measured as the frequency distributions of faecal ARGs, differed between animal species (Figure 4). A high distribution overlap of ARG abundance was found among farm animals and differed from wild boars and dogs. The results of fish are not shown in Figure 4 as there were too many missing values per gene target. In the production chain of pigs and broilers, faeces and production environment samples showed higher relative ARG abundance than human faeces in both farms and slaughterhouses (Figures S3–S6). ARG distribution overlapped obviously between carcass and gloves and differed from human stool samples (Figure S4 and Figure S6).
Figure 4.
PCA biplot of relative ARG abundance in animal faecal samples. ARG targets: aph(3′)-III, erm(B), sul2, tet(W). Relative abundance of gene target was calculated by log10 (gene copies/16S copies). Symmetric scaling was used. Circles indicate 95% confidence ellipses that were computed with the assumption of multivariate normal distribution of the data.
VCA of relative ARG abundance
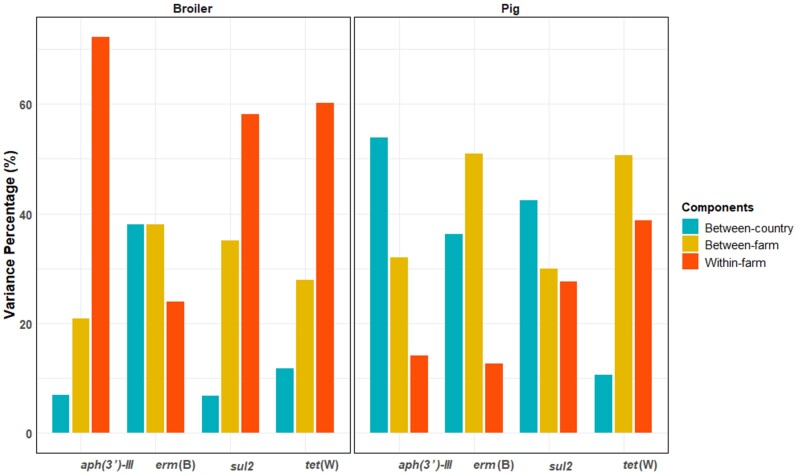
In the null model for pigs, broilers, veal calves and turkeys, we generally found the within-farm variation of AMR was low (absolutely and relatively) in pig faeces but high in faeces of the other three animal species. (Figure 5 and Table S4). In pig faeces, we found high between-country and between-farm variation. In broiler faeces, within-farm variation dominated the relative ARG abundance, while the smallest variation was found between countries (Figure 5 and Table S4).
Figure 5.
Variance component percentages of relative ARG abundance in faecal samples from pigs and broilers. Relative abundance of gene target was calculated by log10 (gene copies/16S copies). Variance percentages of three components (between-country variance, between-farm variance and within-farm variance) were calculated using VCA.
In the adjusted VCA model for veal calves and turkeys (Tables S5 and S6), we found that between-country and between-farm variation could partly be explained by AMU and biosecurity measures. For example, in the adjusted model for sul2 in veal calves, the use of trimethoprim and sulphonamide accounted for 6.78% of the total variation, while the contribution of between-country variation decreased from 11.49% to 6.07%. In the adjusted model for erm(B) in turkeys, the biosecurity measure ‘Visitor access more than once a month’ accounted for 13.70% of the total variation, while the contribution of between-farm variation decreased from 46.56% to 30.66%. For pigs and broilers, the adjusted VCA models are described elsewhere.23
Comparison of ARG abundance between qPCR data and metagenomic data
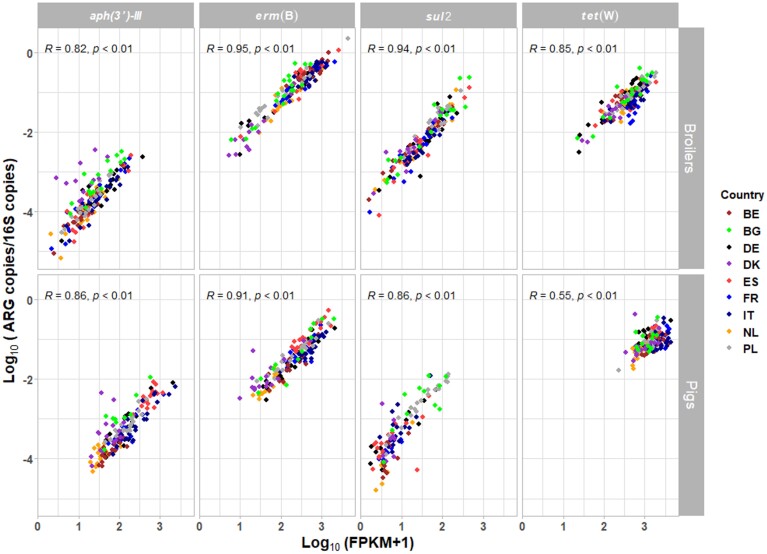
In total, 4, 15, 19 and 5 gene variants in the metagenomic data were allocated to aph(3′)-III, erm(B), sul2 and tet(W), respectively. For pigs and broilers, we found a high correlation (ρ>0.7; P < 0.01) between qPCR data and metagenomic data of pooled faecal samples for the four ARG targets, except for tet(W) (ρ<0.7; P < 0.01) in pigs (Figure 6).
Figure 6.
Spearman’s rank correlation of relative ARG abundance and ARG FPKM abundance in pooled faecal samples from pig and broiler farms. ARG targets: aph(3′)-III, erm(B), sul2, tet(W). Relative abundance of gene target was calculated by log10 (gene copies/16S copies). FPKM was log10 transformed after adding a pseudocount of 1.4 BE, Belgium; BG, Bulgaria; DE, Germany; DK, Denmark; ES, Spain; FR, France; IT, Italy; NL, the Netherlands; PL, Poland.
Discussion
We applied qPCR on a large scale to explore the association between ARG abundance from different sources, animal species and humans in nine European countries in more than 9500 samples. A significant decrease in relative ARG abundance was observed along both pig and broiler production chains. In addition, the between-country and between-farm variation in pigs, broilers, veal calves and turkeys could partially be explained by AMU and biosecurity levels. Furthermore, a high correlation between qPCR and metagenomically assessed ARGs in pooled faecal samples was found in pigs and broilers.
Comparison of ARG abundance among animal species
We speculate that the observed variation of ARGs across animals is partly a result of different AMU exposure. For example, the highest mean relative tet(W) abundance was observed in farm animals, especially in pigs. This could be explained by previous findings in EFFORT,32,33 in which a higher proportion of tetracycline use was found in pig farms (15.3%)33 than in broiler farms (11%)32 among nine countries. Still, as there is a strong association between faecal microbiome and resistome,4,19,21 the difference in ARG abundance among species can also be related to differences in microbiome compositions. In fish samples, we only detected a low concentration of ARGs. This may be due to the fact that fish faecal samples in our study were collected from frozen guts, which made it difficult to separate fish intestines and faecal contents, leading to a high proportion of host DNA. As a result, AMR levels of fish faeces may have been underestimated. In the future, more appropriate approaches for collecting fish faecal samples are worthy of studying.
Wild boars showed the lowest mean ARG abundance among all animal species, which is likely the result of negligible antimicrobial exposure of wild animals.34,35 This is consistent with previous results of European wild-animal AMR studies.36–38 In companion animals, we saw higher relative ARG abundance in cat faeces than in dog faeces. Since the previous EFFORT study showed no AMU–AMR association in companion animals,17 this higher ARG abundance in cat faeces may be related to differences in the gut microbiota, or may be due to the fact that cats roam more freely than dogs,39 through which cats are probably exposed to more environmental sources than dogs.
ARG abundance declines along the pig and broiler production chain
In pig and broiler production chains, a decline in relative ARG abundance was seen from primary production to slaughterhouse and retail meat. The main explanation is that livestock gut bacteria are enriched in AMR genes through AMU, but also through exposure to AMR through animal faeces and dust in farms. We found higher relative ARG abundance in farm dust than in farm animal faeces in pig and broiler farms, which is consistent with previous findings using metagenomic data.19 The explanation may be that microorganisms in dust sources such as animal faeces19,20 and constituents (skin, feather)40–42 are richer in ARGs. The relative ARG abundance in retail pork and chicken meat was found to be significantly lower than that of carcasses and meat samples in pig and broiler slaughterhouses. One potential explanation is that the production steps along the slaughterhouse line, including the cooling process, reduce bacterial loads and related AMR.13,43 Additionally, location of ARGs in bacterial hosts that are unable to survive the production process may lead to a reduction in AMR levels from farm to fork. More research is needed in the future to link these changes to specific production steps.
Sources of AMR variation in pigs, broilers, veal calves and turkeys
For pigs and broilers, substantial between-country variation in ARG levels was previously reported in faecal samples.4–6,23 For veal calves and turkeys, we found high within-farm variation. Considering the large variation in individual characteristics,44,45 it is not surprising that the AMR abundance varies highly among animals per farm. The other explanation is the limited sample size [sample numbers per farm (7/5) and country numbers (3)] in veal calves and turkeys. The differences in farm management practices may also be associated with the distribution of AMR variation.
In our previous adjusted VCA models of pigs and broilers,23 we observed that among farm characteristics, AMU contributed most to total AMR variation. In the present study on veal calves and turkeys, we found that in addition to AMU, farm biosecurity measures also played an important role in AMR variation. Although the small sample size of veal calves and turkeys reduced statistical power in our study, our results provide evidence that AMU and farm biosecurity contributed to both variation sources (country and farm).
Correlation of ARG abundance between qPCR data and metagenomic data
To the best of our knowledge, this study is the first to compare ARG abundance using qPCR and metagenomics in the same animal samples in parallel. In this context, we found a high correlation between pooled metagenomic data46 and pooled qPCR data. This indicates that qPCR with pooled data can be an alternative cost-effective approach for the quantitative analysis of ARG targets when a project budget is limited.
ARG abundance in humans
Previous studies reported that farmers had higher nasal MRSA prevalence than slaughterhouse workers,47 and AMR exposure in pig farms was higher than in pig slaughterhouses.48 Similar results were found in this study where pig/broiler farmers showed significantly higher mean relative aph(3′)-III abundance than pig/broiler slaughterhouse workers. Although the exposure difference, as well as other determinants (working hours, life history etc.) within the production chain, may affect workers’ ARG carriage,13 we argue that people working in pig/broiler farms are more likely to be exposed to ARGs than people working in pig/broiler slaughterhouses, especially considering the high dust levels in farms.19 There is also a possibility that antimicrobial residues in farm dust could impact human gut microbiota by selecting for resistance.49,50 In the future, more in-depth studies are needed to reproduce and confirm these findings.
Conclusions
This study shows that qPCR analysis is a valuable tool to assess the abundances of selected ARGs in a large amount of livestock-associated samples collected across Europe. High variation of ARG abundance assessed using qPCR was found across animal species, environmental samples and humans. A ‘farm to fork’ decreasing trend in ARG abundance was found for both pigs and broilers. The between-country and between-farm variation could be partially attributed to AMU and farm biosecurity levels. Occupational livestock AMR exposure is related to the ARG abundance in human faeces.
Supplementary Material
dkac133_Supplementary_Data
Acknowledgements
We would like to thank all the farmers from Belgium, Bulgaria, Germany, Denmark, Spain, France, Italy, the Netherlands and Poland. We wish to thank all the employees in the slaughterhouses in the Netherlands and Germany, all the field workers, laboratory analysts and data analysts at the Institute for Risk Assessment Sciences, The Netherlands (Daisy de Vries, Nynke Jansen, Janne Heederik). We also wish to acknowledge the services of the Lifelines Cohort Study, the contributing research centres delivering data to Lifelines, and all the study participants.
Contributor Information
Dongsheng Yang, Institute for Risk Assessment Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Dick J J Heederik, Institute for Risk Assessment Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Peter Scherpenisse, Institute for Risk Assessment Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Liese Van Gompel, Institute for Risk Assessment Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Roosmarijn E C Luiken, Institute for Risk Assessment Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Katharina Wadepohl, Außenstelle für Epidemiologie, Tierärztliche Hochschule Hannover, Hannover, Germany.
Magdalena Skarżyńska, Department of Microbiology, National Veterinary Research Institute, Pulawy, Poland.
Eri Van Heijnsbergen, Institute for Risk Assessment Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Inge M Wouters, Institute for Risk Assessment Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Gerdit D Greve, Institute for Risk Assessment Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Betty G M Jongerius-Gortemaker, Institute for Risk Assessment Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Monique Tersteeg-Zijderveld, Institute for Risk Assessment Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Lützen Portengen, Institute for Risk Assessment Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Katharina Juraschek, Department of Biological Safety, German Federal Institute for Risk Assessment, Berlin, Germany.
Jennie Fischer, Department of Biological Safety, German Federal Institute for Risk Assessment, Berlin, Germany.
Magdalena Zając, Department of Microbiology, National Veterinary Research Institute, Pulawy, Poland.
Dariusz Wasyl, Department of Microbiology, National Veterinary Research Institute, Pulawy, Poland.
Jaap A Wagenaar, Department of Infectious Diseases and Immunology, Utrecht University, Utrecht, The Netherlands.
Dik J Mevius, Department of Infectious Diseases and Immunology, Utrecht University, Utrecht, The Netherlands; Department of Bacteriology and Epidemiology, Wageningen Bioveterinary Research, Lelystad, The Netherlands.
Lidwien A M Smit, Institute for Risk Assessment Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Heike Schmitt, Institute for Risk Assessment Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands; National Institute for Public Health and the Environment, Bilthoven, The Netherlands.
Members of the EFFORT consortium
Haitske Graveland (UUVM), Philip Joosten (UGENT), Steven Sarrazin (UGENT), Jeroen Dewulf (UGENT), Alieda Van Essen (WBVR), Bruno Gonzalez-Zorn (UCM), Gabriel Moyano (UCM), Pascal Sanders (ANSES), Julie David (ANSES), Christophe Soumet (ANSES), Antonio Battisti (IZSLT), Andrea Caprioli (IZSLT), Thomas Blaha (TIHO), Maximiliane Brandt (TIHO), Frank Aarestrup (DTU), Tine Hald (DTU), Ana Sofia Ribeiro Duarte (DTU), Andrzej Hoszowski (NVRI), Agnieszka Pękala-Safińska (NVRI), Ewa Pażdzior (NVRI), Hristo Daskalov (NDRVI), Helmut W. Saatkamp (BEC) and Katharina D. C. Stärk (SAFOSO), as well as authors (Dick J. J. Heederik, Dik J. Mevius, Jaap A. Wagenaar, Lidwien A. M. Smit and Heike Schmitt) of this paper.
Funding
This work was part of the Ecology from Farm to Fork Of microbial drug Resistance and Transmission (EFFORT) project, co-funded by the European Commission, 7th Framework Programme for Research and Innovation (FP7-KBBE-2013-7, grant agreement: 613754). Research at the National Veterinary Research Institute (PIWet), Poland, was supported by the Polish Ministry of Science: No. 3173/7PR/2014/2. D.Y. was also funded by the China Scholarships Council (No. 201709110149). Lifelines is a multi-disciplinary prospective population-based cohort study examining, in a unique three-generation design, the health and health-related behaviour of 167729 persons living in the North of the Netherlands. It employs a broad range of investigative procedures in assessing the biomedical, sociodemographic, behavioural, physical and psychological factors that contribute to the health and the disease of the general population, with a special focus on multi-morbidity and complex genetics. The Lifelines Biobank initiative has been made possible by subsidy from the Dutch Ministry of Health, Welfare and Sport, the Dutch Ministry of Economic Affairs, the University Medical Center Groningen (UMCG the Netherlands), University Groningen and the Northern Provinces of the Netherlands.
Transparency declarations
None to declare.
Supplementary data
Supplementary methods, references, Tables S1 to S6 and Figures S1 to S6 are available as Supplementary data at JAC Online.
References
- 1.WHO . Antimicrobial resistance: global report on surveillance. 2014. https://apps.who.int/iris/handle/10665/112642.
- 2.CLSI . Performance Standards for Antimicrobial Susceptibility Testing—Twentieth Edition: M100. 2011.
- 3.CLSI . Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals—Fourth Edition: VET01. 2013.
- 4.Munk P, Knudsen BE, Lukjancenko Oet al. Abundance and diversity of the faecal resistome in slaughter pigs and broilers in nine European countries. Nat Microbiol 2018; 3: 898–908. [DOI] [PubMed] [Google Scholar]
- 5.Van Gompel L, Luiken RE, Sarrazin Set al. The antimicrobial resistome in relation to antimicrobial use and biosecurity in pig farming, a metagenome-wide association study in nine European countries. J Antimicrob Chemother 2019; 74: 865–76. [DOI] [PubMed] [Google Scholar]
- 6.Luiken REC, Van Gompel L, Munk Pet al. Associations between antimicrobial use and the faecal resistome on broiler farms from nine European countries. J Antimicrob Chemother 2019; 74: 2596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huijbers PM, Blaak H, de Jong MCet al. Role of the environment in the transmission of antimicrobial resistance to humans: a review. Environ Sci Technol 2015; 49: 11993–2004. [DOI] [PubMed] [Google Scholar]
- 8.Holmes AH, Moore LS, Sundsfjord Aet al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016; 387: 176–87. [DOI] [PubMed] [Google Scholar]
- 9.Silbergeld EK, Graham J, Price LB. Industrial food animal production, antimicrobial resistance, and human health. Annu Rev Public Health 2008; 29: 151–69. [DOI] [PubMed] [Google Scholar]
- 10.Van Cleef B, Broens E, Voss Aet al. High prevalence of nasal MRSA carriage in slaughterhouse workers in contact with live pigs in the Netherlands. Epidemiol Infect 2010; 138: 756–63. [DOI] [PubMed] [Google Scholar]
- 11.Dorado-Garcia A, Bos MEH, Graveland Het al. Risk factors for persistence of livestock-associated MRSA and environmental exposure in veal calf farmers and their family members: an observational longitudinal study. BMJ Open 2013; 3: e003272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aarestrup FM, Agerso Y, Gerner-Smidt Pet al. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagn Microbiol Infect Dis 2000; 37: 127–37. [DOI] [PubMed] [Google Scholar]
- 13.Van Gompel L, Dohmen W, Luiken REet al. Occupational exposure and carriage of antimicrobial resistance genes (tetW, ermB) in pig slaughterhouse workers. Ann Work Expo Health 2020; 64: 125–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poeta P, Costa D, Rodrigues Jet al. Antimicrobial resistance and the mechanisms implicated in faecal enterococci from healthy humans, poultry and pets in Portugal. Int J Antimicrob Agents 2006; 27: 131–7. [DOI] [PubMed] [Google Scholar]
- 15.Yang D, Van Gompel L, Luiken REet al. Association of antimicrobial usage with faecal abundance of aph (3′)-III, ermB, sul2 and tetW resistance genes in veal calves in three European countries. Int J Antimicrob Agents 2020; 56: 106131. [DOI] [PubMed] [Google Scholar]
- 16.Horie M, Yang D, Joosten Pet al. Risk factors for antimicrobial resistance in turkey farms: a cross-sectional study in three European countries. Antibiotics 2021; 10: 820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joosten P, Ceccarelli D, Odent Eet al. Antimicrobial usage and resistance in companion animals: a cross-sectional study in three European countries. Antibiotics 2020; 9: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stolk RP, Rosmalen JG, Postma DSet al. Universal risk factors for multifactorial diseases. Eur J Epidemiol 2008; 23: 67–74. [DOI] [PubMed] [Google Scholar]
- 19.Luiken RE, Van Gompel L, Bossers Aet al. Farm dust resistomes and bacterial microbiomes in European poultry and pig farms. Environ Int 2020; 143: 105971. [DOI] [PubMed] [Google Scholar]
- 20.Luiken RE, Heederik DJ, Scherpenisse Pet al. Determinants for antimicrobial resistance genes in farm dust on 333 poultry and pig farms in nine European countries. Environ Res 2022; 208: 112715. [DOI] [PubMed] [Google Scholar]
- 21.Van Gompel L, Luiken RE, Hansen RBet al. Description and determinants of the faecal resistome and microbiome of farmers and slaughterhouse workers: a metagenome-wide cross-sectional study. Environ Int 2020; 143: 105939. [DOI] [PubMed] [Google Scholar]
- 22.Wadepohl K, Müller A, Seinige Det al. Association of intestinal colonization of ESBL-producing Enterobacteriaceae in poultry slaughterhouse workers with occupational exposure—a German pilot study. PLoS One 2020; 15: e0232326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang D, Heederik DJ, Mevius DJet al. Risk factors for the abundance of antimicrobial resistance genes aph(3 ′ )-III, erm(B), sul2 and tet(W) in pig and broiler faeces in nine European countries. J Antimicrob Chemother 2022; 77: 969–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oksanen J, Blanchet FG, Friendly Met al. vegan: Community Ecology Package. R package version 2.5-6. 2019. https://cran.r-project.org/src/contrib/Archive/vegan/.
- 25.R Core Team . R: A language and environment for statistical computing, Version 4.0.3. R Foundation for Statistical Computing. 2020. https://cran.r-project.org/bin/windows/base/old/4.0.3/.
- 26.Harrell FE Jr, Dupont C. Hmisc: Harrell miscellaneous. R package version 4.0-3. 2017. https://cran.r-project.org/web/packages/Hmisc/index.html.
- 27.Fox J. Applied Regression Analysis and Generalized Linear Models. Sage Publications, 2015. [Google Scholar]
- 28.Welch BL. On the comparison of several mean values - an alternative approach. Biometrika 1951; 38: 330–6. [Google Scholar]
- 29.Ruxton GD, Beauchamp G. Time for some a priori thinking about post hoc testing. Behav Ecol 2008; 19: 690–3. [Google Scholar]
- 30.Yandell BS. Practical Data Analysis for Designed Experiments. Chapman & Hall/CRC Press, 1997. [Google Scholar]
- 31.Schuetzenmeister A, Dufey F. VCA: variance component analysis. R package version 1.4.3. 2020. https://CRAN.R-project.org/package=VCA.
- 32.Joosten P, Sarrazin S, Van Gompel Let al. Quantitative and qualitative analysis of antimicrobial usage at farm and flock level on 181 broiler farms in nine European countries. J Antimicrob Chemother 2019; 74: 798–806. [DOI] [PubMed] [Google Scholar]
- 33.Sarrazin S, Joosten P, Van Gompel Let al. Quantitative and qualitative analysis of antimicrobial usage patterns in 180 selected farrow-to-finish pig farms from nine European countries based on single batch and purchase data. J Antimicrob Chemother 2019; 74: 807–16. [DOI] [PubMed] [Google Scholar]
- 34.Swift BMC, Bennett M, Waller Ket al. Anthropogenic environmental drivers of antimicrobial resistance in wildlife. Sci Total Environ 2019; 649: 12–20. [DOI] [PubMed] [Google Scholar]
- 35.Skarżyńska M, Leekitcharoenphon P, Hendriksen RSet al. A metagenomic glimpse into the gut of wild and domestic animals: quantification of antimicrobial resistance and more. PLoS One 2020; 15: e0242987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skurnik D, Ruimy R, Andremont Aet al. Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J Antimicrob Chemother 2006; 57: 1215–9. [DOI] [PubMed] [Google Scholar]
- 37.Wasyl D, Zając M, Lalak Aet al. Antimicrobial resistance in Escherichia coli isolated from wild animals in Poland. Microb Drug Resist 2018; 24: 807–15. [DOI] [PubMed] [Google Scholar]
- 38.Schierack P, Römer A, Jores Jet al. Isolation and characterization of intestinal Escherichia coli clones from wild boars in Germany. Appl Environ Microbiol 2009; 75: 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wongsaengchan C, McKeegan DE. The views of the UK public towards routine neutering of dogs and cats. Animals 2019; 9: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baptiste KE, Williams K, Williams NJet al. Methicillin-resistant staphylococci in companion animals. Emerg Infect Dis 2005; 11: 1942–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daum RS. Skin and soft-tissue infections caused by methicillin-resistant Staphylococcus aureus. N Engl J Med 2007; 357: 380–90. [DOI] [PubMed] [Google Scholar]
- 42.Miskiewicz A, Kowalczyk P, Oraibi SMet al. Bird feathers as potential sources of pathogenic microorganisms: a new look at old diseases. Antonie van Leeuwenhoek 2018; 111: 1493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pacholewicz E, Swart A, Schipper Met al. A comparison of fluctuations of Campylobacter and Escherichia coli concentrations on broiler chicken carcasses during processing in two slaughterhouses. Int J Food Microbiol 2015; 205: 119–27. [DOI] [PubMed] [Google Scholar]
- 44.Craven J, Barnum D. Ecology of intestinal Escherichia coli in pigs. Can J Comp Med 1971; 35: 324. [PMC free article] [PubMed] [Google Scholar]
- 45.Hinton M, Hampson D, Hampson Eet al. A comparison of the ecology of Escherichia coli in the intestine of healthy unweaned pigs and pigs after weaning. J Appl Bacteriol 1985; 58: 471–7. [DOI] [PubMed] [Google Scholar]
- 46.Munk P, Andersen VD, de Knegt Let al. A sampling and metagenomic sequencing-based methodology for monitoring antimicrobial resistance in swine herds. J Antimicrob Chemother 2017; 72: 385–92. [DOI] [PubMed] [Google Scholar]
- 47.Sun C, Chen B, Hulth Aet al. Genomic analysis of Staphylococcus aureus along a pork production chain and in the community, Shandong Province, China. Int J Antimicrob Agents 2019; 54: 8–15. [DOI] [PubMed] [Google Scholar]
- 48.Duarte ASR, Röder T, Van Gompel Let al. Metagenomics-based approach to source-attribution of antimicrobial resistance determinants–identification of reservoir resistome signatures. Front Microbiol 2021; 11: 3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marshall BM, Levy SB. Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev 2011; 24: 718–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levy SB, Fitzgerald GB, Macone AB. Spread of antibiotic-resistant plasmids from chicken to chicken and from chicken to man. Nature 1976; 260: 40–2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
dkac133_Supplementary_Data