Multiple Genes for the Last Step of Proline Biosynthesis in Bacillus subtilis (original) (raw)
Abstract
The complete Bacillus subtilis genome contains four genes (proG, proH, proI, and comER) with the potential to encode Δ1-pyrroline-5-carboxylate reductase, a proline biosynthetic enzyme. Simultaneous defects in three of these genes (proG, proH, and proI) were required to confer proline auxotrophy, indicating that the products of these genes are mostly interchangeable with respect to the last step in proline biosynthesis.
The pathway of proline synthesis from glutamate, the most common mechanism of proline biosynthesis, comprises three enzymatic steps (Fig. 1). The corresponding genes of Escherichia coli, proB, proA, and proC, encode γ-glutamyl kinase, γ-glutamyl phosphate reductase, and Δ1-pyrroline-5-carboxylate (P5C) reductase, respectively (21). The _proBA_-dependent pathway of proline synthesis was shown to function also in Bacillus subtilis; mutations within the proBA locus cause auxotrophy for proline (8, 29). While B. subtilis has a single _proA_-like gene, a second _proB_-like gene, proJ of the proHJ locus (B. R. Belitsky and A. L. Sonenshein, GenBank accession number AF006720) has been found. In a manner unique to this bacterium, either ProB-like enzyme can provide enough γ-glutamyl kinase activity to support growth in the absence of exogenous proline (unpublished results). Apparently, previously described mutations to auxotrophy in the proBA locus either affect proA or are proB alleles that are polar on proA expression. No proC mutant of B. subtilis has been described, and four genes have the potential to encode ProC-like proteins with P5C reductase activity: proH (also called orf257 and proC), comER (also called comED), proI (also called yqjO), and ykeA (here renamed proG) (1, 14, 20). The four genes are located at 172.3°, 225.5°, 211.2°, and 116.1° on the chromosomal map (http://genolist.pasteur.fr/SubtiList [26]) and code for proteins of 271, 273, 278, and 272 amino acids, respectively (the originally reported coding region of proH [1, 20] was extended by resequencing the proH 3′ end [GenBank accession number AF006720]). ProH and ProI are 42% identical to each other and up to 35% identical to many other P5C reductases from bacteria, archaea, and eukaryotes. ProG and ComER have more limited similarity to other P5C reductases and to each other. The functions of the four B. subtilis genes are not known. In this work we sought to identify the gene(s) responsible for the last step of proline biosynthesis.
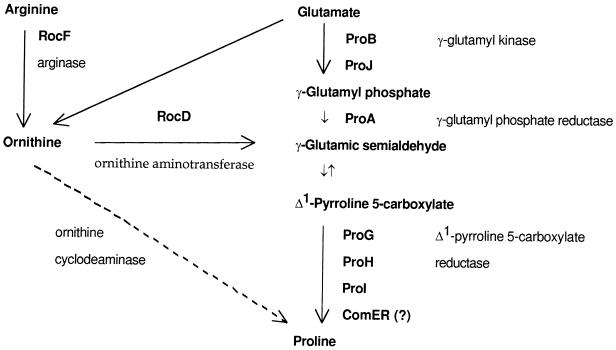
FIG. 1.
Pathways of proline biosynthesis in B. subtilis. The major pathway of proline synthesis from glutamate is shown as a descending series of reactions. Some proline is also apparently synthesized from glutamate via ornithine (an intermediate in arginine synthesis) by the action of the RocD product (unpublished results). Since no ortholog of E. coli ArgE protein is present in B. subtilis, conversion of _N_-acetylglutamic γ-semialdehyde (an intermediate in arginine synthesis) to γ-glutamic semialdehyde (21) apparently does not occur. Details of the anabolic pathway from glutamate to ornithine and the catabolic reaction from γ-glutamic semialdehyde to glutamate have been omitted. The catabolism of citrulline includes its conversion to ornithine (unpublished data) but has not been characterized further. Ornithine cyclodeaminase (dashed line) is not present in B. subtilis. The proG and proI genes have been previously known as ykeA and yqjO, respectively.
Construction and properties of a proG (ykeA) null mutant.
To create pBB1081, the 1.56-kb _Pvu_II-_Eco_RI fragment from pCM103 (23) containing most of the proG gene and the 5′ end of the dppA gene was cloned between the _Pst_I (blunt-ended) and _Eco_RI sites of pJPM1, a derivative of pBS (Stratagene) containing a chloramphenicol resistance marker (27). Methods for plasmid isolation, agarose gel electrophoresis, use of restriction and DNA modification enzymes, DNA ligation, PCR, Southern hybridization with digoxigenin-labeled DNA probes, and electroporation of E. coli JM107 or DH5α cells were as described by Sambrook et al. (30). DNA and protein sequences were analyzed using the DNA Strider (22) or BLAST (2) programs. A deletion-insertion mutation within the proG gene was created by replacing the 0.10-kb _Pst_I-_Sac_I fragment of pBB1081 with a 1.43-kb Pst_I-Sac_I ble cassette determining resistance to phleomycin, excised from pJPM136 (6). The orientation of the ble gene in the resulting Δ_proG::ble plasmid, pBB1082, coincides with that of the proG gene. pBB1082 was introduced into B. subtilis SMY, and phleomycin-resistant, chloramphenicol-sensitive transformants, arising from double-crossover homologous recombination events, were selected. Growth of B. subtilis cells, transformation by chromosomal or plasmid DNA, and isolation of chromosomal DNA were as described previously (6). The replacement of the chromosomal proG gene by the Δ_proG::ble allele in strain BB1951 was confirmed by comparing sizes of the PCR products from the wild-type and mutant proG chromosomal loci. Strain BB1951 (proG::ble) had the growth characteristics of a wild-type strain in the presence and absence of proline.
Construction and properties of a proH null mutant.
The 0.27-kb _Eco_RI-_Pst_I 3′-end fragment of the proH gene was subcloned in several steps from pLS23-17 (7) between the _Eco_RI and _Pst_I sites of pBB544, a derivative of pBluescript SK(−) (Stratagene) containing a neomycin resistance marker (5). The resulting plasmid, pBB575, was integrated via a single-crossover recombination event into the chromosome of B. subtilis strain SMY at the proH locus. To clone DNA adjacent to the site of integration of pBB575, the chromosomal DNA of the resulting strain was digested with _Hin_dIII, self-ligated, and introduced by electroporation into E. coli cells. The isolated plasmid, pBB576, had a 1.32-kb insert of chromosomal DNA carrying most of proH. A deletion-insertion mutation within the proH gene was created by replacing the 0.55-kb _Bcl_I-_Eco_RI fragment of pBB576 with a 1.9-kb _Bam_HI-_Eco_RI tet cassette, excised from pBEST307 (17). The orientation of the tet gene in the resulting plasmid, pBB734, coincides with that of the proH gene. Strain BB286 (proH::tet) was constructed as described above for strain BB734, using pBB734 and selecting for tetracycline-resistant, neomycin-sensitive transformants. Strain BB286 (proH::tet) had the growth characteristics of a wild-type strain in the presence and absence of proline.
Construction and properties of a proI (yqjO) null mutant.
The 1.85-kb ′_yqjP proI yqjN_′ chromosomal region was amplified by PCR using custom synthesized oligonucleotides as primers. To create pJS18, the internal 1.62-kb _Cla_I-_Nsi_I fragment of the PCR product, including the entire proI gene and the flanking regions of the yqjP and yqjN genes, was cloned in pBluescript SK(−) (Stratagene), cleaved with _Cla_I and Pst_I. For construction of pJS20 (Δ_proI::spc), the 0.25-kb _Bcl_I-_Stu_I fragment of pJS18 that is internal to proI was replaced with the 1.3-kb _Bam_HI-Xba_I (filled-in) fragment, excised from plasmid pRMK65 (18), which contains the spc gene. The orientation of the spc gene in this construction is opposite to that of the proI gene. Strain JSB9 (Δ_proI::spc) was isolated by transformation of strain JH642 (trpC2 pheA1) (obtained from J. Hoch) with _Bam_HI-linearized DNA of pJS20. Spectinomycin-resistant transformants were selected, and the correct double-crossover integration event was verified by Southern hybridization using the _Sal_I-_Not_I fragment of pJS18 as a probe. Strain JSB9 (proI::spc) had the growth characteristics of a wild-type strain in the presence and absence of proline.
Construction and properties of multiple mutants.
comER mutants were constructed previously and shown to be prototrophic (14, 16). Strains containing all possible combinations of two or three mutations in the proG, proH, proI, and comER genes and the corresponding quadruple mutant were constructed by transformation of strain SMY with chromosomal DNAs from appropriate mutants. The Pro phenotype of some of the double and triple mutants is shown in Table 1. The growth rate in glucose-ammonia medium of any of the double mutants was identical to the growth rate of a wild-type strain. No contribution of the comER gene to the cells' ability to grow without proline was detected for any of the mutants. The proG proH proI triple mutant required proline for growth, demonstrating that the P5C reductase enzymes are in fact required for proline synthesis in B. subtilis as in other organisms but that this function can be taken over by any one of three proteins, ProG, ProH, or ProI. In some experiments the proG proI mutant exhibited a long lag period before initiating growth in minimal medium without proline. This effect probably reflects the need for proH to be induced in order to provide enough enzyme to support proline synthesis. We cannot exclude the possibility that spontaneous mutations which elevate expression of proH accumulate in the culture of the proG proI mutant.
TABLE 1.
Growth of mutant strains in minimal mediuma
| Strain | Genotype | Nitrogen source | |
|---|---|---|---|
| Ammonium | Arginine ± ammonium | ||
| SMY | Wild type | + | + |
| BB1954 | proG proH | + | + |
| BB1960 | proG proI | +/− | +/− |
| BB1969 | proH proI | + | + |
| BB1973 | proG proH proI | − | − |
| BB1980 | proB proG proH | − | + |
| BB1979 | proB proG proI | − | +/− |
| BB1978 | proB proH proI | − | + |
| BB1983 | proB proG proH proI | − | − |
Role of _proC_-like genes in proline generation through the arginase pathway.
In addition to the anabolic pathway of proline synthesis from glutamate, two catabolic pathways can lead to proline from ornithine; neither of these pathways requires the first two steps of proline synthesis from glutamate (Fig. 1). B. subtilis does not contain any gene that could code for ornithine cyclodeaminase (cyclase) (20, 28, 31) but has a well-characterized arginase pathway for arginine degradation (11). In that pathway, ornithine aminotransferase, the product of the rocD gene, generates γ-glutamic semialdehyde, a substrate of P5C reductase, from ornithine (Fig. 1) (4, 13). B. subtilis cells are able to utilize extracellular ornithine or the related amino acids arginine and citrulline as sources of proline (8), and they failed to do so in a rocD mutant, indicating that the arginase pathway is essential for proline generation under these conditions (Fig. 1). Formation of proline from ornithine, arginine or citrulline was also dependent on the presence of proG, proH, or proI, the same genes that can support proline synthesis from glutamate. The proG proI double mutant, whose only P5C reductase is encoded by proH, again had a small growth defect under these conditions (Table 1). To ensure that no proline was derived from glutamate, which can be formed from ornithine and related amino acids, we introduced a proB::cat mutation into our strains and confirmed the requirement for either proG, proH, or proI (Table 1). Thus, proG, proH, or proI is essential for proline formation, both through the glutamate pathway and through the arginase pathway.
Possible roles of multiple P5C reductases.
Participation of at least three P5C reductase isoenzymes in the last step of proline synthesis is unique to B. subtilis among characterized organisms and may reflect specialized functions or regulation or both. Multiple genes with potential to code for P5C reductase isoenzymes have been detected in the genomes of the gram-positive bacteria Bacillus halodurans (32), Bacillus anthracis (http://www.tigr.org), Enterococcus faecalis (http://www.tigr.org), and Clostridium difficile (http://www.sanger.ac.uk) and the gram-negative bacterium Pseudomonas putida (http://www.tigr.org), but their functions have not been verified.
Though we could not detect a unique role for ProG, ProH, or ProI in proline synthesis from either glutamate or arginine, it is possible that such a role exists under some physiological conditions. Transcription of the proBA and proI genes is increased during proline limitation (unpublished data) and seems to be regulated by a termination-antitermination control mechanism, the T-box system (15); both proBA and proI contain 18-bp T-box-like sequences with the predicted proline specifier codons CCU and CCC (9). Coordinate induction of proI and proBA by proline limitation suggests that ProI is the major P5C reductase under such conditions. The proHJ locus, encoding enzymes for the first and the last steps of proline synthesis, is induced by high concentrations of salt (unpublished data), in keeping with the role of proline as the major endogenously produced osmoprotectant in B. subtilis (19, 25, 33). Finally, multiple P5C reductase isoenzymes may be involved in removal of excess P5C, which was reported to be toxic in Aspergillus nidulans (3) and in human cells (24) and has also been shown to be toxic for B. subtilis cells (unpublished data).
The role of the comER gene remains unknown (16). comER itself and its unusual overlapping, divergent orientation with respect to that of the comEA-EB-EC operon (14) are conserved in B. halodurans, B. anthracis, and Bacillus stearothermophilus, i.e., all Bacillus species for which sequencing information is available. comER expression decreases in stationary phase of growth in competence medium (14); no effect of comER mutations on cell competence was observed in earlier work (14, 16). The comER gene is at least partially under sporulation control, and its putative ςE-dependent promoter has been identified (10). We could not detect any effect of comER mutations on sporulation efficiency in nutrient broth medium or in minimal medium either without proline or in the presence of a limiting amount of proline when the proG proH proI mutant was used.
Acknowledgments
We are grateful to D. Dubnau for a gift of strains.
This work was supported by U.S. Public Health Service grant GM36718, the Deutsche Forschungsgemeinschaft (SFB-395 and Graduiertenkolleg Proteinfunktion auf atomarer Ebene), and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Ahn K S, Wake R G. Variations and coding features of the sequence spanning the replication terminus of Bacillus subtilis 168 and W23 chromosomes. Gene. 1991;98:107–112. doi: 10.1016/0378-1119(91)90111-n. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arst H N, Jones S A, Bailey C R. A method for the selection of deletion mutations in the L-proline catabolism gene cluster of Aspergillus nidulans. Genet Res. 1981;38:171–195. doi: 10.1017/s0016672300020516. [DOI] [PubMed] [Google Scholar]
- 4.Baumberg S, Harwood C R. Carbon and nitrogen repression of arginine catabolic enzymes in Bacillus subtilis. J Bacteriol. 1979;137:189–196. doi: 10.1128/jb.137.1.189-196.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belitsky B R, Gustafsson M C, Sonenshein A L, Von Wachenfeldt C. An lrp-like gene of Bacillus subtilis involved in branched-chain amino acid transport. J Bacteriol. 1997;179:5448–5457. doi: 10.1128/jb.179.17.5448-5457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belitsky B R, Sonenshein A L. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J Bacteriol. 1998;180:6298–6305. doi: 10.1128/jb.180.23.6298-6305.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohannon D E, Rosenkrantz M S, Sonenshein A L. Regulation of Bacillus subtilis glutamate synthase genes by the nitrogen source. J Bacteriol. 1985;163:957–964. doi: 10.1128/jb.163.3.957-964.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buxton R S. Selection of Bacillus subtilis 168 mutants with deletions of the PBSX prophage. J Gen Virol. 1980;46:427–437. doi: 10.1099/0022-1317-46-2-427. [DOI] [PubMed] [Google Scholar]
- 9.Chopin A, Biaudet V, Ehrlich S D. Analysis of the Bacillus subtilis genome sequence reveals nine new T-box leaders. Mol Microbiol. 1998;29:662–664. doi: 10.1046/j.1365-2958.1998.00912.x. [DOI] [PubMed] [Google Scholar]
- 10.Fawcett P, Eichenberger P, Losick R, Youngman P. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 2000;97:8063–8068. doi: 10.1073/pnas.140209597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher S H. Utilization of amino acids and other nitrogen-containing compounds. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 221–228. [Google Scholar]
- 12.Fouet A, Sonenshein A L. A target for carbon source-dependent negative regulation of the citB promoter of Bacillus subtilis. J Bacteriol. 1990;172:835–844. doi: 10.1128/jb.172.2.835-844.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardan R, Rapoport G, Debarbouille M. Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis. J Mol Biol. 1995;249:843–856. doi: 10.1006/jmbi.1995.0342. [DOI] [PubMed] [Google Scholar]
- 14.Hahn J, Inamine G, Kozlov Y, Dubnau D. Characterization of comE, a late competence operon of Bacillus subtilis required for the binding and uptake of transforming DNA. Mol Microbiol. 1993;10:99–111. doi: 10.1111/j.1365-2958.1993.tb00907.x. [DOI] [PubMed] [Google Scholar]
- 15.Henkin T M. tRNA-directed transcription antitermination. Mol Microbiol. 1994;13:381–387. doi: 10.1111/j.1365-2958.1994.tb00432.x. [DOI] [PubMed] [Google Scholar]
- 16.Inamine G S, Dubnau D. ComEA, a Bacillus subtilis integral membrane protein required for genetic transformation, is needed for both DNA binding and transport. J Bacteriol. 1995;177:3045–3051. doi: 10.1128/jb.177.11.3045-3051.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itaya M. Construction of a novel tetracycline resistance gene cassette useful as a marker on the Bacillus subtilis chromosome. Biosci Biotechnol Biochem. 1992;56:685–686. doi: 10.1271/bbb.56.685. [DOI] [PubMed] [Google Scholar]
- 18.Kappes R M, Kempf B, Kneip S, Boch J, Gade J, Meier-Wagner J, Bremer E. Two evolutionarily closely related ABC transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol Microbiol. 1999;32:203–216. doi: 10.1046/j.1365-2958.1999.01354.x. [DOI] [PubMed] [Google Scholar]
- 19.Kempf B, Bremer E. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolarity environments. Arch Microbiol. 1998;170:319–330. doi: 10.1007/s002030050649. [DOI] [PubMed] [Google Scholar]
- 20.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 21.Leisinger T. Biosynthesis of proline. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 434–441. [Google Scholar]
- 22.Marck C. ‘DNA Strider’: a ‘C’ program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathiopoulos C, Mueller J P, Slack F J, Murphy C G, Patankar S, Bukusoglu G, Sonenshein A L. A Bacillus subtilis dipeptide transport system expressed early during sporulation. Mol Microbiol. 1991;5:1903–1913. doi: 10.1111/j.1365-2958.1991.tb00814.x. [DOI] [PubMed] [Google Scholar]
- 24.Maxwell S A, Davis G E. Differential gene expression in p53-mediated apoptosis-resistant vs. apoptosis-sensitive tumor cell lines. Proc Natl Acad Sci USA. 2000;97:13009–13014. doi: 10.1073/pnas.230445997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Measures J C. Role of amino acids in osmoregulation of non-halophilic bacteria. Nature. 1975;257:398–400. doi: 10.1038/257398a0. [DOI] [PubMed] [Google Scholar]
- 26.Moszer I. The complete genome of Bacillus subtilis: from sequence annotation to data management and analysis. FEBS Lett. 1998;430:28–36. doi: 10.1016/s0014-5793(98)00620-6. [DOI] [PubMed] [Google Scholar]
- 27.Mueller J P, Bukusoglu G, Sonenshein A L. Transcriptional regulation of Bacillus subtilis glucose starvation-inducible genes: control of gsiA by the ComP-ComA signal transduction system. J Bacteriol. 1992;174:4361–4373. doi: 10.1128/jb.174.13.4361-4373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muth W L, Costilow R N. Ornithine cyclase (deaminating). II. Properties of the homogeneous enzyme. J Biol Chem. 1974;249:7457–7462. [PubMed] [Google Scholar]
- 29.Ogura M, Kawata-Mukai M, Itaya M, Takio K, Tanaka T. Multiple copies of the proB gene enhance degS-dependent extracellular protease production in Bacillus subtilis. J Bacteriol. 1994;176:5673–5680. doi: 10.1128/jb.176.18.5673-5680.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Sans N, Schindler U, Schroder J. Ornithine cyclodeaminase from Ti plasmid C58: DNA sequence, enzyme properties and regulation of activity by arginine. Eur J Biochem. 1988;173:123–130. doi: 10.1111/j.1432-1033.1988.tb13975.x. [DOI] [PubMed] [Google Scholar]
- 32.Takami H, Nakasone K, Takaki Y, Maeno G, Sasaki R, Masui N, Fuji F, Hirama C, Nakamura Y, Ogasawara N, Kuhara S, Horikoshi K. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 2000;28:4317–4331. doi: 10.1093/nar/28.21.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whatmore A M, Chudek J A, Reed R H. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J Gen Microbiol. 1990;136:2527–2535. doi: 10.1099/00221287-136-12-2527. [DOI] [PubMed] [Google Scholar]